Abstract
Since the emergence of viral resistance of hepatitis B virus (HBV) during treatment is becoming an important issue even with newer drugs, there is a need for alternative treatment options such as, for example, RNA interference (RNAi) technology. While short-term suppression of HBV replication is easily achieved with small interfering RNA oligonucleotides, this is not the case for long-term suppression due to the lack of an optimal vector system. Based on the nonviral scaffold/matrix attachment region (S/MAR)-based vector system pEPI-1, which is free of common side effects and is stably retained as an episome even in the absence of selection, we designed a short hairpin RNA (shRNA) expression vector called pEPI-RNAi for HBV suppression. HBV-replicating HepG2.2.15 cells were transfected with pEPI-RNAi, and the intracellular status of the plasmid was followed by PCR and Southern analysis. HBV replication was measured on the DNA, RNA, and protein level. HBV RNA expression was reduced by almost 85% 3 months posttransfection with pEPI-RNAi. At 8 months posttransfection in the absence of antibiotic selection pressure, the suppression level was still 70% and the vector was retained as an episome. The reduction of total intracellular HBV DNA at this point was 77%, showing a marked suppression of HBV DNA replication. At a comparable level, secretion of viral antigens, as well as progeny HBV virions, was inhibited. The S/MAR-based vector system pEPI-1 allows long-term suppression of HBV replication by the expression of suitable shRNAs. Due to its unique properties compared to commonly used vectors, it provides an interesting option for the treatment of chronically HBV-infected individuals.
Hepatitis B virus (HBV) infection remains a major global health concern, with approximately 300 million people chronically infected worldwide with a high risk of developing liver cirrhosis or hepatocellular carcinoma. At present, antiviral agents like type 1 interferons, lamivudine, adefovir, and entacavir are used to treat a selected population of chronic HBV carriers. Although most of these drugs are effective in suppressing the viral load, long-term therapy is required to avoid viral reactivation and progression of liver disease. Due to the variability of the HBV genome, such long-term treatments are often associated with the emergence of resistant viral strains, which may compromise the initial clinical benefit of the treatment. Since this emergence of viral resistance is becoming an important issue even with newer drugs (26), there is a need for alternative treatment options.
During the last decade, RNA interference (RNAi) (2, 3) has opened new perspectives not only in experimental biology but also in the field of future gene therapy.
The mechanism of RNAi is initiated by the RNase III enzyme Dicer, which processes double-stranded RNA into ∼22-nucleotide oligomers which serve as guide sequences in the RNA-induced silencing complex, thus allowing specific mRNA cleavage. Since the discovery of RNAi as a sequence-specific posttranscriptional gene silencing mechanism (2), many attempts have been made to understand and potentially exploit this highly conserved regulatory mechanism to inhibit the expression of specific genes (19).
Recently, several groups showed that suppression of HBV replication can be achieved with small interfering RNA (siRNA) oligonucleotides. This effect, however, is only transient in mammalian cells, and repeated applications of siRNA oligonucleotides are required. Therefore, expression of short hairpin RNAs (shRNAs) under the control of RNA polymerase III promoters has been evaluated in vitro and in vivo (13, 24, 25).
Conventional vectors currently used for the expression of shRNAs in mammalian cells and organisms all have major disadvantages that limit their application in gene therapy (4). These include integration into the host genome, which may lead to insertional mutagenesis and silencing of the transgene, expression of viral proteins with a subsequent immunological response of the recipient organism, and only transient expression of the transgene (4, 8). Issues concerning the safety of the vector systems used are even more important in patients not suffering from end-stage, life-threatening diseases such as, for example, chronic viral infections like HBV infection.
Recently, we were able to develop a nonviral episomal replicating vector (pEPI-1) whose function relies exclusively on a transcription unit linked to a scaffold/matrix attachment region (S/MAR) (1, 7, 14, 23). This vector is mitotically stable in the absence of selection over hundreds of generations and ensures long-term expression of transgenes (6). Very recently, our in vitro data have been confirmed in vivo (10; O. Argyros, S. Wong, M. Niceta, S. Waddington, C. Coutelle, A. Miller, and R. Harbottle, unpublished data, 2007). In addition, its general capability of specific protein suppression by shRNA expression has been demonstrated in a disease model system (5).
