Abstract
The oxacillinase gene was reported to confer limited resistance to carbapenem in Acinetobacter baumannii. In this study, we have demonstrated that an A. baumannii clinical isolate harboring a plasmid, pTVICU53, has 11,037 bp encoding 13 open reading frames. A blaOXA-58 gene with an upstream insertion of truncated ISAba3 (ΔISAba3) and IS1008 was found in this plasmid. ΔISAba3and IS1008 provided two independent promoters for the transcription control of the blaOXA-58 gene. The transformation of pTVICU53 or a shuttle vector bearing IS1008-ΔISAba3-blaOXA-58 to different A. baumannii recipients can increase their MICs of carbapenem 64- to 256-fold. The deletion of promoters provided by IS1008 resulted in dramatic decreases in blaOXA-58 transcription and a 32- to 64-fold reduction in the carbapenem MIC. These findings highlight that A. baumannii might develop carbapenem resistance with a single transformation step, taking up a plasmid containing a genetic construct with a potentially high level of transcription of the blaOXA-58 gene.
Outbreaks of nosocomial infections caused by multidrug-resistant Acinetobacter baumannii strains have been reported with increasing frequency over the past three decades (1, 10, 28). Carbapenems including imipenem and meropenem have been the drugs of choice for the treatment of severe infection caused by multidrug-resistant A. baumannii strains (10). However, strains that are resistant to these potent antibiotics have evolved worldwide (18, 23, 31) and thereby severely compromised the effective therapeutic options. Therefore, there is an urgent need to understand the mechanisms of carbapenem resistance in A. baumannii.
A number of different molecular mechanisms associated with carbapenem resistance in A. baumannii clinical strains have been reported (31), such as the acquisition of carbapenemases (6, 31), a loss of an outer membrane porin (13, 27), a change of penicillin binding protein (17, 20), and the overexpression of efflux systems (29).
Among these mechanisms, the acquisition of carbapenemases plays the major role in carbapenem resistance in most gram-negative bacilli, including A. baumannii clinical isolates (30, 31, 37). Enzymes with carbapenem-hydrolyzing activity belong to either Ambler class A, class B (metallo-β-lactamases, [MBLs]), or class D (carbapenem-hydrolyzing oxacillinases [CHDLs]) β-lactamases (33). MBLs have the highest level of carbapenem-hydrolyzing activity among these three classes of carbapenemases. They have been identified in many gram-negative bacteria and Acinetobacter genomic species 13 TU but rarely in A. baumannii (25, 26, 31). Class A carbapenemases have been frequently detected in Enterobacter cloacae, Serratia marcescens, and Klebsiella spp. but not in Acinetobacter spp. (33). In fact, the most common carbapenemases detected in A. baumannii were CHDLs (25, 31). Among the nine clusters of CHDLs (33), four have been identified to date in A. baumannii (5). These included members of OXA-23, -24, -51, and -58 families. However, their reported hydrolytic efficiencies for carbapenems were relatively low (37) and were 100- to 1,000-fold less than that of the MBLs (31). Thus, the exact contribution of these enzymes to carbapenem resistance in A. baumannii is uncertain (31). It has been suggested that a combination of antibiotic resistance mechanisms might be needed for a high level of carbapenem resistance in clinical strains of A. baumannii (5, 37), such as the coexistence of carbapenemases and the overexpression of the efflux system (22) or a decreased level of expression of porin (4, 12).
In this study, we have demonstrated that a plasmid-borne CHDL with appropriate upstream insertion sequences (ISs) was enough to confer a high level of carbapenem resistance in A. baumannii. The blaOXA-58 gene with an upstream insertion of a truncated ISAba3 (ΔISAba3) and IS1008 was detected on a plasmid obtained from a clinical carbapenem-resistant isolate. Electrotransformation of the original or recombinant plasmid bearing IS1008-ΔISAba3-blaOXA-58 into different A. baumannii strains conferred a 64- to 256-fold increase in their carbapenem MICs. The deletion of IS1008 resulted in decreases in levels of blaOXA-58 gene expression and carbapenem resistance. This study demonstrates that an efficient mechanism has evolved naturally for A. baumannii to become carbapenem resistant.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A. baumannii clinical isolates TVICU53 and Ab290 were isolated at the Taipei Veterans General Hospital in 2004 and 1999, respectively. Isolate TVICU53 was resistant to most tested antimicrobial agents except aminoglycosides. Isolate Ab290 was susceptible to most of the antimicrobial agents tested but showed intermediate resistance to ceftriaxone (MIC, 16 μg/ml). A. baumannii reference strain ATCC 15151T (CIP 70.10T) was purchased from the Bioresources Collection and Research Center, Taiwan. To exclude contamination during experiments, all A. baumannii isolates including transformants were verified to the genomic species level by a multiplex PCR method (8). Escherichia coli reference strain DH5α was used in the cloning and transformation experiments. The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this work
| Plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| pMAC | 9.5-kb plasmid of A. baumannii (ATCC 19606T) involved in organic peroxide resistance | 15 |
| pTVICU53 | 11-kb plasmid of A. baumannii TVICU53 | This work |
| pYMAb3 | RepM and Ori fragments of pMAC were subcloned into a pET28a-based vector | This work |
| pOXA-58-3 | A fragment containing IS1008-ΔISAba3-blaOXA-58 was subcloned into an E. coli and A. baumannii shuttle vector, pYMAb3 | This work |
| pOXA-58-4 | A fragment containing ΔISAba3-blaOXA-58 was subcloned into an E. coli and A. baumannii shuttle vector, pYMAb3 | This work |
| pET28a | Backbone of pYMAb3; Kmr | Novagen |
| pMAL-c2x | Restriction with HindIII and treatment with alkaline phosphatase; used for cloning of a HindIII-generated fragment of pTVICU53 | New England Biolabs |
General DNA manipulations.
