Abstract
In the sequenced genome of Salmonella enterica serovar Typhimurium strain LT2, an open reading frame (STM0580) coding for a putative regulatory protein of the TetR family is found upstream of the ramA gene. Overexpression of ramA results in increased expression of the AcrAB efflux pump and, consequently, multidrug resistance (MDR) in several bacterial species. The inactivation of the putative regulatory protein gene upstream of ramA in a susceptible serovar Typhimurium strain resulted in an MDR phenotype with fourfold increases in the MICs of unrelated antibiotics, such as quinolones/fluoroquinolones, phenicols, and tetracycline. The inactivation of this gene also resulted in a fourfold increase in the expression of ramA and a fourfold increase in the expression of the AcrAB efflux pump. These results indicated that the gene encodes a local repressor of ramA and was thus named ramR. In contrast, the inactivation of marR, marA, soxR, and soxS did not affect the susceptibilities of the strain. In quinolone- or fluoroquinolone-resistant strains of serovar Typhimurium overexpressing AcrAB, several point mutations which resulted in amino acid changes or an in-frame shift were identified in ramR; in addition, mutations interrupting ramR with an IS1 element were identified in high-level fluoroquinolone-resistant serovar Typhimurium DT204 strains. One serovar Typhimurium DT104 isolate had a 2-nucleotide deletion in the putative RamR binding site found upstream of ramA. These mutations were confirmed to play a role in the MDR phenotype by complementing the isolates with an intact ramR gene or by inactivating their respective ramA gene. No mutations in the mar or sox region were found in the strains studied. In conclusion, mutations in ramR appear to play a major role in the upregulation of RamA and AcrAB and, consequently, in the efflux-mediated MDR phenotype of serovar Typhimurium.
Fluoroquinolones, together with extended-spectrum cephalosporins, are the treatment of choice for nontyphoid salmonellosis, as stable resistance to the most common members of different families of antimicrobial agents (ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline) has developed during the 1990s with the epidemic Salmonella enterica serovar Typhimurium phage type DT104 (8, 10, 21, 31). Emerging resistance to fluoroquinolones in Salmonella spp. has been reported for both human and animal cases and is thus threatening to become a serious public health problem (8, 10, 21, 31).
In Salmonella spp., quinolone and fluoroquinolone resistance has been attributed to point mutations in the quinolone resistance-determining regions (QRDRs) of the target genes gyrA, gyrB, parC, and parE. For the gyrA gene, coding for the A subunit of DNA gyrase, whose complex with DNA is the primary target of quinolones, mutations resulting in amino acid changes at Ser83 (to Phe, Tyr, or Ala) or at Asp87 (to Gly, Asn, or Tyr) are the most frequently observed in nalidixic acid-resistant strains (8, 10, 21, 31). Double mutations at both residues 83 and 87 have been identified in clinical isolates of serovar Typhimurium DT204 showing high-level resistance to fluoroquinolones, together with one mutation leading to the amino acid change Ser464Phe in the QRDR of gyrB, encoding the B subunit of DNA gyrase, and one mutation leading to the amino acid change Ser80Ile in the QRDR of parC, coding for the ParC subunit of topoisomerase IV, the secondary target of quinolones (4, 5).
Fluoroquinolone resistance in serovar Typhimurium has also been attributed to an active efflux mechanism (8, 10, 11, 22), and we have recently reported the participation of the AcrAB-TolC efflux system as an important mechanism of high-level resistance to fluoroquinolones in serovar Typhimurium DT204 as well as an important mechanism of both multidrug resistance (MDR) and quinolone resistance in serovar Typhimurium DT104 (4, 5, 6). High-level resistance to fluoroquinolones in Salmonella is thus essentially explained by the combination of two major resistance mechanisms, i.e., multiple target gene mutations and active efflux.
The expression of acrAB, encoding the major AcrAB efflux pump, is subject to multiple levels of regulation. In Escherichia coli, it is modulated at the lowest level by the local repressor AcrR. At a more global level, acrAB expression is modulated by stress conditions and by global regulators like MarA, SoxS, or Rob (1, 2, 3, 14, 16). The acrAB locus is indeed part of the mar, sox, and rob regulons of E. coli, whose activation confers a low level of resistance to a wide range of antimicrobial agents and organic solvents. Proteins encoded by the mar locus include the transcriptional activator MarA, its local repressor MarR, and two proteins with unknown functions, MarB and MarC. MarR negatively regulates the expression of marRAB by binding to the marO operator region. Proteins encoded by the soxRS locus include the transcriptional activator SoxS and a protein, SoxR, whose oxidized form can activate soxS expression.
While these regulator systems have been well studied in E. coli, less is known about their role in Salmonella spp. Moreover, in Salmonella spp. and in other bacteria, such as Enterobacter aerogenes, Enterobacter cloacae, and Klebsiella pneumoniae, RamA, a homologue of MarA that is absent in E. coli, has been shown to be implicated in MDR, and the overexpression of ramA correlated well with an increased expression of the AcrAB efflux pump (7, 12, 13, 27-30, 33).
In previous studies, we investigated whether mutations in regulatory regions like acrR, marRAB, or soxRS, in addition to target gene mutations (gyrA), could participate in the fluoroquinolone and MDR phenotype of serovar Typhimurium overproducing AcrAB (19, 20). However, only mutations in the acrR gene of in vitro fluoroquinolone-selected serovar Typhimurium mutants were identified (19). Therefore, in this study, we investigated the roles of MarR, MarA, SoxR, SoxS, and RamA in the MDR and quinolone resistance of serovar Typhimurium. We investigated particularly the role of the putative local repressor of RamA, which belongs, according to amino acid sequence homology, to the TetR family of proteins and whose gene is found upstream of ramA in the opposite orientation in the serovar Typhimurium LT2 genome (Fig. 1). This putative local repressor has not yet been reported to occur in Salmonella and other bacteria. Therefore, we also investigated the presence of mutations in the putative repressor gene and in the regulatory region of ramA that could explain the overproduction of RamA and, consequently, of AcrAB in MDR and quinolone-resistant serovar Typhimurium.
FIG. 1.
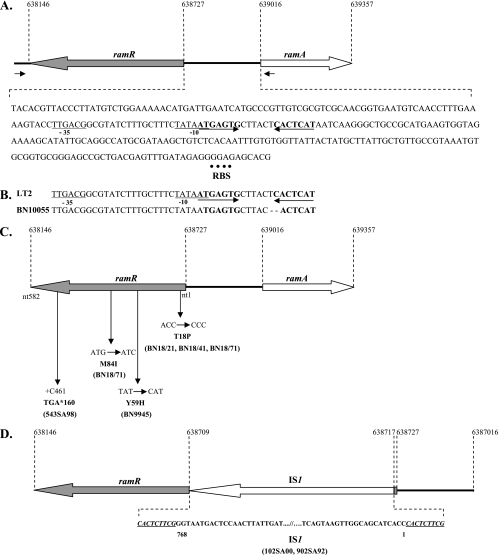
Sequence analysis of the ramR-ramA region in serovar Typhimurium strains. (A) Features of the 288-bp-long ramA-ramR intergenic region in serovar Typhimurium strain LT2. The predicted −10 and −35 promoter regions are underlined. The inverted repeat sequences are bold and indicated by arrows. The putative ribosome-binding site (RBS) is indicated with a dotted line. (B) Sequence alignment of the putative promoter region of serovar Typhimurium strains LT2 and BN10055 showing two nucleotide deletions in the putative RamR binding site of the latter strain. (C) Mutations found in the ramR gene in serovar Typhimurium strains BN18/21, BN18/41, BN18/71, BN9945, and 543SA98. nt, nucleotide. (D) Interruption by an IS1 element of the ramR gene in serovar Typhimurium DT204 strains 102SA00 and 902SA92.
MATERIALS AND METHODS
Bacterial strains.
All strains studied are listed in Table 1. MDR serovar Typhimurium DT104 strains were isolated from cattle in Belgium (strain 543SA98) and France (strains BN10055, BN9945, and BN9181), and MDR serovar Typhimurium DT204 strains, showing a high level of resistance to fluoroquinolones, were isolated from cattle in Belgium (strain 902SA92) and from animal feed imported into Belgium from China (strain 102SA00). Susceptible serovar Typhimurium DT104 control strain S/921495 was isolated from cattle in Scotland. Susceptible serovar Typhimurium strain BN18 was isolated from a pigeon in France, and in vitro-selected, quinolone-resistant clones (strains BN18/21, BN18/41, and BN18/71) derived from this strain (4, 5, 6, 11). Additional strains used in this study were the susceptible serovar Typhimurium reference strain LT2, whose genome has completely been sequenced (GenBank accession number NC_003197) (18), and the E. coli cloning strain TG1.
TABLE 1.
Salmonella enterica serovar Typhimurium strains used in this studya
| Strain | Phage type | Origin | SGI1 | MIC of indicated quinolone (μg/ml)
|
MIC of indicated antibiotic (μg/ml)
|
Substitution(s) in the QRDR of:
|
Mutation in acrR | WB AcrA ratio | Source | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nal | Flu | Enr | Cip | Cm | Ff | Tc | GyrA | GyrB | ParC | |||||||
| MDR strains | ||||||||||||||||
| 902SA92 | DT204 | B | − | >4,096 | 2,048 | 64 | 32 | 1,024 | 16 | 256 | S83A, D87N | S464F | S80I | − | 4 | 5 |
| 902SA92(pRamR) | DT204 | B | − | 4,096 | 512 | 16 | 8 | 256 | 4 | 256 | S83A, D87N | S464F | S80I | − | 1 | This study |
| 902SA92 ΔramA::kan | B | − | 4,096 | 512 | 16 | 8 | 256 | 4 | 256 | S83A, D87N | S464F | S80I | − | 1 | This study | |
| 102SA00 | DT204 | B | − | >4,096 | 2,048 | 64 | 32 | 512 | 16 | 256 | S83A, D87N | S464F | S80I | − | 4 | 5 |
| 102SA00(pRamR) | DT204 | B | − | 4,096 | 512 | 16 | 8 | 256 | 4 | 256 | S83A, D87N | S464F | S80I | − | 1 | This study |
| 102SA00 ΔramA::kan | DT204 | B | − | 4,096 | 512 | 16 | 8 | 256 | 4 | 256 | S83A, D87N | S464F | S80I | − | 1 | This study |
| BN10055 | DT104 | F | + | >4,096 | 64 | 2 | 1 | 1,024 | 128 | 256 | S83Y | − | − | − | 4 | 6 |
| BN10055(pRamR) | DT104 | F | + | >4,096 | 64 | 2 | 1 | 1,024 | 128 | 256 | S83Y | − | − | − | 4 | This study |
| BN10055 ΔramA::kan | DT104 | F | + | 512 | 16 | 0.5 | 0.25 | 128 | 32 | 64 | S83Y | − | − | − | 1 | This study |
| 543SA98 | DT104 | B | + | 1,024 | 32 | 2 | 0.5 | 512 | 512 | 128 | S83F | − | − | − | 4 | 6 |
| 543SA98(pRamR) | DT104 | B | + | 512 | 8 | 0.5 | 0.125 | 128 | 128 | 32 | S83Y | − | − | − | 1 | This study |
| 543SA98 ΔramA::kan | DT104 | B | + | 512 | 8 | 0.5 | 0.125 | 128 | 128 | 32 | S83Y | − | − | − | 1 | This study |
| BN9945 | DT104 | F | + | 1,024 | 32 | 1 | 0.5 | 512 | 128 | 128 | − | − | − | − | 2 | 6 |
| BN9945(pRamR) | DT104 | F | + | 512 | 8 | 0.25 | 0.125 | 128 | 64 | 32 | − | − | − | − | 1 | This study |
| BN9945 ΔramA::kan | DT104 | F | + | 512 | 8 | 0.25 | 0.125 | 128 | 64 | 32 | − | − | − | − | 1 | This study |
| BN9181 | DT104 | F | + | 4 | 0.5 | 0.030 | 0.015 | 128 | 32 | 32 | − | − | − | − | 1 | 6 |
| BN9181(pRamR) | DT104 | F | + | 4 | 0.5 | 0.030 | 0.015 | 128 | 32 | 32 | − | − | − | − | 1 | This study |
| BN9181 ΔramA::kan | DT104 | F | + | 4 | 0.5 | 0.015 | 0.0075 | 128 | 32 | 32 | − | − | − | − | 1 | This study |
| Control strains | ||||||||||||||||
| S/921495 | DT104 | S | − | 4 | 0.5 | 0.030 | 0.015 | 4 | 4 | 1 | − | − | − | − | 1 | 6 |
| S/921495 ΔmarR::kan | DT104 | S | − | 2 | 0.5 | 0.030 | 0.015 | 4 | 4 | 1 | − | − | − | − | 1 | This study |
| S/921495 ΔmarR | DT104 | S | − | 2 | 0.5 | 0.030 | 0.015 | 4 | 4 | 1 | − | − | − | − | 1 | This study |
| S/921495 ΔmarA::kan | DT104 | S | − | 4 | 0.5 | 0.030 | 0.015 | 4 | 4 | 1 | − | − | − | − | 1 | This study |
| S/921495 ΔsoxR::kan | DT104 | S | − | 2 | 0.5 | 0.030 | 0.015 | 4 | 4 | 1 | − | − | − | − | 1 | This study |
| S/921495 ΔsoxS::kan | DT104 | S | − | 4 | 0.5 | 0.030 | 0.015 | 4 | 4 | 1 | − | − | − | − | 1 | This study |
| S/921495(pRamR) | DT104 | S | − | 4 | 0.5 | 0.030 | 0.015 | 4 | 4 | 1 | − | − | − | − | 1 | This study |
| S/921495 ΔramR::kan | DT104 | S | − | 16 | 2 | 0.125 | 0.060 | 16 | 16 | 4 | − | − | − | − | 4 | This study |
| S/921495 ΔramR | DT104 | S | − | 16 | 2 | 0.125 | 0.060 | 16 | 16 | 4 | − | − | − | − | 4 | This study |
| S/921495 ΔramR(pRamR) | DT104 | S | − | 4 | 0.5 | 0.030 | 0.015 | 4 | 4 | 1 | − | − | − | − | 1 | This study |
| S/921495 ΔramA::kan | DT104 | S | − | 4 | 0.5 | 0.030 | 0.015 | 4 | 4 | 1 | − | − | − | − | 1 | This study |
| BN18 | ND | F | − | 4 | 0.5 | 0.030 | 0.015 | 4 | 4 | 1 | − | − | − | − | 1 | 11 |
| BN18(pRamR) | ND | F | − | 4 | 0.5 | 0.030 | 0.015 | 4 | 4 | 1 | − | − | − | − | 1 | This study |
| BN18 ΔramA::kan | ND | F | − | 4 | 0.5 | 0.030 | 0.015 | 4 | 2 | 1 | − | − | − | − | 1 | This study |
| BN18/21 | ND | F | − | 64 | 8 | 0.5 | 0.125 | 16 | 16 | 4 | − | − | − | − | 4 | 11 |
| BN18/21(pRamR) | ND | F | − | 8 | 2 | 0.125 | 0.060 | 4 | 4 | 1 | − | − | − | − | 1 | This study |
| BN18/21 ΔramA::kan | ND | F | − | 8 | 2 | 0.125 | 0.060 | 4 | 4 | 1 | − | − | − | − | 1 | This study |
| BN18/41 | ND | F | − | >4,096 | 256 | 4 | 2 | 32 | 64 | 8 | G81C | − | − | + | 6 | 11 |
| BN18/41(pRamR) | ND | F | − | 4,096 | 64 | 2 | 0.5 | 16 | 8 | 2 | G81C | − | − | + | 1 | This study |
| BN18/41 ΔramA::kan | ND | F | − | 4,096 | 64 | 2 | 0.5 | 16 | 8 | 2 | G81C | − | − | + | 1 | This study |
| BN18/71 | ND | F | − | >4,096 | 512 | 16 | 8 | 64 | 64 | 16 | G81C | − | − | + | 8 | 11 |
| BN18/71(pRamR) | ND | F | − | 4,096 | 256 | 4 | 2 | 16 | 8 | 2 | G81C | − | − | + | 1 | This study |
| BN18/71 ΔramA::kan | ND | F | − | 4,096 | 256 | 4 | 2 | 16 | 8 | 2 | G81C | − | − | + | 1 | This study |
ND, not determined; F, France; B, Belgium; S, Scotland; Nal, nalidixic acid; Flu, flumequine; Enr, enrofloxacin; Cip, ciprofloxacin; Cm, chloramphenicol; Ff, florfenicol; Tc, tetracycline; SGI1, Salmonella genomic island 1; WB, Western blot with anti-AcrA polyclonal antibody; +, presence; −, absence.
All strains were cultivated at 37°C in Luria-Bertani (LB) or brain heart infusion medium. Mutants carrying the kan gene and transformants carrying the pBR1MCS2 vector were grown in the presence of kanamycin (Fluka Sigma-Aldrich, Saint-Quentin Fallavier, France) at 50 μg/ml.
MIC determination.
Susceptibility testing was performed according to the guidelines of the CASFM (http://www.sfm.asso.fr/nouv/general.php?pa=2). The MICs of nalidixic acid (Fluka Sigma-Aldrich, Saint-Quentin Fallavier, France), flumequine (Sigma, St. Louis, MO), enrofloxacin (Vetoquinol, Lure, France), ciprofloxacin (Fluka Sigma-Aldrich, Saint-Quentin Fallavier, France), chloramphenicol (Fluka Sigma-Aldrich, Saint-Quentin Fallavier, France), florfenicol (Schering-Plough Animal Health, Kenilworth, NJ), and tetracycline (Fluka Sigma-Aldrich, Saint-Quentin Fallavier, France) were determined by the standard agar doubling dilution method as described previously (20).
Construction of the marR, marA, soxR, soxS, ramR, and ramA deletion mutants.
The Datsenko and Wanner gene inactivation method (9) was used to create ΔmarR::kan, ΔmarA::kan, ΔsoxR::kan, ΔsoxS::kan, ΔramR::kan, and ΔramA::kan mutants of the susceptible serovar Typhimurium DT104 strain S/921495 as described previously (6). Plasmid pKD4 carrying the kan gene was used as the plasmid template. The 50 nucleotides that are homologous to the gene to be inactivated and that extend to the pKD4-specific primers P1 and P2 (9) are listed in Table 2. The ramA mutation was further introduced into all serovar Typhimurium strains studied (Table 1) by transduction using phage P22 as described previously (4, 5, 6, 20). The resulting ramA::kan mutants were selected on LB plates containing 50 μg/ml of kanamycin. Replacement of the target gene with the kan resistance gene was confirmed by PCR using the k2 and kt primers and primers flanking the deleted regions (Table 2) (9, 19). The kan resistance gene was eliminated from the S/921495 ΔramR::kan strain by using the pCP20 helper plasmid, which acts on the repeated sites flanking the resistance gene (9).
TABLE 2.
Primers used for PCRs
| Primer use and target region | Primer(s) | Nucleotide positiona | Oligonucleotide sequence(s) (5′ to 3′) | Annealing temp (°C) |
|---|---|---|---|---|
| Construction of deletion mutants | ||||
| ramA-kan | ramAH1-P1 | 639041 | ACACGATTGTCGAGTGGATTGATGATAATTTGAATCAGCCGTTAC-GTGTAGGCTGGAGCTGCTTC | 52 |
| ramAH2-P2 | 639285 | ACGATAAGCGCCTGGCGGCAGGTTGAACGTGCGGGTAAAAATGCG-CATATGAATATCCTCCTTAG | 52 | |
| ramR-kan | ramRH1-P1 | 638183 | TCGAATCCCAGCGCAATATATTCGCCAGCGCGAGCGGGATCGCGC-GTGTAGGCTGGAGCTGCTTC | 52 |
| ramRH2-P2 | 638650 | AAGCATTACTGGAAGCGGCAACCCAGGCGAAACGCAATCCGGTAT-CATATGAATATCCTCCTTAG | 52 | |
| marA-kan | marAH1-P1 | 1597108 | GGTTCAGCGGCAGCATATACCGTGATTCGCCATGCATATT-GTGTAGGCTGGAGCTGCTTC | 52 |
| marAH1-P2 | 1597328 | TTCCAAATGGCACCTGCAACGGATGTTTAAAAAAGAGACC-CATATGAATATCCTCCTTAG | 52 | |
| marR-kan | marRH1-P1 | 1597517 | GCAAATACTCAAGCGTTGCCACTTCGTCCGCCGTTAAGTT-GTGTAGGCTGGAGCTGCTTC | 52 |
| marRH2-P2 | 1597796 | TTATCCCCGCTGGATATCACCGCAACACAGTTTAAAGTGC-CATATGAATATCCTCCTTAG | 52 | |
| soxS-kan | soxSH1-P1 | 4503981 | GACGGTAATCGCTGGGAGTGCGATCGAACTCGCGGCGGAA-GTGTAGGCTGGAGCTGCTTC | 52 |
| soxSH2-P2 | 4504210 | TGAACATATCGACCAACCGCTAAACATTGATGTGGTGGCA-CATATGAATATCCTCCTTAG | 52 | |
| soxR-kan | soxRH1-P1 | 4504393 | CTCCCCGTTTAAAAGCCTTACTGACGCCGGGGGAAGTTGC-GTGTAGGCTGGAGCTGCTTC | 52 |
| soxRH2-P2 | 4504767 | CCCGTGTTCGCCAAGCCTGTCGCCTGGATTTCGCAGCGGA-CATATGAATATCCTCCTTAG | 52 | |
| Construction of complementation plasmid pRamR | ||||
| ramR | BamHI-ramR1 | 638092 | CATGGGATCC-CGTGTCGATAACCTGAGCGG | 62 |
| EcoRI-ramR2 | 639026 | CTAGGAATTC-AAGGCAGTTCCAGCGCAAAG | 62 | |
| Detection of mutations | ||||
| ramR-ramA | ramR1 | 638092 | CGTGTCGATAACCTGAGCGG | 62 |
| ramR2 | 639026 | AAGGCAGTTCCAGCGCAAAG | 62 | |
| RT-PCR expression analysis | ||||
| ramA | ramA3 | 639043 | CACGATTGTCGAGTGGATTG | 58 |
| ramA4 | 639275 | AAAATGCGCGTAAAGGTTTG | 58 | |
| gyrB | gyrB3 | 4040174 | TACCTGCTGGAAAACCCATC | 58 |
| gyrB4 | 4039810 | CTTGTCCGGGTTGTACTCGT | 58 |
Nucleotide position according to the complete genome of S. enterica serovar Typhimurium LT2 (GenBank accession number NC_003197).
Complementation with the ramR gene.
The ramR-complementing plasmid was constructed as follows: the 934-bp ramR fragment generated by PCR using primers BamHI-ramR1 and EcoRI-ramR2 (Table 2) from the genomic DNA of strain S/921495 was digested with EcoRI and BamHI (Promega, Madison, WI), purified, and ligated into the EcoRI- and BamHI-digested broad-host-range plasmid vector pBR1MCS2 (17). The resulting recombinant plasmid was then electroporated into E. coli TG1 cells, with selection on LB agar plates containing 50 μg/ml of kanamycin. The cloned wild-type ramR gene was transferred into the serovar Typhimurium strains by transformation with the recombinant plasmid. The effect of complementation with the wild-type ramR gene was examined by determining antibiotic susceptibilities.
Detection of mutations in the ramR region.
The presence of mutations in the region ranging from the 3′ end of ramR to the 5′ end of ramA was assessed by PCR (Fig. 1). The sequences of the primers used are shown in Table 2. PCR was performed with a 0.2 μM concentration of each primer, a 200 μM concentration of the deoxynucleoside triphosphates (dNTPs), 1× Taq buffer, and 1.25 U of Taq DNA polymerase. A single colony of each serovar Typhimurium strain was used as the template DNA. After a 5-min denaturation at 95°C, amplification was performed for 30 cycles of 1 min at 95°C, 2 min at 60°C, and 2 min at 72°C, with a final extension of 10 min at 72°C. The PCR products were sent for nucleotide sequencing to Genome Express (Meylan, France). Mutations in this amplified region were analyzed by using BLASTN (http://www.ncbi.nlm.nih.gov/BLAST/) and with multiple sequence alignments using CLUSTALW (http://www.ebi.ac.uk/clustalw/).
AcrA expression analysis by dot blotting.
The dot blotting used in this study was adapted from a Western blot method previously described (11). Bacteria were grown at late exponential phase at 37°C in LB medium, harvested by centrifugation, and resuspended at an A600 of 10.0. Cells were diluted to one-half in the sample buffer of Laemmli and were heated for 10 min at 100°C. Whole-cell proteins were spotted onto a nitrocellulose membrane. The membrane was washed three times with Tris-buffered saline (TBS; 0.15% NaCl, 10 mM Tris-HCl [pH 7.5]), saturated for 30 min at room temperature with TBS containing 1% skim milk, and incubated overnight at room temperature with an anti-AcrA polyclonal antibody diluted 1/2,000 in TBS containing 0.33% skim milk. After three washes in TBS, the membrane was incubated for 1 h with peroxidase conjugated to protein A (Sigma, St. Louis, MO) diluted 1/1,000 in TBS. Finally, after three washes in TBS, the blot was revealed with the ECL detection system (GE Healthcare, Chalfont, United Kingdom). The capture of the chemiluminescence image was done by the Chemi-Smart system (Vilber-Lourmat, Marne-la-Vallée, France). The density of each dot was compared to that of the susceptible S/921495 control strain with the Bio1D++ software (Vilber-Lourmat, Marne-la-Vallée, France).
RT-PCR.
Reverse transcription-PCR (RT-PCR) was used to assess the expression of ramA. Total RNA (1 μg), dNTPs (500 μM), and 50 ng of random hexamers (Promega, Madison, WI) were incubated for 5 min at 65°C, chilled on ice, and then reverse transcribed in a volume of 20 μl containing 0.01 M of dithiothreitol, 40 U of RNaseOUT RNase inhibitor (Invitrogen, Cergy-Pontoise, France), 200 U of Superscript II reverse transcriptase (Invitrogen, Cergy-Pontoise, France), and 1× first-strand buffer for 50 min at 42°C and then for 15 min at 70°C. Generated cDNA was incubated for 20 min at 37°C with 1 μl of RNase A (500 μg/ml; Qbiogene, Illkirch, France) and stored at −20°C until it was used. Differences in ramA gene expression were estimated by PCR, using the target-specific primers ramA3 and ramA4 (Table 2). Total cDNA (1 μl) was amplified in a 20-μl final volume containing a 0.5 μM concentration of each target-specific primer, a 250 μM concentration of the dNTPs, 1× Taq buffer, and 0.5 U of Taq DNA. Amplifications were performed with an initial step of 3 min at 95°C, followed by 35 cycles of 20 s at 95°C, 20 s at 58°C, and 20 s at 72°C. The constitutive expression of gyrB assessed in the same cDNA preparation was used as a control, using primers gyrB3 and gyrB4 (Table 2). PCR products were detected on 1.5% agarose gel containing ethidium bromide, and the level of gene overexpression was estimated by a comparison of the band intensities relative to those of twofold serial dilutions of cDNAs.
RESULTS
Lack of a role of MarR, MarA, SoxR, and SoxS in the MDR of serovar Typhimurium.
Genes marR, marA, soxR, and soxS were inactivated in susceptible serovar Typhimurium DT104 strain S/921495. According to the MICs, none of these genes' susceptibilities to the quinolones nalidixic acid and flumequine, to the fluoroquinolones enrofloxacin and ciprofloxacin, to the phenicols chloramphenicol and florfenicol, and to tetracycline were affected (Table 1). The inactivation of these genes also did not affect acrAB expression, according to the dot blot results with an anti-AcrA polyclonal antibody (Table 1). These results suggest that the mar and sox regions are not involved in the regulation of the expression of acrAB and, consequently, of MDR in serovar Typhimurium.
Identification of ramR encoding a putative regulatory protein in serovar Typhimurium.
Upstream of ramA in the serovar Typhimurium LT2 genome sequence (GenBank accession number NC_003197), the open reading frame STM0580, coding for a protein of 194 amino acids which, according to its amino acid sequence homology, belongs to the TetR family of transcriptional repressors (Fig. 1A), was detected. The gene is located in the orientation opposite to that of ramA, as is the case for acrR, the local repressor gene of acrAB. Since open reading frame STM0580 might be the local repressor of ramA, it was named ramR in the present study. A putative RamR binding site was found upstream of ramA in the 288-bp-long ramA-ramR intergenic region corresponding to inverted repeat sequences downstream of the putative promoter region (Fig. 1A).
Characterization of ramR, coding for the local repressor of ramA in serovar Typhimurium.
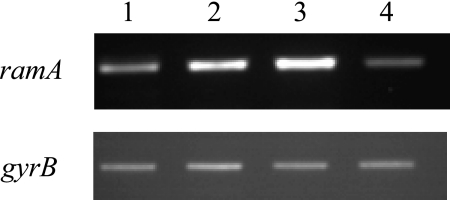
Inactivation of the putative repressor gene upstream of ramA in the susceptible serovar Typhimurium DT104 strain S/921495 resulted in an MDR phenotype, with fourfold increases in the MICs of nalidixic acid, flumequine, enrofloxacin, ciprofloxacin, chloramphenicol, florfenicol, and tetracycline. Complementation with a plasmid containing the wild-type gene restored the initial susceptibilities of the strain. The inactivation of the putative repressor gene also resulted in a fourfold-increased expression of ramA, as shown by RT-PCR (Fig. 2), and a fourfold-increased expression of the AcrAB efflux pump, according to the AcrA dot blot results (Table 1). Complementation with the wild-type gene restored the basal levels of expression of ramA (Fig. 2) and AcrA (Table 1). These results indicated that the gene encodes a local repressor of ramA, and thus, the designation ramR was justified.
FIG. 2.
RT-PCR analysis of ramA and gyrB (control) expression in serovar Typhimurium wild-type strain S/921495 (lane 1), deletion mutant strains S/921495 ΔramR::kan (lane 2) and S/92/1495 ΔramR (lane 3), and the complemented strain S/921495 ΔramR(pRamR) (lane 4).
The inactivation of ramA in serovar Typhimurium DT104 strain S/921495 did not affect its susceptibilities to nalidixic acid, flumequine, enrofloxacin, ciprofloxacin, chloramphenicol, florfenicol, and tetracycline and also did not affect the expression of the AcrAB efflux pump (Table 1). This can be explained by the fact that the expression of ramR is the major cause of the downregulation of ramA expression and, consequently, of acrAB expression.
Identification of mutations in ramR and in the regulatory region of ramA participating in the MDR of serovar Typhimurium.
To date, we have not found any mutations in the mar or sox region that may explain the overexpression of acrAB in a set of previously studied MDR serovar Typhimurium strains that are also quinolone or fluoroquinolone resistant (Table 1) (4, 5, 6, 11, 19, 20). Since, in contrast to mar and sox, the ram region appears to play a major role in the regulation of acrAB expression, we looked for mutations in ramR and in the ramR-ramA intergenic region in this set of strains. As shown in Fig. 1, in all strains studied, several point mutations which resulted in amino acid changes or in a frameshift were identified in ramR (Fig. 1C). In addition, the interruption of ramR by an IS1 element was seen in high-level fluoroquinolone-resistant serovar Typhimurium DT204 strains (Fig. 1D). One serovar Typhimurium DT104 isolate had a 2-nucleotide deletion in the putative RamR binding site found upstream of ramA (Fig. 1B). These mutations were confirmed to play a role in the MDR phenotype by complementation with the wild-type ramR gene or inactivation of their respective ramA genes (Table 1). Interestingly, in MDR DT104 strains overexpressing acrAB and carrying the Salmonella genomic island 1-borne floR and tet(G) phenicol and tetracycline efflux pump genes, respectively, the inactivation of the ramA gene resulted in two- to fourfold decreases in levels of resistance to chloramphenicol, florfenicol, and tetracycline. In a previous study, we had shown that there was interplay between the FloR, Tet(G), and AcrAB efflux pumps to obtain high levels of resistance to phenicols and tetracycline (6).
DISCUSSION
Following a study using the salicylate induction of marA, it has been suggested that efflux-mediated MDR resistance in serovar Typhimurium may occur by both mar-dependent and mar-independent pathways (25). In another study on the bile salt-mediated induction of antimicrobial and bile resistance in serovar Typhimurium, it was concluded that while the transcription of acrAB was activated by bile, this activation was independent of marRAB (23). It was also observed that there was no correlation between cyclohexane tolerance in the multiple-antibiotic-resistant mutants of 14 different S. enterica serovars and the overexpression of acrB, soxS, or marA (32). In a more recent study on the selection of ciprofloxacin-resistant serovar Typhimurium, it was suggested that neither marA nor soxS is critical for S. enterica to generate a multiple-antibiotic-resistant mutant (26). Besides, no mutations in the mar region that would explain the overexpression of acrAB in serovar Typhimurium have been described to date (19, 24). In the case of the sox region, Koutsolioutsou et al. have shown that a clinical isolate of serovar Typhimurium became resistant by a point mutation in soxR (15). However, the mechanism by which this MDR resistance was conferred was not examined, and whether soxR influenced the expression of AcrAB or another efflux pump was not explored (21). For all these reasons, we reexamined the roles of the mar and sox regions by inactivating the marR, marA, soxR, and soxS genes in serovar Typhimurium DT104 strain S/921495. The inactivation of these genes did not alter the strain's susceptibilities to quinolones, fluoroquinolones, phenicols, and tetracycline. There was also no effect on AcrA production. Our results thus indicated that in contrast to the situation in E. coli, the mar or sox region likely does not play a significant role in the efflux-mediated MDR phenotype via the overexpression of acrAB in serovar Typhimurium.
In Salmonella spp. and in other bacteria, such as E. aerogenes, E. cloacae, and K. pneumoniae, RamA, a homologue of MarA that is absent in E. coli, has been shown to be implicated in MDR, and the overexpression of ramA correlated well with an increased AcrAB efflux pump expression (7, 12, 13, 27-30, 33). In this study, the inactivation of ramR and complementation experiments with DT104 strain S/921495 confirmed that it codes for the local repressor of ramA, as we observed fourfold increases in the MICs of the antibiotics tested, which correlated well with the fourfold overexpression of ramA and also with the fourfold overproduction of AcrA. However, the inactivation of ramA in the control strain S/921495 did not affect antibiotic susceptibilities or AcrA production. Most probably, the expression of ramA is basal, and to affect antibiotic susceptibilities, the local repressor RamR must be affected either by an as-yet-unknown mechanism or by mutations in the ramR gene or in the RamR binding region. Therefore, we investigated the presence and role of mutations in the ramR region of a set of quinolone- or fluoroquinolone-resistant serovar Typhimurium strains previously shown to overproduce AcrAB (4, 5, 6, 11, 19, 20). Complementation experiments with the wild-type ramR gene confirmed that the mutations identified were critical for the RamR repressor function according to the decreased MICs of quinolones and fluoroquinolones observed as well as to the decreased AcrA production observed (Table 1). On the other hand, the inactivation of ramA in these strains also resulted in two- to fourfold reductions in the MICs of quinolones and fluoroquinolones (Table 1). All these data indicate that mutations in the ramR region may result in an up-to-fourfold increase in levels of resistance to unrelated antibiotics via ramA and acrAB overexpression. With additional mutations in acrR, as in serovar Typhimurium strains BN18/41 and BN18/71, the resistance levels and acrAB overexpression may increase up to eightfold (Table 1).
In conclusion, efflux-mediated MDR resistance in serovar Typhimurium via the overexpression of acrAB is assumed to be due mainly to what we can now call the ram regulon, whereas the mar and sox regions do not seem to play a significant role in this resistance. Probably, as with the mar regulon in E. coli, the ram regulon in serovar Typhimurium may be involved in the regulation of other genes; the first example is the flavohemoglobin (hmp) gene (12), and this should be further investigated by global transcriptomic or proteomic approaches.
Acknowledgments
We thank C. Mouline for his expert technical assistance.
Footnotes
Published ahead of print on 28 April 2008.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410-413. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baucheron, S., E. Chaslus-Dancla, and A. Cloeckaert. 2004. Role of TolC and parC mutation in high-level fluoroquinolone resistance in Salmonella enterica serotype Typhimurium DT204. J. Antimicrob. Chemother. 53:657-659. [DOI] [PubMed] [Google Scholar]
- 5.Baucheron, S., H. Imberechts, E. Chaslus-Dancla, and A. Cloeckaert. 2002. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type DT204. Microb. Drug Resist. 8:281-289. [DOI] [PubMed] [Google Scholar]
- 6.Baucheron, S., S. Tyler, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. AcrAB-TolC directs efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium DT104. Antimicrob. Agents Chemother. 48:3729-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chollet, R., J. Chevalier, C. Bollet, J.-M. Pages, and A. Davin-Regli. 2004. RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob. Agents Chemother. 48:2518-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloeckaert, A., and E. Chaslus-Dancla. 2001. Mechanisms of quinolone resistance in Salmonella. Vet. Res. 32:291-300. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraud, E., S. Baucheron, and A. Cloeckaert. 2006. Resistance to fluoroquinolones in Salmonella: emerging mechanisms and resistance prevention strategies. Microbes Infect. 8:1937-1944. [DOI] [PubMed] [Google Scholar]
- 11.Giraud, E., A. Cloeckaert, D. Kerboeuf, and E. Chaslus-Dancla. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez-Urzua, E., D. S. Zamorano-Sanchez, J. Ponce-Coria, E. Morett, S. Grogan, R. K. Poole, and J. Membrillo-Hernandez. 2007. Multiple regulators of the flavohaemoglobin (hmp) gene of Salmonella enterica serovar Typhimurium include RamA, a transcriptional regulator conferring the multidrug resistance phenotype. Arch. Microbiol. 187:67-77. [DOI] [PubMed] [Google Scholar]
- 13.Keeney, D., A. Ruzin, and P. A. Bradford. 2007. RamA, a transcriptional regulator, and AcrAB, an RND-type efflux pump, are associated with decreased susceptibility to tigecycline in Enterobacter cloacae. Microb. Drug Resist. 13:1-6. [DOI] [PubMed] [Google Scholar]
- 14.Kern, W. V., M. Oethinger, A. S. Jellen-Ritter, and S. B. Levy. 2000. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 44:814-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koutsolioutsou, A., E. A. Martins, D. G. White, S. B. Levy, and B. Demple. 2001. A soxRS-constitutive mutation contributing to antibiotic resistance in a clinical isolate of Salmonella enterica (serovar Typhimurium). Antimicrob. Agents Chemother. 45:38-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koutsolioutsou, A., S. Peña-Llopis, and B. Demple. 2005. Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob. Agents Chemother. 49:2746-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 18.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 19.Olliver, A., M. Vallé, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Role of an acrR mutation in multidrug resistance of in vitro-selected fluoroquinolone-resistant mutants of Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 238:267-272. [DOI] [PubMed] [Google Scholar]
- 20.Olliver, A., M. Vallé, E. Chaslus-Dancla, and A. Cloeckaert. 2005. Overexpression of the multidrug efflux operon acrEF by insertional activation with IS1 or IS10 elements in Salmonella enterica serovar Typhimurium DT204 acrB mutants selected with fluoroquinolones. Antimicrob. Agents Chemother. 49:289-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piddock, L. J. V. 2002. Fluoroquinolone resistance in Salmonella serovars isolated from humans and food animals. FEMS Microbiol. Rev. 26:3-16. [DOI] [PubMed] [Google Scholar]
- 22.Piddock, L. J. V. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prouty, A. M., I. E. Brodsky, S. Falkow, and J. S. Gunn. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150:775-783. [DOI] [PubMed] [Google Scholar]
- 24.Randall, L. P., S. W. Cooles, L. J. V. Piddock, and M. J. Woodward. 2004. Effect of triclosan or a phenolic farm disinfectant on the selection of antibiotic-resistant Salmonella enterica. J. Antimicrob. Chemother. 54:621-627. [DOI] [PubMed] [Google Scholar]
- 25.Randall, L. P., and M. J. Woodward. 2001. Multiple antibiotic resistance (mar) locus in Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 67:1190-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ricci, V., P. Tzakas, A. Buckley, N. C. Coldham, and L. J. V. Piddock. 2006. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob. Agents Chemother. 50:38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruzin, A., M. A. Visalli, D. Keeney, and P. A. Bradford. 2005. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 49:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneiders, T., S. G. B. Amyes, and S. B. Levy. 2003. Role of AcrR and RamA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob. Agents Chemother. 47:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Straaten, T., R. Janssen, D. J. Mevius, and J. T. van Dissel. 2004. Salmonella gene rma (ramA) and multiple-drug-resistant Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 48:2292-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Straaten, T., L. Zulianello, A. van Diepen, D. L. Granger, R. Janssen, and J. T. van Dissel. 2004. Salmonella enterica serovar Typhimurium RamA, intracellular oxidative stress response, and bacterial virulence. Infect. Immun. 72:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velge, P., A. Cloeckaert, and P. Barrow. 2005. Emergence of Salmonella epidemics: the problems related to Salmonella enterica serotype Enteritidis and multiple antibiotic resistance in other major serotypes. Vet. Res. 36:267-288. [DOI] [PubMed] [Google Scholar]
- 32.Webber, M., A. M. Buckley, L. P. Randall, M. J. Woodward, and L. J. V. Piddock. 2006. Overexpression of marA, soxS and acrB in veterinary isolates of Salmonella enterica rarely correlates with cyclohexane tolerance. J. Antimicrob. Chemother. 57:673-679. [DOI] [PubMed] [Google Scholar]
- 33.Yassien, M. A., H. E. Ewis, C.-D. Lu, and A. T. Abdelal. 2002. Molecular cloning and characterization of the Salmonella enterica serovar Paratyphi B rma gene, which confers multiple drug resistance in Escherichia coli. Antimicrob. Agents Chemother. 46:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]