In this study, we inserted shRNA expression cassettes targeting different regions of the HBV mRNAs under the control of the human RNA polymerase III promoter U6 into the vector pEPI-1. In HepG2.2.15 cells transfected with this vector, HBV mRNA expression, as well as secretion of viral antigens and progeny HBV virions, was significantly suppressed for as long as 8 months, even in the absence of selection pressure.
MATERIALS AND METHODS
Transfection of HepG2.2.15 cells.
HepG2.2.15 cells were cultured as described previously (20). The cells were grown in Williams E medium supplemented with 20% fetal calf serum, 1% nonessential amino acid, 1 mM sodium pyruvate, 2 mM l-glutamine, 100 U/liter penicillin, 100 mg/ml streptomycin, and 100 mg/ml G418. Cells (106) were transfected by lipofection (FuGene; Roche) with 2.5 μg vector DNA. Selection pressure was applied 48 to 72 h after transfection with blasticidin at an initial dose of 5 μg/ml for the first 10 days, reduced to 3 μg/ml for an additional 10 days, and then completely withdrawn.
HepG2.2.15 cells were then cultured in Williams E medium supplemented with 10% fetal calf serum, 2.25 mM l-glutamine, 0.06% glucose, 23 mM HEPES (pH 7.4), 4.8 μg of hydrocortisone/ml, 1 μg of inosine/ml, 1.5% dimethyl sulfoxide, 50 μg of streptomycin/ml, 50 IU of penicillin/ml, and 50 mg/ml G418. Once the cells were confluent, 1.75% DMSO was added to the medium and the serum content was reduced to 5% to stimulate HBV replication (16).
Vector construction.
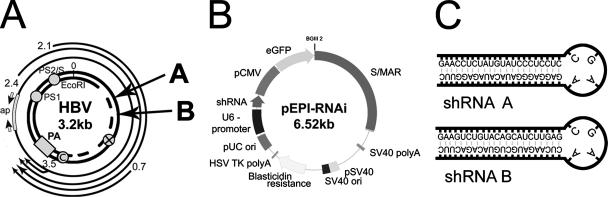
The shRNA expression cassette was designed with the BlockIT-siRNA kit from Invitrogen. The shRNA cassette was inserted into the vector upstream of the cytomegalovirus-green fluorescent protein expression cassette with the Gateway System from Invitrogen (Fig. 1). Three different shRNAs were designed targeting different HBV mRNAs, as well as one nonsense sequence.
FIG. 1.
Experimental design. (A) Locations of RNAi target sites. Arrows indicate the locations of RNAi target sites within the HBV genome. Open reading frames: PS1, preS1; PS2/S, preS2; C, core; ×, protein X; PA, polyadenylation site; ap, sequence amplified by real-time PCR. (B) Vector used in this study (pEPI-RNAi). eGFP, enhanced green fluorescent protein; pCMV, cytomegalovirus promoter; SV40, simian virus 40; HSV, herpes simplex virus; TK, thymidine kinase. (C) Predicted folding of shRNA sequences RNAi-A and RNAi-B.
Sequences were as follows: A, 5′-CACCGAACCTCTATGTATCCCTCCTCCGAAGAGGAGGGATACATAGAGGTTC-3′; B, 5′-CACCGAAGTCTGTACAGCATCTTGAGCGAACTCAAGATGCTGTACAGACTTC-3′; C, 5′-CACCGGACATTGACCCGTATAAAGACCGAAGTCTTTATACGGGTCAATG TCC-3′; NSSQ, 5′-CACCAATTCTACGTACGTGTCACGTGCGAACACGTGACACGTACGTAGAATT-3′.
DNA isolation and Southern analysis.
For Southern analysis, total DNA, as well as episomal DNA obtained by Hirt extraction (9), was isolated. It was digested with an appropriate restriction enzyme if indicated, separated on 0.8% agarose gels, blotted onto nylon membranes, and hybridized with a 32P-labeled pEPI-1 probe. Hybridization was done in Church buffer (0.5 M sodium phosphate [pH 7.2], 7% sodium dodecyl sulfate, 1 mM EDTA) at 65°C for 14 h.
pEPI-1 long PCR.
Total DNA and DNA obtained by Hirt extraction were digested with HindIII and ApaII. PCR amplification was performed as follows: 94°C for 3 min; 25 cycles of 94°C for 1 min, 60°C for 1 min, and 68°C for 7 min; and a final extension at 68°C for 10 min. The primers used were as follows: pEPIlong-for, 5′-AGATACCTACAGCGTGAGCTATGAG-3′; pEPIlong-back, 5′-GGTAAGACACGACTTATCGCCACT-3′.
ELISA.
HBsAg concentrations were quantified with an enzyme-linked immunosorbent assay (ELISA) from Abbott (Wiesbaden, Germany).
Dot blot analysis of progeny HBV.
The total, HBV progeny-containing supernatant of cultured cells that was obtained upon stimulation of HBV expression was blotted onto nylon membranes and hybridized with 32P-labeled HBV DNA (Rediprime II Random Prime Labeling System; Amersham Pharmacia) in Church buffer at 68°C for 12 h. HBV DNA concentrations were determined relative to that of an external HBV DNA standard with a PhosphorImager (15).
Quantitative PCR analysis.
For PCR analysis, DNA was extracted with a Qiagen DNA purification kit. RNA was extracted with Trizol (Invitrogen, Paisley, United Kingdom) and reverse transcribed into cDNA with the Synthesis Kit from Super Array (Biomol) according to the manufacturer's instructions. Real-time PCR was then performed with a LightCycler and SYBR green detection (Roche, Mannheim, Germany). The primers used for intracellular HBV DNA and RNA were as follows: HBV-RTfor, 5′-CTCCTCCAGCTTATAGACC-3′; HBV-RTrev, 5′-GTGAGTGGGCCTACAAA-3′.
Amplification was done at 94°C for 2 min and then at 94°C for 1 min, 51°C for 30 s, and 72°C for 15 s for a total of 30 cycles, followed by 72°C for 2 min for total HBV DNA and HBV RNA.
The amount of total HBV DNA or RNA was analyzed in relation to that of intracellular β-actin DNA or RNA, respectively. The primers used for β-actin were as follows: bActinfor, 5′-CGTCTTCCCCTCCATCG-3′; bActinrev, 5′-CTCGTTAATGTCACGCAC-3′.
β-Actin amplification was done at 94°C for 2 min and then at 94°C for 30 s, 54°C for 30 s, and 72°C for 12 s for a total of 30 cycles, followed by 72°C for 2 min.
RESULTS
Vector design.
Three different RNAi target sequences were chosen on the basis of previous reports (see Material and Methods) (11, 21, 24) according to their efficiency in terms of suppression of HBV replication and on the basis of their conservation among the major HBV genotypes.
In all cases, overlapping reading frames were targeted such that multiple viral RNAs were potentially inhibited by each shRNA, and in all cases, the sequences exactly matched the HBV1 genome subtype ayw integrated into the HepG2.2.15 cells (20; GenBank accession number EF103275). These cells contain four tandem dimers of the HBV genome chromosomally integrated and constitutively express HBV. shRNA expression cassettes driven by the human U6 promoter were designed and cloned into pEPI-1. In addition, the original kanamycin resistance gene in pEPI-1 was replaced with a blasticidin resistance cassette as depicted in Fig. 1B.
Transfection and analysis of the intracellular state of the vector system.
pEPI-RNAi vectors were transferred as superhelical plasmids by lipofection into HepG2.2.15 cells. As controls, vectors containing either a nonsense shRNA cassette or no shRNA cassette at all were used. After an initial selection period of 2 weeks, the selection pressure with blasticidin was reduced and completely withdrawn 4 weeks after transfection. To determine HBV gene expression and RNAi efficiency, the HBsAg concentration in the supernatant was measured at this point by quantitative ELISA and was shown to be reduced between ∼90% for siRNA-A, ∼70% for siRNA-B, and ∼55% for siRNA-C. The target regions of the two most efficient sequences in the HBV genome are shown in Fig. 1A. All further experiments were only made for the siRNA-A and siRNA-B sequences and were done in total cell populations rather than individual clones at 3 and 8 months posttransfection.
The data presented are the results of three subsequent individual transfection experiments.