Plasmid DNA was extracted with a plasmid DNA Miniprep kit (Bioman, Taipei, Taiwan) or a Qiagen (Taipei, Taiwan) plasmid Maxi kit. PCR was performed with annealing temperatures appropriate for the primers used (Table 2), and the products were isolated with a PCR purification kit (Geneaid, Taipei, Taiwan). DNA digestions were achieved with selected restriction enzymes according to protocols suggested by the manufacturer (New England Biolabs, Taipei, Taiwan). DNA sequencing was carried out by Mission Biotech, Taipei, Taiwan.
TABLE 2.
Oligonucleotide primers used in PCR or Southern blot analysis
| Primer | Sequence (5′-3′) | Description |
|---|---|---|
| PMAC-ori-XbaI-F | TCTAGAGCTCAGTTGGTTTTATCCAAAG | ori and repM of pMAC |
| PMAC-ori-SphI-R | GCATGCCTATGCAGCTTTGAAGCCTAC | |
| XbaIIS1008 | TCTAGATCGGCACTTTCAAGGTGAAAT | IS1008 |
| XbaIΔISAba3 | TCTAGAGCTAGAGTTATTTGCATTTCTC | ΔISAba3 |
| NcoIOXA58 | CCATGGTTATAAATAATGAAAAACACCCA | End of blaOXA-58 |
| OXA58-Fa | AAGTATTGGGGCTTGTGCTG | Internal fragment of blaOXA-58; used for Southern blot analysis and PCR |
| OXA58-Ba | CCCCTCTGCGCTCTACATAC | |
| OXA51-Ba | TGGATTGCACTTCATCTTGG | blaOXA-51 |
| OXA-58-ext-1 | GTGGCTTTCCATCCCACTTA | Used for 5′ RACE |
| OXA-58-ext-2 | GCGCTTGAACATTCTGATCG | Used for nested PCR; upstream of OXA-58-ext-1 |
| ISAba1Bb | CATGTAAACCAATGCTCACC | tnpA of ISAba1 |
| ISAba2Ab | AATCCGAGATAGAGCGGTTC | tnpA of ISAba2 |
| ISAba3Cb | AGCAATATCTCGTATACCGC | tnpA of ISAba3 |
| IS18Bb | ACCAGCCATAACTTCACTCG | tnpA of IS18 |
| Oligo(dG) | AAATCTAGAGGGGGGGGGGG | Used for PCR of dC tailed cDNA |
| PEU-7 | GCAAACAGGATTAGATACCC | 16S rRNA; used as internal control in RT-PCR |
| PEU-8 | CGTCATCCCCACCTTCCTCC |
PCR detection of carbapenemase genes and ISs and phenotypic assay for MBL production.
The MBL and CHDL genes were detected by PCR methods suggested previously by Ellington et al. (16) and Woodford et al. (38), respectively. A phenotypic assay for MBL production was carried out using a double-disk synergy test and a combined-disk test with imipenem and EDTA according to methods described previously by Franklin et al. (19). Detection of the IS preceding the bla gene was performed by PCR using a forward primer of the IS (ISAba1B, ISAba2A, ISAba3C, and IS18B) (32) and a reverse primer of the bla gene (OXA58-B and OXA51-B) (Table 2).
Electroporation of plasmids into A. baumannii.
A highly efficient electroporation system for A. baumannii using plasmids was established in this study. The electrocompetent cells were prepared according to BTX (San Diego, CA) protocols. Briefly, bacteria grown to exponential phase (optical density at 600 nm of 0.5 to 0.7) were collected at 4°C, washed and resuspended with 15% ice-cold glycerol, and frozen at −70°C until use. The electroporation was performed with an ECM 630 electroporator (BTX, San Diego, CA) in 2-mm-wide cuvettes. Plasmids were electroporated into cells at 2.5 kV, 25 μF, and 200 Ω, and the transformed cell mix was suspended in 1 ml of SOC broth (35) and then incubated at 37°C for 1 h with shaking. Different amounts of electroporated cells were plated and selected with appropriate antibiotics. The transformants were verified by PCR and Southern blot analyses.
Southern blot analyses.
Southern blot analyses were conducted using standard protocols (35) with high-stringency conditions (21). Hybridization probes were labeled with PCR in the reaction mix containing [α-32P]dCTP. The radioactive bands were exposed to Kodak X-OMAT-AR film (Eastman Kodak, NY).
Full-length cloning and sequencing of plasmid pTVICU53.
pTVICU53 was digested into several pieces with HindIII in order to subclone it into vector pMAL-c2x and transformed into E. coli DH5α cells for sequencing. The cloned fragments of pTVICU53 were sequenced from both strands. Sequences were examined and assembled using the published sequence of pMAC (GenBank accession number AY541809) as a template. pMAC is a 9.5-kb plasmid isolated from A. baumannii (ATCC 19606T) (15). Gaps and junctions between fragments of pTVICU53 were filled and verified by PCR and sequencing. The integrity and reliability of pTVICU53 were examined by PCR using primers from different cloned fragments, restriction mapping, and both-direction sequencing. Nucleotide and deduced amino acid sequences were analyzed by software available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). The IS was analyzed using IS Finder (http://www-is.biotoul.fr). The map of pTVICU53 was constructed by BVTech plasmid-drawing software (http://www.biovisualtech.com).
Predictions of transcription initiation sites and promoter regions.
The transcription initiation site of the blaOXA-58 gene was determined by 5′-end rapid amplification of cDNA ends (RACE)-PCR. Total RNA was isolated by RNA TRIzol reagent (Sigma) according to the manufacturer's protocols and treated with RNase-free DNase (Qiagen, Taipei, Taiwan). The first-strand cDNA was synthesized with Moloney murine leukemia virus reverse transcriptase (Epicenter, WI) using 5 μg of total RNA and primer OXA-58-ext-1. An oligo(dC) tail was terminally transferred to the 5′ end of the primer-extended cDNA, which was then PCR amplified using primers oligo(dG) and OXA-58-ext-1. The 5′ RACE-PCR products resolved by electrophoresis were Southern transferred onto a nylon membrane and hybridized with a radiolabeled blaOXA-58 gene probe. The DNA fragments corresponding to the positive signals in the Southern hybridization were eluted from the gel and served as templates in the seminested PCR using primers oligo(dG) and OXA-58-ext-2. The specifically amplified products were purified, subcloned, and sequenced. The transcription start site and the putative promoters were therefore determined based on the sequences of the RACE fragments and conserved motifs. Promoter sequences were identified using the BPROM program.
RT-PCR.
Bacterial RNA was isolated from 0.5-ml portions of bacterial cultures grown to an optical density at 600 nm of 0.5 using RNAprotect bacterial reagent and an RNeasy Mini kit (Qiagen, Valencia, CA) according to instructions provided by the manufacturer. Genomic DNA was eliminated by RNase-free DNase (Qiagen, Taipei, Taiwan) treatment during the isolation procedure. Two micrograms of total RNA was reverse transcribed at 42°C for 60 min using reverse primers (OXA58-B and PEU8) with Moloney murine leukemia virus reverse transcriptase (Epicenter, WI). Expression of 16S rRNA using primers PEU7 and PEU8 (34) was used as an internal control. PCR of total RNA without reverse transcription (RT) was used to examine the DNA contamination of RNA samples. The PCRs were performed under the following conditions: (i) 95°C for 4 min; (ii) 95°C for 30 s, 60°C for 30 s, and 72°C for 45 s for 25 cycles; and (iii) 72°C for 7 min. All experiments were done in duplicate.
Determination of antimicrobial susceptibility.
The MICs for A. baumannii were determined by the broth dilution test using an automated Sensititre susceptibility plate (TREK Diagnostic Systems Ltd., United Kingdom) or using an agar dilution test (9). Antimicrobial agents used on clinical isolates TVICU53 and Ab290 in the tests were gentamicin, tobramycin, amikacin, piperacillin, piperacillin-tazobactam, ticarcillin-clavulanate, ampicillin-sulbactam, aztreonam, ceftriaxone, ceftazidime, cefepime, imipenem, meropenem, ciprofloxacin, chloramphenicol, and trimethoprim-sulfamethoxazole. The MICs of carbapenems were confirmed by the Etest (AB Biodisk, Solna, Sweden). The results were interpreted according to guidelines of the CLSI (formerly NCCLS) (9). Two control strains, an E. coli strain (ATCC 25922) and a Pseudomonas aeruginosa strain (ATCC 27853), were included in the tests as quality controls. All tests were duplicated.
RESULTS
A plasmid (pTVICU53) isolated from a clinical isolate conferring a very high increase in the level of carbapenem resistance in a recipient host.
An outbreak of carbapenem-resistant A. baumannii infection occurred in an intensive care unit of the Taipei Veterans General Hospital in 2004. Effort was undertaken to study the mechanism of carbapenem resistance in these isolates, and isolate TVICU53 was one of them. Based on the preliminary screening analyses using PCR, TVICU53 lacked the MBL genes encoding IMP-, VIM-, GIM-, SPM-, and SIM-hydrolyzing enzymes (data not shown). Phenotypic assays (described in Materials and Methods) also failed to identify any MBL production in TVICU53 (data not shown). In addition, TVICU53 also lacked blaOXA-23-like and blaOXA-24-like genes but contained blaOXA-51- and blaOXA-58-like genes (data not shown). A plasmid, pTVICU53, of around 11 kb was found in the isolate, which contained the blaOXA-58-like gene but not the blaOXA-51-like gene, as analyzed by Southern blot analysis (data not shown). Well-known ISs (ISAba1, ISAba2, ISAba3, and IS18) were not detected in the upstream region of blaOXA-58. However, an electrotransformation of purified pTVICU53 into a carbapenem-susceptible strain, Ab290, resulted in a high level of carbapenem resistance of the recipient cell (see Table 4). The outcome indicated that unidentified factors or unexpected functions of blaOXA-58 carried in this plasmid were probably contributing to the high level of carbapenem resistance. Full-length sequencing was therefore essential for examining the possible mechanism of drug resistance of this plasmid.
TABLE 4.
MICs for parent strains and their transformants carrying different recombinant plasmids
| Bacterium | MIC (μg/ml) of β-lactama:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MEM | IPM | AMX | TIC | CFZ | CXM | CTX | CAZ | ATM | |
| E coli | |||||||||
| DH5α | <0.25 | 0.25 | 8 | 8 | 4 | 4 | <0.12 | <0.12 | <0.12 |
| DH5α(pOXA-58-3)b | <0.25 | 2 | >256 | >1,024 | 8 | 4 | <0.12 | <0.12 | <0.12 |
| DH5α(pOXA-58-4)b | <0.25 | 0.25 | >256 | 256 | ND | ND | ND | ND | ND |
| A. baumannii | |||||||||
| TVICU53 | 16 | 64 | >256 | >1,024 | >128 | >128 | 64 | 128 | 64 |
| Ab290 | 0.25 | 0.25 | 32 | 16 | >128 | 32 | 8 | 4 | 16 |
| Ab290(pTVICU53)b | 16 | 32 | >256 | >1,024 | >128 | 32 | 8 | 4 | 16 |
| Ab290(pOXA-58-3)b | 16 | 64 | >256 | >1,024 | >128 | 32 | 8 | 4 | 16 |
| Ab290(pOXA-58-4)b | 0.5 | 1 | >256 | 256 | ND | ND | ND | ND | ND |
| ATCC 15151T | 0.5 | 0.5 | >256 | 8 | >128 | 128 | 16 | 8 | 64 |
| ATCC 15151T(pOXA-58-3)b | 32 | 32 | >256 | >1,024 | >128 | >128 | 16 | 8 | 128 |
AMX, amoxicillin; TIC, ticarcillin; CFZ, cefazolin, CXM, cefuroxime CAZ, ceftazidime; CTX, cefotaxime; ATM, aztreonam; IPM, imipenem; MEM, meropenem; ND, not done.
The plasmid in parentheses was transformed into the host cell indicated.
Genetic organization of pTVICU53.
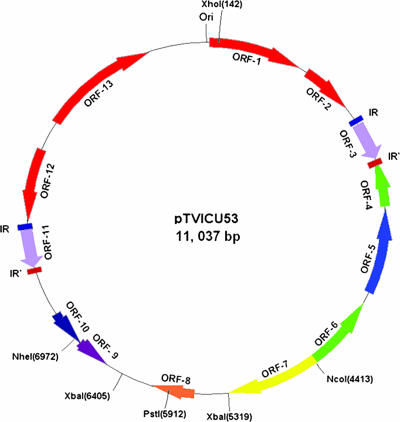
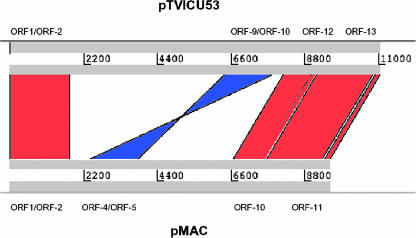
Plasmid pTVICU53 was subjected to restriction digestion, subcloning, sequencing, and assembling. The full-length DNA sequence of pTVICU53 is an 11,037-bp circular plasmid (Fig. 1). The nucleotide sequences of pTVICU53 and pMAC were compared by use of WebACT software (Fig. 2). The plasmid DNA was double digested with restriction enzymes XhoI and either NcoI, NheI, PstI, or XbaI, and the sizes of the restricted fragments coincided with the expected sizes (data not shown), indicating the reliability of assembly. Open reading frame (ORF) scanning revealed 13 ORFs with more than 100 amino acids (Table 3 and Fig. 1). Two perfect inverted repeats (IRs) of 39 bp (5′-ATAAGGTGTATTATGTTAATTTTAGGGAAACTTAATTCT-3′) were found in the sequence of pTVICU53. Between these two IRs, four ORFs (ORF-1, -2, -12, and -13) and a plasmid origin (ori) were identified (Fig. 1). These gene structures (marked in red) covering 4,635 bp are almost identical (except 4 bp) to plasmid pMAC (GenBank accession number AY541809). Two repeated integrase genes (int) (ORF-3 and -11) were flanked by a 39-bp IR and an IR′ (46 bp) (5′-ATTTTCTAATCTTGTACTTAGAGTCCCCTACTATTTAACATAATGG-3′). In this plasmid, the β-lactamase blaOXA-58 gene revealed 2 bp that were different from a previously published blaOXA-58 sequence (GenBank accession number AY665723) but identical to the prototype OXA-58 (GenBank accession number AAW57529) at the amino acid level. The blaOXA-58 gene is bracketed by two copies of an ISAba3-like element that are in opposite orientations. However, the expected functional ISAba3-like element upstream of blaOXA-58 is truncated (ΔISAba3) by an insertion of IS1008 (GenBank accession number AJ251307) but has transitional substitutions at nucleotides 177 and 426. IS1008, belonging to the IS6 family, is 820 bp long, encoding an ORF of a 234-amino-acid transposase. It is flanked by two IRs, IR"L and IR"R. At the 5′ end of IS1008, no transposase gene (tnpA) of the ISAba3-like element could be detected, but instead, there was a truncated putative iron-regulated outer membrane protein (OMP) gene (ORF-7). On the other side, the downstream ISAba3-like element is linked to a putative int gene (ORF-3). Except for the blaOXA-58 gene, no other genes on this plasmid were found to have a known association with carbapenem resistance.
FIG. 1.
Genetic organization of pTVICU53. Position 1 (Ori) is assigned to a G located within an intergenic region flanked by the predicted ORF-1 and ORF-13, as that assigned on pMAC (15). The colored arrows indicate the predicted ORFs and their directions of transcription. Arrows in red indicate a cassette gene originating from pMAC. Two light purple arrows are identical integrases. Two green arrows are transposases flanking blaOXA-58 (the light blue arrow). Two blue bars and two red bars indicate the IR (39 bp) and IR′ (46 bp), respectively. The details of predicted proteins are described in Table 3. The locations of the selected restriction enzyme digestion sites are marked in parentheses.
FIG. 2.
Synteny map of pTVICU53 and pMAC. Sequences of pTVICU53 and pMAC were compared by WebACT (http://www.webact.org/WebACT/home) and visualized by ACT (7). Red indicates similar organization, whereas blue represents inversions. Corresponding ORFs are indicated above and below nucleotide number lines.
TABLE 3.
Sequence analysis of predicted ORFs of pTVICU53a
| ORF | Protein size (aa) | Similaritya | E valueb | % Identity | GenBank accession no. |
|---|---|---|---|---|---|
| 1 | 311 | Replication protein (pMAC of ATCC 19606T) | 3e−176 | 100 | YP_213946 |
| 2 | 172 | Hypothetical protein 1 (pMAC of ATCC 19606T) | 2e−92 | 100 | YP_213947 |
| 3 | 99 | Putative integrase | 1.3 | 35 | YP_879283 |
| 4 | 148 | TnpA of ISAba3 | 1e−68 | 100 | AAW57533 |
| 5 | 280 | OXA-58 | 3e−158 | 100 | AAW57529 |
| 6 | 234 | TnpA of IS1008 | 1e−139 | 100 | CAC80879 |
| 7 | 302 | Putative iron-regulated OMP (pAB2 of ATCC 17978) | 2e−161 | 100 | YP_001083098 |
| 8 | 153 | Hypothetical protein 2 (ATCC 17978 genome) | 2e−48 | 74 | YP_001085297 |
| 9 | 100 | Cro-like protein (pMAC of ATCC 19606T) | 23−47 | 99 | YP_213950 |
| 10 | 118 | Hypothetical protein 3 (pMAC of ATCC 19606T) | 5e−62 | 100 | YP_213949 |
| 11 | 99 | Putative integrase | 1.3 | 35 | YP_879283 |
| 12 | 254 | Hypothetical protein 4 (pMAC of ATCC 19606\T) | 5e−139 | 100 | YP_213955 |
| 13 | 389 | Mobilization protein (pMAC of ATCC 19606T) | 0.0 | 100 | YP_213956 |
aa, amino acids.
The E values of the highest and more significant matches obtained during the BLASTp analyses are shown.
Promoter analysis of the blaOXA-58 gene.
Using the 5′ RACE method, two transcripts of the blaOXA-58 gene were visualized. The +1 transcription start site of the shorter transcript is located 30 bp upstream of the initiation codon of blaOXA-58 (Fig. 3). The −35 and −10 promoters are located within the truncated ISAba3 (ΔISAba3)-like element. The +1 start site of the longer transcript is located 132 bp upstream of the initiation codon of blaOXA-58. This promoter is a hybrid, as the −10 promoter is within the tnpA gene of the ISAb3-like element, but the −35 promoter is located in the right IR (IR"R) of IS1008 (Fig. 3).
FIG. 3.
Transcript start sites and promoter regions of the blaOXA-58 gene predicted based on results of sequence analysis and 5′ RACE-PCR. Two transcripts were detected by 5′ RACE-PCR. For the long transcript, the −10 promoter is provided by tnpA of the ΔISAba3-like element, but the −35 promoter is provided by the right IR of IS1008. For the short transcript, both the −35 and −10 promoters were within the ΔISAba3-like element. Boldface-typed and asterisk-marked characters are the initiation sites of the long and short transcripts. The boxed characters are potential promoters. Three gray blocked characters are genes coding for the transposase tnpA of IS1008, ΔISAba3, and blaOXA-58, respectively.
The IS1008-ΔISAba3-blaOXA-58 gene contributes a high level of carbapenem resistance.
The marked increase in carbapenem MICs in strain Ab290 was obtained through a transformation of intact plasmid pTVICU53. Based on sequence analyses, several putative peptides or uncharacterized factors were found in the plasmid. Among them, blaOXA-58 was the only well-known gene related to limited carbapenem resistance. To examine the function of blaOXA-58 of pTVICU53 against antibiotics, the coding gene of blaOXA-58, including its promoter region (IS1008-ΔISAba3), was subcloned into a pYMAb3 shuttle vector containing A. baumannii plasmid RepM-Ori, namely pOXA-58-3. The transformed Ab290 (with pOXA-58-3) exhibited a level of carbapenem resistance similar to that of Ab290 harboring the whole plasmid of pTVICU53 (Table 4), indicating that IS1008-ΔISAba3 with the blaOXA-58 gene was enough to confer a 64- to 256-fold increase in carbapenem MICs in A. baumannii. The transformation of pOXA-58-3 into a carbapenem-susceptible reference strain, ATCC 15151T, also resulted in a 64-fold increase in carbapenem MICs, indicating that the contribution of IS1008-ΔISAba3-blaOXA-58 to drug resistance was not strain specific. The transformation of the shuttle plasmid itself (pYMAb3), used as a control, did not increase the carbapenem resistance (data not shown), thus excluding the possibility that the carbapenem resistance came from genes in the shuttle plasmid. The transformation of pOXA-58-3 into E. coli also increased resistance to amino- and carboxypenicillin to a high level but only slightly decreased the susceptibility to imipenem (MIC from 0.25 to 2 μg/ml).
Deletion of IS1008 resulted in decreases in levels of blaOXA-58 expression and carbapenem resistance.
A fragment that contained the promoter of ΔISAba3 and the blaOXA-58 gene but without the hybrid promoter provided by IS1008 was obtained by amplification using primers XbaIΔISAba3 and NcoIOXA58. This fragment was subcloned into the pYMAb3 shuttle vector to make pOXA-58-4. Transformation of pOXA-58-4 into Ab290 did not confer significant carbapenem resistance to the recipient (Table 4). The transcription level of the blaOXA-58 gene in pOXA-58-4-harboring Ab290 cells was much lower than that of pOXA-58-3-harboring Ab290 cells investigated by RT-PCR (Fig. 4).
FIG. 4.
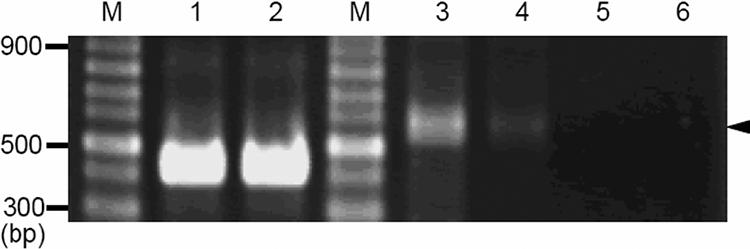
The level of expression of the blaOXA-58 gene was dramatically decreased when the IS1008 motif was deleted. The same amount of total RNA was used for RT (lanes 1, 2, 3, and 4). Total RNA extracted from bacteria without RT was included in the PCR to demonstrate no contamination of bacterial genomic DNA (lanes 5 and 6). Total RNAs extracted from Ab290 transformed by pOXA-58-3 are in lanes 1, 3, and 5; total RNAs extracted from Ab290 transformed by pOXA-58-4 are in lanes 2, 4, and 6. PCR amplifications using primers for 16S rRNA (416 bp) were used as internal controls (lanes 1 and 2). The expression levels of the blaOXA-58 gene examined by PCR (599 bp, indicated by an arrowhead) are presented in lanes 3, 4, 5, and 6. Lanes M, 100-bp DNA ladders.
DISCUSSION
The gene organization of plasmid pTVICU53 is unique, containing ORFs from a number of origins, namely plasmid pMAC (of ATCC 19606T), plasmid pAB2 (of ATCC 17978 [GenBank accession number CP000523]), plasmid pKLH201 (of Acinetobacter calcoaceticus KHW14 [GenBank accession number AJ251307]), and the A. baumannii genome (of ATCC 17978 [NC_009085]). Thirteen ORFs were observed in pTVICU53, and four of them were almost identical to pMAC of ATCC 19606T with regard to gene organization, which includes ORF-1 (replicase), ORF-2 (hypothetical protein 1), ORF-12 (hypothetical protein 3), and ORF-13 (mobilization protein). These four ORFs are flanked by two IRs, and this region (the red-marked ORFs in Fig. 1) contains a predicted OriT fragment (AGGGCAGTCTGA). The DNA replication and mobilization functions in this region were verified previously by Dorsey et al. (15). Plasmid pTVICU53 should therefore possess the same characteristics of replication and transconjugation as pMAC. This is a core function in order for a plasmid to be maintained and spread in A. baumannii. The aligned comparison by the WebACT program also revealed a high level of similarity between pTVICU53 and pMAC, including two inverted ORFs. There were four hypothetical proteins detected in pTVICU53. Three (OFR-2, ORF-10, and ORF-12) of them are almost identical to that of pMAC and have unknown functions. The fourth protein, ORF-8, may originate from the genome of ATCC 17978, with which it has 74% identity. Two identical ORFs (ORF-3 and ORF-11) encode a phage integrase-like protein (3) or the ATP/ADP translocase of Rickettsia bellii (GenBank accession number YP_001496131). Two transposases (ORF-4 and ORF-6) enclosed the oxacillinase OXA-58 (ORF-5), which may allow pTVICU53 to obtain carbapenem resistance from other microorganisms. ORF-7 (302 amino acids) has 100% identity to the C-terminal end of a putative OMP (803 amino acids) of pAB2 isolated from ATCC 17978 (36). This protein was speculated to be a Fe-related transporter with a transmembrane domain at the N-terminal end. The truncated pTVICU53-OMP lacked the N-terminal end but had an intact C-terminal end. Class D blaOXA-51-like β-lactamases are weak carbapenemases (5). It was suggested that blaOXA-51-like genes may confer carbapenem resistance if an ISAba1 insertion in the upstream region provides an enhanced promoter function (24, 31). However, ISAba1 was shown to be absent in the upstream region of a blaOXA-51-like gene in TVICU53 (data not shown) that minimized the contribution of blaOXA-51 to carbapenem resistance in this isolate. Although this study did not evaluate any possible role of penicillin binding proteins, porins, or efflux systems in TVICU53, the high MIC levels of carbapenems resulting from the transformation of a plasmid containing only the IS1008-ΔISAba3-blaOXA-58 fragment indicated that this genetic structure is enough to confer a high level of carbapenem resistance in A. baumannii.
Many review articles previously concluded that CHDLs themselves were not enough to confer a high level of carbapenem resistance in A. baumannii strains because of the low carbapenem hydrolysis activity of class D β-lactamases (5); however, these enzymes might still able to hydrolyze the carbapenem efficiently in vivo when they are present in high periplasmic concentrations (37). The association between a higher copy number of blaOXA-58 genes and stronger carbapenem resistance was examined previously (2). In pTVICU53, a partial ISAba3 element was located upstream of the blaOXA-58 gene, but the ISAba3 promoter did not alone account for the high level of carbapenem resistance (22), which was also demonstrated in our study. When the IS1008 motif including the −10 and −35 promoters was deleted, the transcription of the blaOXA-58 gene and carbapenem resistance dramatically declined. Therefore, an additional IS1008 fragment at the upstream region of the blaOXA-58 gene generating a second set of promoters is likely the determinant of high-level carbapenem resistance.
The majority of the blaOXA genes in a number of different genera are flanked by mobile cassettes of class 1 integrons. Some oxacillinases lacking carbapenem-hydrolyzing activity in A. baumannii (e.g., blaOXA-20, blaOXA-21, and blaOXA-37) have the same characteristics (14, 37). The expressions of these blaOXA genes may be driven by the promoter within the integrons. In contrast, many genes encoding CHDLs in A. baumannii have been detected downstream of ISs (11, 24, 31, 32), which might provide a strong promoter for high-level expression. In our 5′ RACE experiments, two RNA transcripts of blaOXA-58 were observed. For the short transcript, both the −35 and −10 promoters were in the ISAba3-like element. However, for the long transcript, the −10 promoter was in the ISAba3-like element, but the −35 promoter was in the IS1008 domain, which confers better expression activity. An investigation of 10 non-clonally-related OXA-58-producing isolates was performed (32). All of them had ISAba3 downstream of the blaOXA-58 gene. The upstream of ISAba3 was interrupted by other ISs of ISAba1, ISAba2, and IS18 that resulted in new promoters or hybrid promoters (32). Interestingly, our hybrid promoter of IS1008-ΔISAba3 is very similar to two of them, which are also hybrid promoters, ISAba2-ΔISAba3 and IS18-ΔISAba3 (32). Transformation of the original plasmid harboring the ISAba2-ΔISAba3-blaOXA-58 gene (pMAD) into A. baumannii CIP 70.10 (ATCC 15151T), however, only slightly increased the imipenem and meropenem MICs (from 0.25 to 2 μg/ml) (22). This result might be attributed to the lower activity of the −35 promoter provided by ISAba2 than that provided by IS1008. In addition, Bertini et al. (2) previously showed that extra copies of ISAba2-blaOXA-58 increased the level of resistance to carbapenem. The blaOXA-58 gene of TVICU53 is located on a plasmid instead of a chromosome. The copy number of a plasmid gene is usually much higher than that of the same gene construct on a chromosome. Taken together, our results indicate that the promoters provided by IS1008 and the high copy number of the blaOXA-58 gene may confer a high level of resistance to carbapenem.
The IS1008-ΔISAba3-blaOXA-58 gene was able to confer carbapenem resistance to different A. baumannii strains (Ab290 and ATCC 15151T), indicating that IS1008-ΔISAba3-blaOXA-58 carried a carbapenem resistance determinant, and it was functional in nonspecific A. baumannii strains. However, a direct transformation of IS1008-ΔISAba3-blaOXA-58-containing plasmid pOXA-58-3 into E. coli cells provided only a mild increase in carbapenem MICs (<2 μg/ml). Similar results were also demonstrated in other studies (22). Southern blot analysis revealed that A. baumannii blaOXA-58 was stably transformed in E. coli (data not shown), but resistance to carbapenem was not significant. This suggests that A. baumannii plasmid promoters had species specificity at transcriptional/translational levels or that other cofactors from A. baumannii were required for full function of carbapenem resistance.
In conclusion, with the use of a shuttle vector derived from an A. baumannii native plasmid and A. baumannii cells as transformation recipient cells, our study demonstrated that a single genetic structure comprising plasmid-borne IS1008-ΔISAba3-blaOXA-58 is enough to confer a high level of carbapenem resistance in A. baumannii. The insertion of IS1008 provided a new hybrid promoter and increased the transcription level of the blaOXA-58 gene. This study highlights that A. baumannii may become carbapenem resistant by simple means, and physicians should be alert to this fact. The identification of clinical isolates harboring these genetic structures and the implementation of strict infection control measures are important to prevent its further dissemination.
Acknowledgments
We thank Stephen Phillips (Division of Infection and Immunity, University of Glasgow, Glasgow, United Kingdom) for his critical reading of the manuscript.
This work was supported by grants from the Ministry of Education, Aim for the Top University Plan 95A-C-D01-PPg-18, and NSC96-2320-B-010-004.
There was no conflict of interest with any commercial company.
Footnotes
Published ahead of print on 28 April 2008.
REFERENCES
- 1.Bergogne-Berezin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertini, A., L. Poirel, S. Bernabeu, D. Fortini, L. Villa, P. Nordmann, and A. Carattoli. 2007. Multicopy blaOXA-58 gene as a source of high-level resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:2324-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettegowda, C., X. Huang, J. Lin, I. Cheong, M. Kohli, S. A. Szabo, X. Zhang, L. A. Diaz, Jr., V. E. Velculescu, G. Parmigiani, K. W. Kinzler, B. Vogelstein, and S. Zhou. 2006. The genome and transcriptomes of the anti-tumor agent Clostridium novyi-NT. Nat. Biotechnol. 24:1573-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bou, G., G. Cervero, M. A. Dominguez, C. Quereda, and J. Martinez-Beltran. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38:3299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, S., and S. Amyes. 2006. OXA (beta)-lactamases in Acinetobacter: the story so far. J. Antimicrob. Chemother. 57:1-3. [DOI] [PubMed] [Google Scholar]
- 6.Canduela, M. J., L. Gallego, E. Sevillano, C. Valderrey, F. Calvo, and J. Perez. 2006. Evolution of multidrug-resistant Acinetobacter baumannii isolates obtained from elderly patients with respiratory tract infections. J. Antimicrob. Chemother. 57:1220-1222. [DOI] [PubMed] [Google Scholar]
- 7.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 8.Chen, T. L., L. K. Siu, R. C. Wu, M. F. Shaio, L. Y. Huang, C. P. Fung, C. M. Lee, and W. L. Cho. 2007. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:801-806. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing, 6th informational supplement. M100-S17. CLSI, Wayne, PA.
- 10.Coelho, J., N. Woodford, J. Turton, and D. M. Livermore. 2004. Multiresistant Acinetobacter in the UK: how big a threat? J. Hosp. Infect. 58:167-169. [DOI] [PubMed] [Google Scholar]
- 11.Corvec, S., L. Poirel, T. Naas, H. Drugeon, and P. Nordmann. 2007. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa, S. F., J. Woodcock, M. Gill, R. Wise, A. A. Barone, H. Caiaffa, and A. S. Levin. 2000. Outer-membrane proteins pattern and detection of beta-lactamases in clinical isolates of imipenem-resistant Acinetobacter baumannii from Brazil. Int. J. Antimicrob. Agents 13:175-182. [DOI] [PubMed] [Google Scholar]
- 13.del Mar Tomas, M., A. Beceiro, A. Perez, D. Velasco, R. Moure, R. Villanueva, J. Martinez-Beltran, and G. Bou. 2005. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:5172-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donald, H. M., W. Scaife, S. G. Amyes, and H. K. Young. 2000. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorsey, C. W., A. P. Tomaras, and L. A. Actis. 2006. Sequence and organization of pMAC, an Acinetobacter baumannii plasmid harboring genes involved in organic peroxide resistance. Plasmid 56:112-123. [DOI] [PubMed] [Google Scholar]
- 16.Ellington, M. J., J. Kistler, D. M. Livermore, and N. Woodford. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob. Chemother. 59:321-322. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Cuenca, F., L. Martinez-Martinez, M. C. Conejo, J. A. Ayala, E. J. Perea, and A. Pascual. 2003. Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 51:565-574. [DOI] [PubMed] [Google Scholar]
- 18.Fournier, P. E., and H. Richet. 2006. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 42:692-699. [DOI] [PubMed] [Google Scholar]
- 19.Franklin, C., L. Liolios, and A. Y. Peleg. 2006. Phenotypic detection of carbapenem-susceptible metallo-β-lactamase-producing gram-negative bacilli in the clinical laboratory. J. Clin. Microbiol. 44:3139-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gehrlein, M., H. Leying, W. Cullmann, S. Wendt, and W. Opferkuch. 1991. Imipenem resistance in Acinetobacter baumannii is due to altered penicillin-binding proteins. Chemotherapy 37:405-412. [DOI] [PubMed] [Google Scholar]
- 21.Graber, K. R., L. M. Smoot, and L. A. Actis. 1998. Expression of iron binding proteins and hemin binding activity in the dental pathogen Actinobacillus actinomycetemcomitans. FEMS. Microbiol. Lett. 163:135-142. [DOI] [PubMed] [Google Scholar]
- 22.Heritier, C., L. Poirel, T. Lambert, and P. Nordmann. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsueh, P. R., L. J. Teng, C. Y. Chen, W. H. Chen, C. J. Yu, S. W. Ho, and K. T. Luh. 2002. Pandrug-resistant Acinetobacter baumannii causing nosocomial infections in a university hospital, Taiwan. Emerg. Infect. Dis. 8:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, W. S., S.-M. Yao, C.-P. Fung, Y.-P. Hsieh, C.-P. Liu, and J.-F. Lin. 2007. An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3844-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, J. H., C. H. Choi, H. Y. Kang, J. Y. Lee, J. Kim, Y. C. Lee, S. Y. Seol, D. T. Cho, K. W. Kim, D. Y. Song, and J. C. Lee. 2007. Differences in phenotypic and genotypic traits against antimicrobial agents between Acinetobacter baumannii and Acinetobacter genomic species 13TU. J. Antimicrob. Chemother. 59:633-639. [DOI] [PubMed] [Google Scholar]
- 26.Lim, Y. M., K. S. Shin, and J. Kim. 2007. Distinct antimicrobial resistance patterns and antimicrobial resistance-harboring genes according to genomic species of Acinetobacter isolates. J. Clin. Microbiol. 45:902-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limansky, A. S., M. A. Mussi, and A. M. Viale. 2002. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 40:4776-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahgoub, S., J. Ahmed, and A. E. Glatt. 2002. Underlying characteristics of patients harboring highly resistant Acinetobacter baumannii. Am. J. Infect. Control 30:386-390. [DOI] [PubMed] [Google Scholar]
- 29.Marchand, I., L. Damier-Piolle, P. Courvalin, and T. Lambert. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48:3298-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirel, L., and P. Nordmann. 2002. Acquired carbapenem-hydrolyzing beta-lactamases and their genetic support. Curr. Pharm. Biotechnol. 3:117-127. [DOI] [PubMed] [Google Scholar]
- 31.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 32.Poirel, L., and P. Nordmann. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothman, R. E., M. D. Majmudar, G. D. Kelen, G. Madico, C. A. Gaydos, T. Walker, and T. C. Quinn. 2002. Detection of bacteremia in emergency department patients at risk for infective endocarditis using universal 16S rRNA primers in a decontaminated polymerase chain reaction assay. J. Infect. Dis. 186:1677-1681. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Smith, M. G., T. A. Gianoulis, S. Pukatzki, J. J. Mekalanos, L. N. Ornston, M. Gerstein, and M. Snyder. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walther-Rasmussen, J., and N. Hoiby. 2006. OXA-type carbapenemases. J. Antimicrob. Chemother. 57:373-383. [DOI] [PubMed] [Google Scholar]
- 38.Woodford, N., M. J. Ellington, J. M. Coelho, J. F. Turton, M. E. Ward, S. Brown, S. G. Amyes, and D. M. Livermore. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351-353. [DOI] [PubMed] [Google Scholar]