Abstract
In bacterial photosynthetic reaction centers, the protonation events associated with the different reduction states of the two quinone molecules constitute intrinsic probes of both the electrostatic interactions and the different kinetic events occurring within the protein in response to the light-generated introduction of a charge. The kinetics and stoichiometries of proton uptake on formation of the primary semiquinone QA− and the secondary acceptor QB− after the first and second flashes have been measured, at pH 7.5, in reaction centers from genetically modified strains and from the wild type. The modified strains are mutated at the L212Glu and/or at the L213Asp sites near QB; some of them carry additional mutations distant from the quinone sites (M231Arg → Leu, M43Asn → Asp, M5Asn → Asp) that compensate for the loss of L213Asp. Our data show that the mutations perturb the response of the protein system to the formation of a semiquinone, how distant compensatory mutations can restore the normal response, and the activity of a tyrosine residue (M247Ala → Tyr) in increasing and accelerating proton uptake. The data demonstrate a direct correlation between the kinetic events of proton uptake that are observed with the formation of either QA− or QB−, suggesting that the same residues respond to the generation of either semiquinone species. Therefore, the efficiency of transferring the first proton to QB is evident from examination of the pattern of H+/QA− proton uptake. This delocalized response of the protein complex to the introduction of a charge is coordinated by an interactive network that links the Q− species, polarizable residues, and numerous water molecules that are located in this region of the reaction center structure. This could be a general property of transmembrane redox proteins that couple electron transfer to proton uptake/release reactions.
Keywords: proton transfer, site-specific mutagenesis, membrane protein, water channel
Reaction centers (RCs) from photosynthetic bacteria convert light energy into chemical free energy. These transmembrane complexes are composed of three protein subunits. Two of them, L and M, are fully embedded in the photosynthetic membrane and carry all of the pigments and cofactors present in the RC. The three-dimensional structures of the RCs from two species, Rhodobacter sphaeroides and Rhodopseudomonas viridis, are known at 2.2 Å (1) and 2.0 Å (2) resolution, respectively. The initial capture of energy is achieved through a transmembrane charge separation between the primary electron donor—a bacteriochlorophyll dimer (“P”), situated near the periplasmic side of the protein, and a system of two quinones located near the cytoplasmic side. The primary quinone acceptor, QA, is bound in a relatively hydrophobic region of the M subunit. The formation of the transient P+QA− state occurs by electron transfer from P via HA (an intermediate bacteriopheophytin electron carrier) and is followed by electron donation to QB, the secondary quinone acceptor. QA functions as a one-electron acceptor and is never protonated. In contrast, QB becomes doubly reduced and doubly protonated (QBH2) after the transfer of two successive electrons from QA and two protons from the cytoplasm. In RCs of Rb. sphaeroides and Rhodobacter capsulatus, both QA and QB are ubiquinone10. Their different functional properties are attributable to differences in their respective protein environments. Unlike QA, QB is surrounded by many charged and polar residues that may determine the energetic properties of QB− and/or be involved in the process of proton transfer to it.
In Rb. sphaeroides RCs, the activity of L213Asp (3, 4) and L223Ser (5, 6) in the transfer of the first proton to QB after the second flash has been demonstrated. Similarly, L212Glu was shown to be involved in the donation of the second proton to QB in RCs of Rb. sphaeroides (7, 8) and Rb. capsulatus (9); that pathway may also involve L213Asp (reviewed in ref. 10).
In isolated RCs, the formation of QB− or QA− (in the absence of QB) after the first flash results in partial proton uptake by residues whose pKas are shifted because of their coulombic interaction with the particular semiquinone species that is formed (11, 12). The groups that are active in this partial proton uptake are not necessarily the same as those that are directly responsible for the last steps of proton transfer to QB.
Two main features of the RC’s response to light-induced generation of semiquinones have recently been suggested both by experimental and theoretical approaches. The first feature that has emerged is the electrostatic connection between the protein environments of the two quinone binding pockets. Indeed, proton uptake measurements have shown that, in RCs of Rb. capsulatus, L212Glu becomes ionized above pH 8.5 on formation of QA− (13, 14); this observation has also been made in Rb. sphaeroides RCs (15). The analysis of the kinetics of proton uptake on QA− formation recently led Maróti and Wraight (16) to suggest that the uptake process is limited by protein dynamics that change the protonation states of residues that respond to the charge, rendering them accessible or inaccessible (see also ref. 17). At neutral pH, theoretical calculations have suggested that groups in the QB environment are the ones that respond to the formation of QA− (18, 19). This last hypothesis has not yet been confirmed by experimental data.
The second feature, suggested by calculations, is that clusters of strongly interacting groups, rather than isolated residues, are responsible for the uptake of protons at neutral pH. In RCs of Rb. sphaeroides, a cluster of acidic residues (L212Glu, L210Asp, and L213Asp) and L223Ser have been proposed to be the main components that buffer the electrostatic environment of QB and QB− (18). Strong electrostatic interactions between these groups increase the pH range in which individual groups would take up protons and therefore allow the RCs of these bacteria to function in a wide pH range. It has previously been suggested (20, 21) that the protein has an important role in maintaining optimal electrostatic potential in the neighborhood of QB to ensure fast proton transfer rates. This proposed role for the protein has been demonstrated by the recovery of the proton transfer capabilities of RCs in which distant electrostatic suppressor mutations (M43Asn → Asp, M231Arg → Leu or Cys) have compensated for the lack of L213Asp in Rb. capsulatus (9, 20, 22) and Rb. sphaeroides (23). The propagation of the electrostatic effects of the mutations to the immediate environment of QB has been proposed to be caused by an “electrostatic dominoes” mechanism (24). This process could result from mutation-induced realignments of salt bridges within the network of acidic and basic residues located in the cytoplasmic region of the protein. Support for this hypothesis has recently been obtained from crystallographic (25) and FTIR data (26).
In the present work, we investigate further the degree of functional electrostatic connection between the QA and QB protein environments. We have determined, in parallel, the stoichiometries and kinetics of proton uptake, at pH 7.5, that are induced by the formation of the QA−, QB−, and QBH− states in various strains modified at the L212Glu and/or L213Asp positions; we have also examined RCs carrying second-site suppressor mutations that compensate for loss of L213Asp. Our data demonstrate that, at neutral pH, these reaction centers respond in a similar manner to the formation of either QA− or QB−. The data also show that the efficiency with which the first proton will be transferred to QB is already indicated by the nature of the H+/QA− proton uptake signals.
Materials and Methods
Mutant Strains and Biochemical Preparations.
The construction of the L212Glu-L213Asp → Ala-Ala [“AA”; (27)] and L212Glu → Gln (14) mutants has been described previously, as have the isolation and genotypic characterization of the L212Ala strain (27) and suppressor strains carrying compensatory mutations at the M231 (27), M43 (27), or M5 (28) sites in conjunction with the L212Ala-L213Ala substitutions. The M247Ala → Tyr mutation was recently constructed by oligonucleotide-directed mutagenesis, according to directions from a kit (Chameleon, Stratagene), and this M-gene mutation was coupled to the AA substitutions in the L gene via restriction endonuclease digestion and ligation of the appropriate fragments (29). All strains, including the wild type, were grown under chemoheterotropic conditions (dark, semiaerobic) at 34°C on RPYE medium (30) containing 30 μg/ml kanamycin to ensure the presence of the plasmid. RCs were prepared as described (31).
Proton Transfer Measurements.
The kinetics of proton uptake were determined at pH 7.5 by measuring the absorbance changes at 585 nm (isosbestic point of the P+ absorbance changes) of the pH-sensitive dye o-cresol red. The RCs were extensively dialyzed to keep the buffer concentration below 5–10 μM. The assay solution routinely contained 1–2 μM RCs, 50 mM NaCl, 0.03% Triton X-100, 100 μM ferrocene, 500 μM potassium ferrocyanide, and 40 μM o-cresol red. The H+/QA− proton uptake measurements were done in the presence of 100 μM terbutryn, which blocks the binding of QB. The H+/QB− proton uptake measurements were completed in the presence of 60 μM UQ6. The calibrations of the stoichiometries of proton uptake were performed by additions of known amounts of HCl (1 M stock; Merck). The amplitude of proton uptake by the PQAQB− state (ΔHQAQB−+) was corrected for the observed proton uptake after the first flash (ΔHobs+) as previously demonstrated (32):
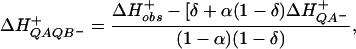 |
1 |
where ΔH+/QA− represents the proton uptake by the RC in the absence of QB, δ is the occupancy of the QB site, and α is the partition coefficient between the QA−QB and QAQB− states. This correction is not applied to the amplitude of proton uptake observed after the second flash because an external calibration is used and the proton uptake by the QA− state has been taken into account after the first flash. The net proton uptake was obtained by subtracting the signal obtained with buffered samples from the signal obtained with unbuffered samples.
Results
Mutant RCs Studied.
Rb. capsulatus RCs carrying two families of mutations were the subjects of these experiments. The first family is mutated at the L212 site: The photosynthetically incompetent (PS−) L212Glu → Gln (L212Q) mutant has previously been described (9), as has the photocompetent (PS+) L212Glu → Ala (L212A) mutant (9). The second family of RCs carries the L212Glu-L213Asp → Ala-Ala mutations (AA) that also cause the PS− phenotype. Photocompetent phenotypic revertants AA+M231L (AA+M231Arg → Leu) and AA+M43D (AA+M43Asn → Asp) carry an additional mutation that compensates for some of the deleterious effects of the AA mutations. Residues M231 and M43 are distant from QB—15 and 9 Å, respectively; some other properties of these RCs have been previously reported (20, 27). Another phenotypic revertant, AA+M5D (AA+M5Asn → Asp), has been described recently (33); M5Asn is equidistant from both QA and QB (≈16 Å) in the RC structure. Finally, we have constructed the AA+M247Y strain (AA+M247Ala → Tyr; PS−), which is the only strain that carries a mutation near QA. M247Ala is located ≈5 Å from QA, and previous results had suggested that the observation of increased rates of transfer of the second electron to QB in revertant RCs could be linked to a tyrosine substitution at this site (34).
The Wild-Type and L212 Mutant RCs.
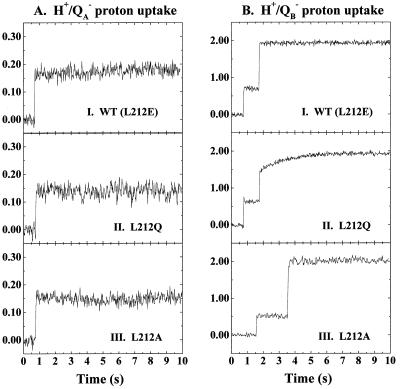
The H+/QA− and H+/QB− proton uptake kinetics, measured at pH 7.5 in the RCs from the wild type (L212E), the L212Q, and the L212A mutants, are presented in Fig. 1. The stoichiometries are listed in Table 1, and the kinetic parameters are presented in Table 2.
Figure 1.
H+/QA− (A) and H+/QB− (B) proton uptake kinetics in the wild-type (L212E, I), L212Q (II), and L212A (III) mutant RCs. The stoichiometries of proton uptake were calibrated as described in Materials and Methods. Conditions: 50 mM NaCl, 0.03% Triton X-100, <10 mM Tris⋅Cl (pH 7.5), 100 μM ferrocene, 500 μM potassium ferrocyanide, and 40 μM o-cresol red. The H+/QA− proton uptake measurements were done in the presence of 100 μM terbutryn.
Table 1.
Amplitudes of proton uptake measured at pH 7.5 on formation of QA− and QB−
| Strains | H+/QA−*
|
H+/QB−, first flash†
|
H+/QB, second flash†
|
Total first and second flashes | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fast | Slow | Total | Fast | Slow | Total | Fast | Slow | Total | ||
| Wild type‡ | 0.16 | — | 0.16 | 0.70 | — | 0.70 | 1.30 | — | 1.30 | 2.00 |
| L212Q | 0.16 | — | 0.16 | 0.63 | — | 0.63 | 0.90 | 0.42 | 1.32 | 1.95 |
| L212A | 0.14 | — | 0.14 | 0.55 | — | 0.55 | 1.45 | — | 1.45 | 2.00 |
| L212A-L213A | 0.15 | 0.15 | 0.30 | 0.45 | 0.55 | 1.00 | 0.20 | 0.75 | 0.95 | 1.95 |
| AA+M231L§ | 0.18 | — | 0.18 | 0.90 | — | 0.90 | 1.05 | — | 1.05 | 1.95 |
| AA+M43D | 0.15 | — | 0.15 | 1.00 | — | 1.00 | 0.75 | 0.15 | 0.90 | 1.90 |
| AA+M5D | 0.15 | 0.10 | 0.25 | 0.30 | 0.75 | 1.05 | 0.20 | 0.85 | 1.05 | 2.10 |
| AA+M247Y | 0.30 | 0.18 | 0.48 | 0.25 | 0.70 | 0.95 | 0.15 | 1.00 | 1.15 | 2.10 |
The standard deviation is 0.03 for the H+/QA− measurements and 0.05 on each flash for the H+/QB measurements. “Fast” refers to components faster than ≈25 ms.
Terbutryn added.
† Excess quinone present.
‡ Wild-type residues are L212E, L213D, M5N, M43N, M231R, and M247A.
§ AA = L212A-L213A.
Table 2.
Lifetimes of proton uptake measured at pH 7.5 on formation of QA− or QB−
| Strains | H+/QA−*
|
H+/QB−, first flash†
|
H+/QB, second flash†
|
|||
|---|---|---|---|---|---|---|
| Fast | Slow | Fast | Slow | Fast | Slow | |
| Wild type‡ | ≈ms | — | ≈ms | — | ≈ms | — |
| L212Q | ≈ms | — | ≈ms | — | ≈ms | 1.5 s |
| L212A | ≈ms | — | ≈ms | — | ≈ms | — |
| L212A-L213A | ≈ms | 0.99 s | <25 ms | 0.78 s | <25 ms | 0.80 s |
| AA+M231L | ≈ms | — | <25 ms | — | <25 ms | — |
| AA+M43D | ≈ms | — | <25 ms | — | <25 ms | 0.12 s |
| AA+M5D | ≈ms | 0.40 | <25 ms | 0.31 s | <25 ms | 0.35 s |
| AA+M247Y | ≈ms | 0.56 | <25 ms | 0.67 s | <25 ms | 0.60 s |
The standard deviations for the slow components are 10% of the values. “≈ms” holds for lifetime values in the millisecond time range (or very few milliseconds; data not shown). “<25 ms” is a higher limit for slightly longer components seen in a different time window (not shown here). These differences are not resolvable on the time scale of the present experiments.
Terbutryn added.
† Excess quinone present.
‡ See Table 1 for wild-type residues and mutant designations.
H+/QA− proton uptake.
In the wild type, ≈0.16 protons are rapidly taken up with formation of QA− (≅ms kinetics). The situation is very similar in the L212Q and L212A RCs. In the L212Q RC, ≈0.14 protons are taken on the same time scale (≅ ms) as is seen for the wild type. Similarly, in the L212A RC, H+/QA− is ≈0.16, and the kinetics are also fast.
H+/QB− proton uptake.
In QB-containing wild-type RCs, 0.7 protons are rapidly taken up with QB− formation after the first flash, and the complement to two protons is taken up after the second flash (9). The situation is similar after the first flash in the L212Q mutant, in which 0.63 H+ are taken up in the millisecond time range. After the second flash, the kinetics are strongly biphasic, as noted previously (9). Rapid (≅ms) uptake of 0.90 H+/QB− is followed by a very slow (≅1.5 s) uptake of ≈0.42 H+/QB−. In the L212A mutant, the amplitude of proton uptake after the first flash is slightly smaller (0.55 H+/QB−), but the kinetics for uptake of the complement to two protons after the second flash are entirely fast (≅ms) (9).
The L212Ala-L213Ala Family.
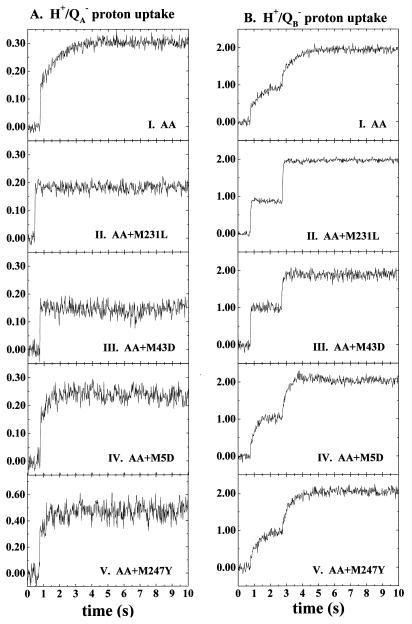
The H+/QA− and H+/QB− proton uptake kinetics, measured at pH 7.5 in RCs from the AA, AA+M231L, AA+M43D, AA+M5D, and AA+M247Y, strains are shown in Fig. 2. The stoichiometries are listed in Table 1 and the kinetic parameters are presented in Table 2.
Figure 2.
H+/QA− (A) and H+/QB− (B) proton uptake kinetics in RCs from the AA (I), AA+M231L (II), AA+M43D (III), AA+M5D (IV), and AA+M247Y (V) mutants. The stoichiometries of proton uptake have been calibrated as described in the text. Conditions are as in Fig. 1. Wild-type residues are M5N, M43N, M231R, M247A; “AA” = L212Glu-L213Asp → Ala-Ala.
H+/QA− proton uptake.
The H+/QA− proton uptake pattern measured in the AA RCs (Fig. 2A, I) is quite different than that observed in the wild type. As was seen for the wild type (Fig. 1A, I), 0.15 protons are also taken up rapidly by the AA RC, but the total amplitude is doubled by the slow uptake (0.99 s) of an additional 0.15 protons. Interestingly, two of the suppressor mutations, M231L or M43D, decrease both the lifetime and the amplitude of proton uptake to native levels when they are added to the AA RC (Fig. 2, II and III, respectively). Indeed, in both the AA+M231L and AA+M43D RCs, the slow phase disappears, and the amplitude of the fast (≅ms) phase is roughly equal to that of the wild type: H+/QA− equals 0.15 in the AA+M231L RC and 0.18 in the AA+M43D RC. The kinetics and amplitude of H+/QA− uptake in the AA+M5D RC are intermediate between those of the AA and native RCs (Fig. 2A, IV). In the AA+M5D RC, fast uptake of 0.15 protons is coupled with slow (0.40 s) uptake of 0.10 H+/QA−. The pattern is different still in the RCs from the AA+M247Y mutant in which the total amplitude of H+/QA− uptake increases to 0.44, nearly three-fold the amount of uptake that is seen for the wild-type RCs (Fig. 2A, V). In the AA+M247Y RC, 0.30 H+ are taken up in the fast phase, and 0.14 protons are taken up with slower (≅0.36 s) kinetics.
H+/QB− proton uptake.
In general, the RCs of the AA family display first-flash amplitudes of H+/QB− proton uptake in the range of 0.90–1.00 H+, which are notably higher than those observed for the wild-type, L212Q, and L212A RCs (0.55–0.70 H+). In the AA mutant, we previously reported that the kinetics are strongly biphasic (34). In the fast (<25 ms) phase, 0.45 H+ are taken up whereas the amplitude of the very slow phase (≈0.78 s) is equivalent to 0.55 H+ (Fig. 2B, I).
In RCs of the AA+M231L and AA+M43D phenotypic revertants, proton uptake kinetics after the first and the second flashes are remarkably accelerated relative to those of the AA RC (Fig. 2B, II and III, respectively). In the RC of the AA+M231L strain, proton uptake after both the first and second flashes displays entirely fast (<25 ms) kinetics, with amplitudes of 0.90 H+ and 1.05 H+, respectively. In the AA+M43D RC, the first flash generates the fast uptake of 1.00 H+ whereas the second flash induces the fast uptake of 0.75 H+. A relatively slow (≈0.12 s) phase of 0.15 H+ increases the final amplitude to 1.9 H+.
As was seen for the H+/QA− kinetics, the H+/QB− pattern measured in the AA+M5D mutant displays biphasic kinetics (Fig. 2B, IV). After the first flash, a small (≅0.30 H+) contribution of a fast phase of proton uptake is observed whereas 0.75 H+ are taken up with a lifetime of 0.31 s. After the second flash, 0.20 H+ are taken up on a fast (<25 ms) time range and 0.85 H+ are taken up on a slower time scale (≅0.35 s).
A similar behavior is observed for the AA+M247Y mutant (Fig. 2B, V). After the first flash, 0.25 H+ are taken up on a fast time scale, and 0.70 H+ are taken up with a lifetime of 0.67 s. After the second flash, the kinetics are essentially slow; a small contribution (0.15 H+) comes from the fast phase, with the majority of the amplitude (1.00 H+) being attributable to a component with a lifetime of 0.60 s.
Discussion
Proton Uptake with the Formation of QA−.
The protein portion of the RC reacts to the arrival of an electron on QA by the rapid uptake of protons. Earlier work (11, 12) documented the nonstoichiometric, pH-dependent uptake of 0.2–0.6 protons by the PQA− state in Rb. sphaeroides RCs in which QB had been removed or replaced by terbutryn. No protonation of the semiquinone is observed; rather, the uptake is caused by shifts in pKas of a set of residues that responds to formation of the semiquinone.
The QA− proton uptake stoichiometries measured at pH 7.5 in the L212 mutants are similar to those measured in the wild-type reaction center (Fig. 1A). Likewise, the H+/QA− proton uptake kinetics in the strains carrying mutations of L212Glu are fast (≈ ms), resembling the wild-type kinetics. The H+/QA− proton uptake does not depend on the amino acid present at the L212 site, e.g., Glu vs. Gln vs. Ala (Fig. 1A). Therefore, the absence of L212Glu within the cluster of strongly interacting groups near QB (L212Glu, L213Asp, L210Asp, H173Glu in the Rb. sphaeroides structure) does not influence the proton uptake that is induced by the formation of QA− at pH 7.5. Thus, these data agree with the hypothesis that L212Glu is essentially protonated at neutral pH. This hypothesis had been suggested by earlier experimental data obtained in RCs of Rb. capsulatus (13, 14) and in Rb. sphaeroides (15) and also by theoretical calculations (18, 35). L212Glu is active in proton uptake at higher pH. It was demonstrated previously (7) that L212Glu begins to ionize above pH 8.5 and is directly involved in the H+/QA− proton uptake above pH 9 in the wild-type RC (9, 14, 36).
In contrast, the H+/QA− proton uptake at pH 7.5 is notably modified in the AA RCs, where both L212Glu and L213Asp are relaced by Ala (Fig. 2A, I). The initial fast phase of ≈0.15 H+/QA− is still present in this mutant; thus, it is clear that the rapid proton uptake is accomplished by groups that do not interact with L212Glu or L213Asp. The total amplitude of proton uptake in the AA RC is doubled by the addition of 0.15 H+/QA− that occurs with very slow kinetics (0.99 s). Because the L212Glu → Ala mutation by itself does not result in any change in H+/QA− uptake at pH 7.5 (Fig. 1A, III), the effect that we observe here is caused by the L213Asp → Ala mutation. In the wild-type RC, any acidic residue that interacts strongly with L213Asp will have its pKa shifted to higher than normal values (37). It is likely that the loss of L213Asp in the AA mutant causes shifts of the pKas of some of these interacting residues to lower values such that they are now active in proton uptake at pH 7.5 in response to the formation of the PQA− state.
The slow phase that doubles the amplitude of H+/QA− proton uptake in the AA mutant is remarkably eliminated by the addition of either the M231Arg → Leu or M43Asn → Asp suppressor mutation to the AA RC (Fig. 2A, III and IV). Indeed, in the RCs of these two strains, the H+/QA− patterns are similar, in both amplitude and rate, to what is observed in the wild-type RC. Because the amplitude of proton uptake in the AA+M231L and AA+M43D RCs drops to the level seen in the wild-type RC, this observation is consistent with the idea that these distant suppressor mutations shift the pKas of residues that were rendered active by the loss of L213Asp back to higher values, making them inactive in proton uptake at pH 7.5. These data further support the idea that the suppressor mutations at the M43 and M231 sites fill this role that is normally assigned to L213Asp. Although they are relatively distant from QB, M43 and M231 are well connected to it and the L213 site via the interactive network of ionizable residues and water molecules that is located in this region of the RC.
The addition of the M5Asn → Asp suppressor mutation to the AA RC leads to a pattern of H+/QA− uptake (Fig. 2A, IV) that is intermediate between those of the wild-type and the AA mutant RCs. Like the other RCs, the initial uptake of 0.15 H+/QA− in the AA+M5D RCs is fast. The slow phase that follows is accelerated (0.4 s) compared with the AA strain (Tables 1 and 2), and its amplitude is slightly diminished, to 0.10 H+/QA−. Although this strain is also photocompetent, the M5Asp substitution has a lesser effect than the M231Arg → Leu or M43Asn → Asp substitutions on the pKas of residues that react to the loss of L213Asp by taking up protons with the formation of the QA− state. These data are consistent with previous observations (28, 38) (discussed further below), all of which suggest that M5Asp may not be as well connected to the interactive network as are residues M231 and M43.
Substitution of a tyrosine near QA increases proton uptake.
In the AA+M247Y RC in which a tyrosine has been substituted for an alanine at M247 near QA (≈5 Å), the amplitude of the fast phase of proton uptake is doubled in comparison to any of the above native or modified RCs (Fig. 2A, V). This result points to the unexpected activity of this Tyr in producing the additional fast proton uptake that is measured in this mutant (≈0.15 H+/QA−).
The slow phase of H+/QA− proton uptake is still present in the kinetics from the AA+M247Y RC. It has a similar amplitude as is seen in the AA mutant (≈0.18), and it is slightly accelerated (0.56 s). Thus, although the tyrosine at M247 results in the uptake of more protons, its predominant effect is more probably localized to the neighborhood of QA. Its effect on the pKas of residues that react to the loss of L213Asp is minimal in comparison to the M231Arg → Leu or M43Asn → Asp substitutions discussed above. This result is not unexpected because tyrosine, whose pKa is normally 10 or greater, is not expected to be charged at pH 7.5. However, the pKa of a tyrosine can be modified by its environment (see below), and its pKa at the M247 position is not known. Although its effects are not likely to be electrostatic, clearly the tyrosine is influencing both the rate and stoichiometry of proton uptake in the AA+M247Y RC.
Because the pKa of Tyr is normally high, the mechanisms by which tyrosines participate in proton transfer/abstraction reactions are as yet unclear. Tyrosine residues have been shown to be important for proton transfer in manganese superoxide dismutase (39) and carbonic anhydrase V (40, 41), and in proton abstraction from a steady-state intermediate in thymidylate synthase (42). In superoxide dismutase, the tyrosine is part of a hydrogen-bonded network that connects the active site with water molecules and ionizable residues, and mutational studies suggest that it is a proton donor within this network (39). Tyr131 in carbonic anhydrase V participates as a proton shuttle between the solution and a zinc-bound water molecule at the active site; in this case, its functional pKa is reduced to ≈9 by virtue of its location in an electropositive region of the protein (40, 41). In thymidylate synthase, the data suggested that the tyrosine could abstract a proton from the reaction intermediate or serve to polarize or orient a water molecule to perform this reaction (42). Previously, we suggested that Tyr at M247 could connect to the network of water molecules in the cavity “under” QA and in this way help to conduct protons to the region of the RC near QB (34).
Correlation of H+/QB− Proton Uptake with H+/QA− Proton Uptake.
There is a general correspondence between the presence of a slow phase in the H+/QA− kinetics (e.g., AA, AA+M5D, AA+M247Y; Fig. 2A, I, IV, and V) and its presence in the H+/QB− traces (Fig. 2B, panels I, IV, and V). Furthermore, the H+/QA− kinetic values roughly match those for H+/QB− uptake (Table 2). Indeed, the kinetics of the slow phase of the H+/QB− proton uptake observed after the first flash in the AA mutant (≈0.78 s) are similar to those measured for the slow phase of the H+/QA− uptake (0.99 s). In the AA+M231L and AA+M43D RCs, both the H+/QA− and the H+/QB− kinetics are fast. Again, the kinetics observed for the AA+M5D RC are intermediate between those of the AA and native-like RCs, and the 0.31-s lifetime measured for its H+/QB− proton uptake is close to the 0.40 s lifetime detected in the H+/QA− uptake data. Finally, in the AA+M247Y RC, the slow phase of the first-flash H+/QB− proton uptake (0.67 s) is in good agreement with the 0.56 s measured for the H+/QA− uptake.
These trends that were observed for the first flash also hold for the kinetics of H+/QB− proton uptake measured after the second flash. Except for the L212Q mutant in which only the second proton is taken up slowly, all of the RCs that are impaired in the delivery of the first proton also display the same kind of H+/QB− proton uptake kinetics after the second flash.
Relative to the photosynthetically incompetent L212Glu-L213Asp → Ala-Ala mutant RC, we have shown previously that suppressor mutations increase the negative electrostatic environment near QB and increase the rate of transfer of the second electron to QB by restoring efficient proton transfer (20, 24, 28, 30, 34, 38). The different suppressor mutations restore the electrostatic potential near QB to varying levels, although all phenotypic revertants were selected in screens for photocompetence. The M231Arg → Leu and the M43Asn → Asp substitutions are very efficient at restoring native-like patterns of proton uptake for both the QA− and QB− states (Fig. 2A, II and III and Fig. 2B, II and III). These substitutions accelerate the rate of transfer of the second electron 15- to 25-fold over that found for the AA RC (24). Unlike the situation observed for the amplitude of H+/QA− uptake, the addition the M231L or M43D suppressor mutations to the AA RC does not diminish the amplitudes of the H+/QB− uptake to the native level. Thus, the correspondence between native-like stoichiometries and improved proton transfer capabilities seen in the H+/QA− uptake kinetics is not observed here. This could be fortuitous because we probe the system at pH 7.5. Since the pKas of residues that are active here in proton uptake may have been shifted to higher values if they interact more strongly with QB− than with QA−, such a correspondence might be seen only at higher pH.
The addition of the M5Asn → Asp mutation is less effective in suppressing the effects of the AA mutations on proton uptake by any of the quinone anion states (Fig. 2A, IV, and Fig. 2B, IV). The directly measured rates of proton transfer to QB in the AA+M5D RC are accelerated compared with the AA mutant, yet they are much slower than the rates observed in the AA+M231L and AA+M43D RCs (Table 2). This result correlates with our previous observations that the M5D mutation has a lesser effect on the electrostatic environment of QB—the rate of transfer of the second electron to QB in RCs of the AA+M5D strain is only ≈3 s−1 at neutral pH (28, 38). This rate is accelerated only six-fold over that of the AA RCs. Likewise, in the AA+M247Y mutant, the second electron transfer rate is very slow (1.5 s−1 at pH 7.5; data not shown), as are its proton uptake kinetics (Fig. 2A, V, and Fig. 2B, V).
Conclusions.
Taken together, these data strongly indicate that, for the most part, the same groups respond to the formation of either QA− or QB−, underscoring the idea that the system is reacting as a whole when a charge is introduced. We cannot identify specific groups that are responsible for the differences in proton uptake that were measured, but in general our data suggest that there are three different categories of residues (and residues may belong to more than one category). There are residues that: (i) take up protons regardless of the mutations that are present, (ii) take up extra protons when L213Asp is absent, and (iii) influence the rate of proton uptake in the absence of L213Asp. The compensatory mutations, which are located at several different positions in the RC, specifically affect the pKas of the above types of residues, which in turn influence both the rate and amount of proton uptake. Most importantly, our data show that both the kinetics and amplitude of proton uptake in a particular mutant RC are influenced in the same manner when either quinone anion is formed. Because all of the mutant RCs described here behave in the same way, it is very likely that this is also a property of the wild-type complex and could be a general property of transmembrane redox proteins that couple electron transfer to proton uptake/release reactions.
Theoretical calculations have suggested that, at neutral pH, the main response of the RC to formation of QA− is changes in ionization states of residues that are located near QB (18). Because the data presented here show that distant residues can affect this response, “near” becomes a relative term that is more accurately applied to describe residues that interact with the Q− species, each other, with a Tyr near QA, and numerous water molecules within a network that is a structural feature of the cytoplasmic region of the RC. Because of this general response, it is possible to judge from the pattern of H+/QA− proton uptake kinetics whether the RC is capable of transferring the first proton to QB in an efficient manner. One may therefore consider that the formation of QA− in the reaction centers may be an essential functional step, during which much of the preparation for efficient proton delivery to QB is already accomplished. This hypothesis can be tested by monitoring the coupling between electron transfer and proton transfer in mutant RCs in which QB is reduced solely by activity of B-branch cofactors (43).
Acknowledgments
Drs. L. Baciou and J. Tandori, and A. Taly are thanked for helpful discussions. This work was supported by the Human Frontier of Science Organization RG-329-95M, by the Centre National de la Recherche Scientifique, and by the U.S. Department of Energy, Office of Biological and Environmental Research, under Contract W-31-109-ENG-38.
Abbreviation
- RC
reaction center
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Stowell M H, McPhillips T M, Rees D C, Soltis S M, Abresch E, Feher G. Science. 1997;276:812–816. doi: 10.1126/science.276.5313.812. [DOI] [PubMed] [Google Scholar]
- 2.Lancaster C R D. In: Photosynthesis: Mechanisms and Effects. Garab G, editor. II. Dordrecht, the Netherlands: Kluwer; 1999. pp. 673–678. [Google Scholar]
- 3.Takahashi E, Wraight C A. Biochim Biophys Acta. 1990;31:855–866. [Google Scholar]
- 4.Paddock M L, Rongey S H, McPherson P H, Juth A, Feher G, Okamura M Y. Biochemistry. 1994;33:734–745. doi: 10.1021/bi00169a015. [DOI] [PubMed] [Google Scholar]
- 5.Paddock M L, McPherson P H, Feher G, Okamura M Y. Proc Natl Acad Sci USA. 1990;87:6803–6807. doi: 10.1073/pnas.87.17.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lancaster C R, Michel H. Structure (London) 1997;5:1339–1359. doi: 10.1016/s0969-2126(97)00285-2. [DOI] [PubMed] [Google Scholar]
- 7.Paddock M L, Rongey S H, Feher G, Okamura M Y. Proc Natl Acad Sci USA. 1989;86:6602–6606. doi: 10.1073/pnas.86.17.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi E, Wraight C A. Biochemistry. 1992;31:855–866. doi: 10.1021/bi00118a031. [DOI] [PubMed] [Google Scholar]
- 9.Miksovska J, Kálmán L, Schiffer M, Maróti P, Sebban P, Hanson D K. Biochemistry. 1997;36:12216–12226. doi: 10.1021/bi970442w. [DOI] [PubMed] [Google Scholar]
- 10.Okamura M Y, Feher G. Annu Rev Biochem. 1992;61:861–896. doi: 10.1146/annurev.bi.61.070192.004241. [DOI] [PubMed] [Google Scholar]
- 11.Maróti P, Wraight C A. Biochim Biophys Acta. 1988;934:329–347. [Google Scholar]
- 12.McPherson P H, Okamura M Y, Feher G. Biochim Biophys Acta. 1988;934:348–368. doi: 10.1016/0005-2728(93)90116-w. [DOI] [PubMed] [Google Scholar]
- 13.Maróti P, Hanson D K, Schiffer M, Sebban P. Nat Struct Biol. 1995;2:1057–1059. doi: 10.1038/nsb1295-1057. [DOI] [PubMed] [Google Scholar]
- 14.Miksovska J, Maróti P, Tandori J, Schiffer M, Hanson D K, Sebban P. Biochemistry. 1996;35:15411–15417. doi: 10.1021/bi961299u. [DOI] [PubMed] [Google Scholar]
- 15.McPherson P H, Schönfeld M, Paddock M L, Okamura M Y, Feher G. Biochemistry. 1994;33:1181–1193. doi: 10.1021/bi00171a018. [DOI] [PubMed] [Google Scholar]
- 16.Maróti P, Wraight C A. Biophys J. 1997;73:367–381. doi: 10.1016/S0006-3495(97)78077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riistama S, Hummer G, Puustinen A, Dyer R B, Woodruff W H, Wikström M. FEBS Lett. 1997;414:275–280. doi: 10.1016/s0014-5793(97)01003-x. [DOI] [PubMed] [Google Scholar]
- 18.Alexov E G, Gunner M R. Biochemistry. 1999;38:8253–8270. doi: 10.1021/bi982700a. [DOI] [PubMed] [Google Scholar]
- 19.Lancaster C R, Michel H, Honig B, Gunner M R. Biophys J. 1996;70:2469–2492. doi: 10.1016/S0006-3495(96)79820-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maróti P, Hanson D K, Baciou L, Schiffer M, Sebban P. Proc Natl Acad Sci USA. 1994;91:5617–5621. doi: 10.1073/pnas.91.12.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi E, Wraight C A. Proc Natl Acad Sci USA. 1996;93:2640–2645. doi: 10.1073/pnas.93.7.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miksovska J, Valerio-Lepiniec M, Schiffer M, Hanson D K, Sebban P. Biochemistry. 1998;37:2077–2083. doi: 10.1021/bi972696n. [DOI] [PubMed] [Google Scholar]
- 23.Rongey S H, Paddock M L, Feher G, Okamura M Y. Proc Natl Acad Sci USA. 1993;90:1325–1329. doi: 10.1073/pnas.90.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sebban P, Maróti P, Schiffer M, Hanson D K. Biochemistry. 1995;34:8390–8397. doi: 10.1021/bi00026a021. [DOI] [PubMed] [Google Scholar]
- 25.Paddock M L, Axelrod H L, Abresch E C, Yeh A P, Rees D C, Feher G, Okamura M Y. Biophys J. 1999;76:141. (abstr.). [Google Scholar]
- 26.Nabedryk E, Breton J, Okamura M Y, Paddock M L. Photosynth Res. 1998;55:293–299. [Google Scholar]
- 27.Hanson D K, Tiede D M, Nance S L, Chang C H, Schiffer M. Proc Natl Acad Sci USA. 1993;90:8929–8933. doi: 10.1073/pnas.90.19.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valerio-Lepiniec M, Delcroix J D, Schiffer M, Hanson D K, Sebban P. FEBS Lett. 1997;407:159–163. doi: 10.1016/s0014-5793(97)00328-1. [DOI] [PubMed] [Google Scholar]
- 29.Bylina E J, Ismail S, Youvan D C. Plasmid. 1986;16:175–181. doi: 10.1016/0147-619x(86)90055-7. [DOI] [PubMed] [Google Scholar]
- 30.Hanson D K, Baciou L, Tiede D M, Nance S L, Schiffer M, Sebban P. Biochim Biophys Acta. 1992;1102:260–265. doi: 10.1016/0005-2728(92)90108-e. [DOI] [PubMed] [Google Scholar]
- 31.Baciou L, Bylina E J, Sebban P. Biophys J. 1993;65:652–660. doi: 10.1016/S0006-3495(93)81114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McPherson P H, Okamura M Y, Feher G. Biochim Biophys Acta. 1993;1144:309–324. doi: 10.1016/0005-2728(93)90116-w. [DOI] [PubMed] [Google Scholar]
- 33.Hanson D K, Schiffer M. Photosynth Res. 1998;55:275–280. [Google Scholar]
- 34.Valerio-Lepiniec M, Miksovska J, Schiffer M, Hanson D K, Sebban P. Biochemistry. 1999;38:390–398. doi: 10.1021/bi980500t. [DOI] [PubMed] [Google Scholar]
- 35.Beroza P, Fredkin D R, Okamura M Y, Feher G. Biophys J. 1995;68:2233–2250. doi: 10.1016/S0006-3495(95)80406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sebban P, Maróti P, Hanson D K. Biochimie. 1995;77:677–694. doi: 10.1016/0300-9084(96)88183-1. [DOI] [PubMed] [Google Scholar]
- 37.Edsall J T, Wyman J. Biophysical Chemistry. Vol. 1. New York: Academic; 1958. [Google Scholar]
- 38.Delcroix J-D, Schiffer M, Hanson D K, Sebban P. In: Photosynthesis: From Light to Biosphere. Mathis P, editor. Dordrecht, the Netherlands: Kluwer; 1995. pp. 463–466. [Google Scholar]
- 39.Guan Y, Hickey M J, Borgstahl E O, Hallewell R A, Lepock J R, O’Connor D, Hsieh Y, Nick H S, Silverman D N, Tainer J A. Biochemistry. 1998;37:4722–4730. doi: 10.1021/bi972394l. [DOI] [PubMed] [Google Scholar]
- 40.Earnhardt J N, Qian M, Tu C, Laipis P J, Silverman D N. Biochemistry. 1998;37:7649–7655. doi: 10.1021/bi9729209. [DOI] [PubMed] [Google Scholar]
- 41.Boriack-Sjodin P A, Heck R W, Laipis P J, Silverman D N, Christianson D W. Proc Natl Acad Sci USA. 1995;92:10949–10953. doi: 10.1073/pnas.92.24.10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Barrett J E, Schultz P G, Santi D V. Biochemistry. 1999;38:848–852. doi: 10.1021/bi9822877. [DOI] [PubMed] [Google Scholar]
- 43.Laible P D, Kirmaier C, Holten D, Tiede D M, Schiffer M, Hanson D K. In: Photosynthesis: Mechanisms and Effects. Garab G, editor. Dordrecht, the Netherlands: Kluwer; 1998. pp. 849–852. [Google Scholar]