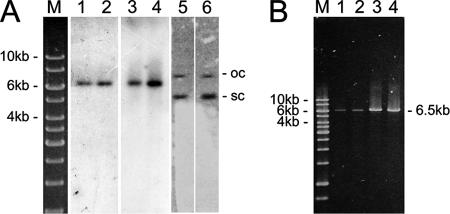
To determine the intracellular state of the vector, Southern analyses of total cellular DNA and Hirt extracts were performed, as well as PCR amplification of the complete vector. As shown in Fig. 2, the hybridization pattern clearly shows that the vector was retained as an episome in the multiclonal cell population for at least 8 months. From the intensity of the signal obtained, we estimated that the vector was maintained at a copy number below 10 per cell, as observed for other cell lines (1, 14, 22).
FIG. 2.
Analysis of the intracellular state of the vector system. (A) Southern blot analyses of total DNA and DNA obtained by Hirt extraction from 107 HepG2.2.15 cells transfected with pEPI-RNAi 8 months after transfection. Lanes 1 and 2, DNA obtained by Hirt extraction digested with the single cutter BglII; lanes 3 and 4, total DNA digested with the single cutter BglII; lanes 5 and 6, total DNA digested with the noncutter HindIII. The DNA was from HepG2.2.15 cells transfected with pEPI-RNAi-A (lanes 1, 3, and 5) or pEPI-RNAi-B (lanes 2, 4, and 6). Lane M, molecular size markers. The linearized form, as well as the open-circle (oc) and supercoiled (sc) forms, of the vector is visualized. Membranes were hybridized with 32P-labeled pEPI-DNA. (B) Amplification of pEPI-RNAi by long PCR in HepG2.2.15 cells at 8 months posttransfection. Lanes 1 and 2, DNA obtained by Hirt extraction; lanes 3 and 4, total DNA. The DNA was from HepG2.2.15 cells transfected with pEPI-RNAi-A (lanes 1 and 3) or pEPI-RNAi-B (lanes 2 and 4). Lane M, molecular size markers.
Analysis of the supernatant.
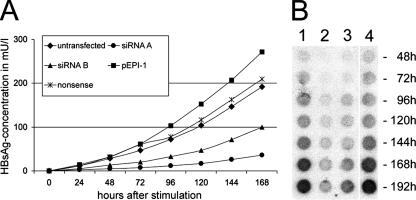
To analyze whether the suppression of HBV gene expression observed at first was sustained, the HBsAg concentration in the supernatant was again measured 8 months posttransfection upon stimulation of HBV gene expression (Fig. 3A). At this time point, the HBsAg concentrations in the supernatant were reduced by about 82% for siRNA-A and 51% for siRNA-B compared to those in untransfected cells, with a standard deviation below 10%.
FIG. 3.
Analysis of the supernatant. (A) Quantitative measurement by ELISA of HBsAg in cell supernatant (8 months after transfection) collected at the indicated time points after stimulation of HBV replication. The HBsAg levels in cell culture medium of cells transfected with pEPI-RNAi-A or pEPI-RNAi-B were compared to that of cells transfected with pEPI containing either no or nonsense siRNA and to that of untransfected cells. All measurements were done in triplicate, and the standard deviation was found to be below 10% in all cases. (B) Dot blot analysis, with a 32P-labeled HBV DNA probe, of the supernatant of HepG2.2.15 cells collected at the indicated time points. Cells were left untransfected (lane 1) or transfected with pEPI-RNAi-A (lane 2), pEPI-RNAi-B (lane 3), or pEPI containing a nonsense interfering RNA (lane 4).
Remarkably, transfection with a vector containing nonsense or no shRNA expression cassette at all even resulted in a slight increase in HBsAg secretion. This phenomenon is not unexpected since HBV replication in HepG2.2.15 cells is very sensitive and transfection with plasmids often leads to the stimulation of HBV replication. Compared to those cells, suppression of the HBsAg concentration was about 87% for siRNA-A and 65% for siRNA-B.
In parallel, the release of progeny HBV particles into the supernatant was determined by dot blot analysis (15) as shown in Fig. 3B. The release of DNA-containing HBV particles was suppressed as efficiently as the expression of the HBV S protein and thus the secretion of HBsAg.
Analysis of intracellular HBV mRNA and DNA.
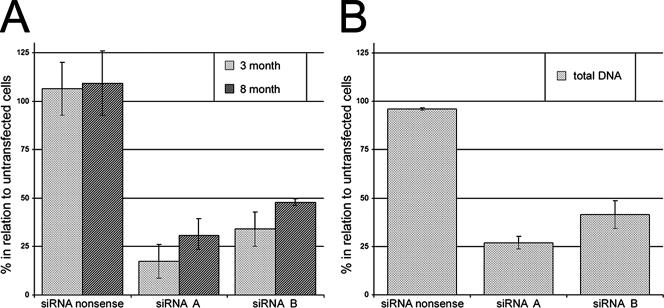
To study the RNAi effect at the RNA level, total RNA was extracted 3 and 8 month posttransfection. cDNA was synthesized with random primers, followed by real-time PCR with primers specific for β-actin and the 3.5-kb HBV RNA which encompasses the HBV pregenome. As shown in Fig. 4, pregenomic HBV RNA levels were reduced by about 90% by siRNA-A and 70% by siRNA-B 3 months after pEPI transfection. This difference between siRNA-A and siRNA-B was found to be reproducible in three independent transfections, while the copy number per cell as estimated by Southern analysis, as well as the stability of the episome, did not differ.
FIG. 4.
Analysis of intracellular HBV mRNA and DNA. (A) Suppression of total HBV RNA at 3 and 8 months after transfection in relation to untransfected cells. Relative HBV RNA expression was determined by real-time reverse transcription-PCR with β-actin expression as a control. The HBV RNA expression levels of cells transfected with pEPI-siRNA-A, pEPI-siRNA-B, and pEPI-siRNA-nonsense were compared with that of untransfected cells. The results shown are the means and standard errors of three (at 3 months) and two (at 8 months) independent transfections. HBV RNA expression was significantly reduced in cells transfected with pEPI-siRNA-A and pEPI-siRNA-B (P < 0.01 for both [Student's t test]). All measurements were done in triplicate. (B) Total intracellular HBV DNA 8 months after transfection of pEPI-siRNA-A and -B into HepG2.2.15 cells in relation to untransfected cells. The relative HBV DNA concentration was determined by real-time reverse transcription-PCR with the β-actin DNA concentration as a control. The HBV DNA levels of cells transfected with pEPI-siRNA-A, pEPI-siRNA-B, and pEPI-siRNA-nonsense were compared with that of untransfected cells. The results shown are the mean and standard error of two independent transfections. The level of intracellular HBV DNA was significantly reduced in cells transfected with pEPI-siRNA-A and pEPI-siRNA-B (P < 0.01 for both [Student's t test]). All measurements were done in triplicate.
The suppression compared to control cells only slightly decreased (70 to 75% for siRNA-A and 55% for siRNA-B) 8 months after transfection (Fig. 4A). This decline is not unexpected considering previous observations that the mitotic stability of this vector system—in the absence of selection pressure—is slightly below 1 due to a maldistribution of the plasmids during mitosis (18). Based on our data, we calculated a 2 to 4% probability for an unequal distribution of each individual plasmid between two daughter cells. This stability is fairly comparable to that of other large mammalian artificial chromosomes. Similar observations were obtained previously with other cell lines (18, 22).
To demonstrate that RNAi-mediated degradation of HBV RNA resulted in the inhibition of intracellular HBV DNA replication, we analyzed total intracellular HBV DNA by real-time PCR (Fig. 4B). This was done after a 7-day stimulation of HBV expression so that the influence of HBV DNA integrated into the HepG2.2.15 genome compared to free HBV DNA can be assumed to be insignificant (12). As shown in Fig. 4B, at 8 months after transfection, intracellular HBV DNA concentrations were reduced by 77% and 60% for siRNA-A and siRNA-B, respectively.
DISCUSSION
In this study, we showed that expression of siRNAs by the S/MAR-based, episomally retained vector pEPI-1 suppresses HBV gene expression, intracellular HBV DNA replication, and release of progeny HBV very efficiently for up to 8 months in the absence of continuous selection pressure when it is transfected into stably HBV-replicating hepatoma cells.
Due to the emergence of viral resistance to conventional therapy (26), new alternative therapeutic approaches have to be established. In the past, various groups have shown that disruption of the viral life cycle by RNAi-mediated RNA degradation is possible in principle. However, this approach currently relies on either repeated treatment with synthetic RNAi oligonucleotides or expression of shRNA from virus-based vectors (11, 13, 21, 24). Both approaches still suffer from either practical or safety problems and are consequently only of very limited use for the future treatment of chronic HBV infection. Therefore, we made an attempt to disrupt the HBV life cycle by shRNA expression encoded on the nonviral vector system pEPI-1. The function of this vector relies exclusively on chromosomal elements (7), is stably retained intracellularly as an episome, and seems to be free of safety limitations (10) observed with commonly used vector systems.
Our data clearly show that this vector system is suitable as a tool for long-term HBV suppression mediated by shRNA expression. Expression of pregenomic, as well as subgenomic, HBV RNAs was significantly silenced. In addition, analysis of total intracellular HBV DNA, as well as the secretion of progeny HBV particles into the supernatant of HepG2.2.15 cells, demonstrated successful disruption of the viral life cycle. The observed difference in effectiveness between various siRNA sequences is most likely sequence dependent since we did not observe differences in copy number or plasmid stability.
This suppressive effect on HBV expression was long lasting in the absence of blasticidin selection pressure, with only a slow decline attributed to the fact that the mitotic stability of pEPI-1 is slightly below 1 (18, 22). However, in vivo, any cells with sufficient suppression of HBV replication will most probably have a significant survival advantage because cells expressing HBV antigens will be targeted by HBV-specific cytotoxic T lymphocytes. Therefore, the liver most likely will be repopulated by cells in which HBV replication is suppressed (17).
In addition, reduction of HBV antigenemia by RNAi-mediated degradation of viral RNA may restore an effective T-cell response. There is strong evidence that chronic antigen stimulation has a negative impact on the T-cell response (17). It also might be that HBV antigens negatively influence antigen presentation by dendritic cells. Therefore, RNAi-mediated disruption of the viral life cycle, in combination with established antiviral treatments such as, for example, lamivudine, might help the immune system to ultimately clear the infection. Even though these considerations are very promising, one major obstacle affecting all known vector systems still remains unsolved, i.e., how to deliver these constructs to hepatocytes in vivo.
In summary, the novel S/MAR-based vector system used in this study might open new perspectives on gene therapy of hepatitis B. As a nonviral vector approach, it most likely fulfils the requirements for a safe genetic therapy that allows long-term suppression of HBV replication.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (AJ491/2 to A.J., Li231/25 to H.J.L., and PR618/4 to U.P.) and by a grant from the European Union to H.J.L.
We especially thank Sina Ronzon and Lyudmila Glushkova.
Footnotes
Published ahead of print on 12 May 2008.
REFERENCES
- 1.Baiker, A., C. Maercker, C. Piechaczek, S. B. Schmidt, J. Bode, C. Benham, and H. J. Lipps. 2000. Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nat. Cell Biol. 2:182-184. [DOI] [PubMed] [Google Scholar]
- 2.Fire, A. 1999. RNA-triggered gene silencing. Trends Genet. 15:358-363. [DOI] [PubMed] [Google Scholar]
- 3.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 4.Jackson, D. A., S. Juranek, and H. J. Lipps. 2006. Designing nonviral vectors for efficient gene transfer and long-term gene expression. Mol. Ther. 14:613-626. [DOI] [PubMed] [Google Scholar]
- 5.Jenke, A. C., T. Eisenberger, A. Baiker, I. M. Stehle, S. Wirth, and H. J. Lipps. 2005. The nonviral episomal replicating vector pEPI-1 allows long-term inhibition of bcr-abl expression by shRNA. Hum. Gene Ther. 16:533-539. [DOI] [PubMed] [Google Scholar]
- 6.Jenke, A. C., M. F. Scinteie, I. M. Stehle, and H. J. Lipps. 2004. Expression of a transgene encoded on a non-viral episomal vector is not subject to epigenetic silencing by cytosine methylation. Mol. Biol. Rep. 31:85-90. [DOI] [PubMed] [Google Scholar]
- 7.Jenke, A. C., I. M. Stehle, F. Herrmann, T. Eisenberger, A. