Abstract
Plasmodium vivax mdr1 gene amplification, quantified by real-time PCR, was significantly more common on the western Thailand border (6 of 66 samples), where mefloquine pressure has been intense, than elsewhere in southeast Asia (3 of 149; P = 0.02). Five coding mutations in pvmdr1, independent of gene amplification, were also found.
An increase in the copy number of the Plasmodium falciparum mdr1gene is the most important determinant of mefloquine resistance in vitro and in vivo (3, 6-8) and is inversely correlated with chloroquine resistance (1). To assess Plasmodium vivax mdr1 gene amplification, we developed a real-time PCR method and evaluated copy numbers and polymorphisms in P. vivax samples from Laos, Myanmar, and Thailand, areas with considerable differences in antimalarial drug usage and P. falciparum antimalarial drug susceptibility.
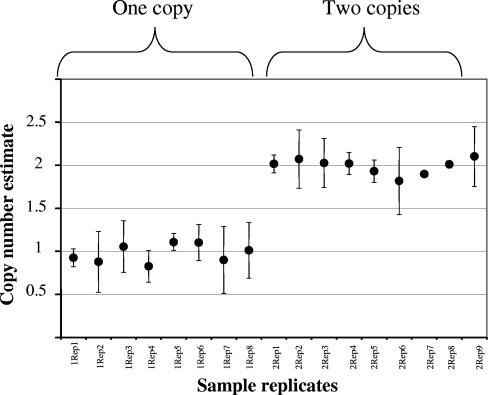
Dry blood samples were collected before treatment from 215 patients with acute vivax malaria from three areas: 66 came from Tak province on the Thai-Myanmar border, 50 were from elsewhere in Thailand, 50 were from Laos, and 49 were from Myanmar. The pvmdr1 copy number was assessed by a novel real-time PCR method on a Corbett Rotor-Gene 3000 (Corbett Research, Australia). The primers and probes were PVMDR1F (CAGCCTGAAAGATTTAGAAGCCTT), PVMDR1R (CGGCTGTTGGAATCACTTTGA), PVMDR1probe (FAM-CGGAGGAGTCGAACGAAGATGGTTTTTCTT-TAMRA), PVTUBULINF (TCGCTTAACGACGTCCCC), PVTUBULINR (TGGAATGTCACAAACGCTGG), and PVTUBULINprobe (VIC-TTCCGCTTCCCCCTCCACAGG-TAMRA). A QuantiTect Multiplex PCR NoROX (Qiagen, Germany) was used, and the temperature profile was prepared according to the manufacturer's instructions. The calibrator, a single-copy control, is a plasmid that was constructed by the insertion of pvmdr1 (nucleotides [nt] 1102 to 1993) and pvtubulin (nt 14393 to 2354) fragments in a ratio of 1:1 into the pCR2.1 vector using a TOPO TA cloning kit (Invitrogen U.S.A.). β-Tubulin served as an internal control to normalize the amount of sample DNA added to the reactions. The relative amounts of the target genes were calculated by using the comparative Ct method. Copy numbers were calculated as follows: copy number = 2−ΔΔCt. All assays with samples containing two copy numbers pvmdr1 were repeated five times, and 33% of the single copy number pvmdr1 analyses were repeated twice. The results were consistent (Fig. 1). Three fragments (496, 590, and 541 bp) of pvmdr1, which covered nt 158 to 653, 2752 to 3341, and 3683 to 4223, respectively, were amplified by PCR. Direct sequencing from PCR products was performed by using an ABI automated sequencer.
FIG. 1.
Copy number estimates (with indicated confidence intervals) for parasites with one or two copies.
Double pvmdr1 copies were significantly more common in Tak province (6 of the 66 samples) than elsewhere in Thailand (0 of 50; Fisher's exact test, P = 0.03). In the samples from Laos (n = 50) and Myanmar (n = 49), a double copy number pvmdr1 was found in only two and one P. vivax isolates, respectively. Comparison of pvmdr1 sequences with the published wild-type sequence pfmdr1 (M29154) revealed that the polymorphisms found in P. falciparum (at codons 86, 184, 1034, 1042, and 1246) corresponded to homologous mutations in pvmdr1 (AY618622) at codons 91, 189, 1071, 1079, and 1291, respectively (Table 1). pvmdr1 sequences were obtained for the 21 different isolates and the Belem laboratory strain. Compared to Sal1 as the reference wild type, 48 mutations were distributed among five positions, but none of these mutations corresponded to those in pfmdr1. Four of the mutations in pvmdr1 were nonsynonymous. These were at codons 133, 139, 976, 1076, and 1261, which are equivalent to codons 128, 134, 940, 1039, and 1216 in pfmdr1 (Table 1). The recent sequencing of pvmdr1 has revealed a single open reading frame of 4,392 bp encoding a deduced protein of 1,464 amino acids, with 12 transmembrane segments (2). Brega et al. reported two SNPs (976 and 1076) in the pvmdr1 gene, which were not associated with chloroquine resistance in P. falciparum. Sa et al. compared pvmdr1 from 10 isolates with different levels of chloroquine sensitivity and did not find a correlation between codon mutations and resistance (9).
TABLE 1.
Mutations present in the pvmdr1 genes amplified from P. vivax isolates
| Isolate | Origin | Residue at codona:
|
Pvmdr1 copy no. | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 86N/91N | 128*K/133K | 134*S/139S | 184Y/189Y | 940*Y/976Y | 1034S/1071S | 1039*L/1076F | 1042N/1079N | 1216Q/1261K | 1246D/1291D | |||
| Belem | Brazil | . | . | . | . | . | . | . | . | E | . | 1 |
| PV3217 | Thailand | . | . | . | . | . | . | . | . | E | . | 1 |
| PV3247 | Thailand | . | . | . | . | . | . | . | . | E | . | 1 |
| PV3224 | Thailand | . | N | R | . | . | . | . | . | E | . | 1 |
| PV3265 | Thailand | . | . | . | . | . | . | L | . | E | . | 1 |
| PV3251 | Thailand | . | . | . | . | F | . | L | . | E | . | 1 |
| L20 | Laos | . | . | . | . | . | . | L | . | E | . | 1 |
| Lao 2003 15 | Laos | . | N | R | . | . | . | L | . | E | . | 1 |
| Lao 2003 32 | Laos | . | N | R | . | . | . | L | . | E | . | 1 |
| P122 | Myanmar | . | . | . | . | . | . | . | . | E | . | 1 |
| P137 | Myanmar | . | . | . | . | . | . | . | . | E | . | 1 |
| P90 | Myanmar | . | . | . | . | F | . | L | . | E | . | 1 |
| P143 | Myanmar | . | . | . | . | F | . | L | . | E | . | 1 |
| P152 | Myanmar | . | . | . | . | F | . | L | . | E | . | 1 |
| PV3230 | Thailand | . | . | . | . | . | . | L | . | E | . | 2 |
| PV3222 | Thailand | . | . | . | . | . | . | L | . | E | . | 2 |
| PV2677 | Thailand | . | . | . | . | . | . | L | . | E | . | 2 |
| PV2667 | Thailand | . | . | . | . | . | . | L | . | E | . | 2 |
| PV3232 | Thailand | . | . | . | . | . | . | L | . | E | . | 2 |
| PV3246 | Thailand | . | . | . | . | . | . | L | . | E | . | 2 |
| L10 | Laos | . | . | . | . | . | . | L | . | E | . | 2 |
| L27 | Laos | . | . | . | . | . | . | L | . | E | . | 2 |
*, No mutant type was found at codons 128, 134, 940, and 1039 in pfmdr1, Wild-type residues (position number and residue) are as indicated in the subheadings in the form: P. falciparum (accession no. M29154)/P. vivax (accession no.618622). A period (.) in the table indicates no change from the wild-type sequence.
Gene amplification is a potentially important resistance mechanism. This study confirms mdr1 amplification does occur in P. vivax, and this varied between geographical locations differing in their use of antimalarial drugs. In Tak province, on the Thai-Burmese border, mefloquine alone or in combination has been used for 23 years. In P. falciparum intense and sustained mefloquine pressure has been associated with high rates of selection for pfmdr1 amplification (13). In other areas of Thailand, Myanmar, and Laos, there has been less exposure of parasites to mefloquine. Gene amplifications of pvmdr1 were significantly more common in isolates from Tak (6 of the 66 samples) than in those from patients from other provinces of Thailand, Myanmar, and Laos (P = 0.02), suggesting that there is a relationship between the pvmdr1 copy number and widespread deployment of mefloquine. The degree of amplification is low currently (no more than two copies), whereas in P. falciparum up to five copies of pfmdr1 have been observed in this region. Point mutations at codons 133, 139, 976, 1076, and 1261 of the pvmdr1 gene (corresponding to codons 128, 134, 940, 1039, and 1216, respectively, in pfmdr1) were found in this study. Mutations at codons 133, 139, and 1261 are located outside the transmembrane segment. Two point mutations at codons 976 and 1076 are located in the putative transmembrane segments X and XI. Double mutations (976 and 1076) in these segments were found in the samples from three areas (31%, n = 13). The Y976F mutation in P. vivax has been correlated with reduced susceptibility to chloroquine (10). This mutation, seen in 1 of 11 Thai and 3 of 5 Myanmar isolates examined here, may affect chloroquine efficacy against P. vivax in this region and is worthy of further exploration. Mutations in pvmdr1 were independent of pvmdr1 amplification. Sal1, a laboratory-adapted P. vivax strain usually regarded as the wild type, was found to possess residue F at position 1076, while the residue L was identified at the correspond codon in pfmdr1 (position 1039, accession no M29154; strain FC27, D10) and pcmdr1 (AY123625), suggesting that the residue L might be a neutral allele. None of the corresponding mutations in pfmdr1 were observed in pvmdr1. This suggests that there may be different mechanisms conferring chloroquine resistance in P. vivax compared to P. falciparum.
Amplification of pfmdr1 has occurred multiple times in P. falciparum (5, 12). This is evident from the ready intrahost selection, the patterns of polymorphism in flanking microsatellite markers, and from the different-sized fragments of chromosome 5 regions containing pfmdr1 that are amplified in different isolates. In contrast, the fact that all P. vivax isolates carrying multiple copies of pvmdr1 have the same point mutations suggests that this gene amplification has a single origin.
Acknowledgments
This work was supported by the Thailand research Fund-the Commission on Higher Education (CHE; MRG 4980091) and the Wellcome Trust, United Kingdom (grant GR080867).
We thank Pakorn Pengin, Naowarat Tanomsing, Ric N. Price, Mayfong Mayxay, Apichart Nontprasert, and Jean-Paul Guthmann for their help.
Footnotes
Published ahead of print on 28 April 2008.
REFERENCES
- 1.Barnes, D. A., S. J. Foote, D. Galatis, D. J. Kemp, and A. F. Cowman. 1992. Selection for high-level chloroquine resistance results in deamplification of the pfmdr1 gene and increased sensitivity to mefloquine in Plasmodium falciparum. EMBO J. 11:3067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brega, S., B. Meslin, F. de Monbrison, C. Severini, L. Gradoni, R. Udomsangpetch, I. Sutanto, F. Peyron, and S. Picot. 2005. Identification of the Plasmodium vivax mdr-like gene (pvmdr1) and analysis of single-nucleotide polymorphisms among isolates from different areas of endemicity. J. Infect. Dis. 191:272-277. [DOI] [PubMed] [Google Scholar]
- 3.Cowman, A. F., D. Galatis, and J. K. Thompson. 1994. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc. Natl. Acad. Sci. USA 91:1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayxay, M., S. Pukrittayakamee, P. N. Newton, and N. J. White. 2004. Mixed-species malaria infections in humans. Trends Parasitol. 20:233-240. [DOI] [PubMed] [Google Scholar]
- 5.Nair, S., D. Nash, D. Sudimack, A. Jaidee, M. Barends, A. C. Uhlemann, S. Krishna, F. Nosten, and T. J. Anderson. 2007. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol. Biol. Evol. 24:562-573. [DOI] [PubMed] [Google Scholar]
- 6.Peel, S. A., P. Bright, B. Yount, J. Handy, and R. S. Baric. 1994. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 51:648-658. [DOI] [PubMed] [Google Scholar]
- 7.Price, R. N., C. Cassar, A. Brockman, M. Duraisingh, M. van Vugt, N. J. White, F. Nosten, and S. Krishna. 1999. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob. Agents Chemother. 43:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price, R. N., A. C. Uhlemann, A. Brockman, R. McGready, E. Ashley, L. Phaipun, R. Patel, K. Laing, S. Looareesuwan, N. J. White, F. Nosten, and S. Krishna. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sa, J. M., T. Nomura, J. Neves, J. K. Baird, T. E. Wellems, H. A. del Portillo. 2005. Plasmodium vivax: allele variants of the mdr1 gene do not associate with chloroquine resistance among isolates from Brazil, Papua, and monkey-adapted strains. Exp. Parasitol. 109:256-259. [DOI] [PubMed] [Google Scholar]
- 10.Suwanarusk, R., B. Russell, M. Chavchich, F. Chalfein, E. Kenangalem, V. Kosaisavee, B. Prasetyorini, K. A. Piera, M. Barends, A. Brockman, U. Lek-Uthai, N. M. Anstey, E. Tjitra, F. Nosten, Q. Cheng, and R. N. Price. 2007. Chloroquine-resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS ONE 2:e1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thaithong, S., L. C. Ranford-Cartwright, N. Siripoon, P. Harnyuttanakorn, N. S. Kanchanakhan, A. Seugorn, K. Rungsihirunrat, P. V. Cravo, and G. H. Beale. 2001. Plasmodium falciparum: gene mutations and amplification of dihydrofolate reductase genes in parasites grown in vitro in presence of pyrimethamine. Exp. Parasitol. 98:59-70. [DOI] [PubMed] [Google Scholar]
- 12.Triglia, T., S. Foote, D. J. Kemp, and A. F. Cowman. 1991. Amplification of the multidrug resistance gene pfmdr1 in Plasmodium falciparum has arisen as multiple independent events. Mol. Cell. Biol. 11:5244-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uhlemann, A. C., M. Ramharter, B. Lell, P. G. Kremsner, and S. Krishna. 2005. Amplification of Plasmodium falciparum multidrug resistance gene 1 in isolates from Gabon. J. Infect. Dis. 192:1830-1835. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe, J., and J. Inselberg. 1994. Establishing a physical map of chromosome no. 4 of Plasmodium falciparum. Mol. Biochem. Parasitol. 1:189-199. [DOI] [PubMed] [Google Scholar]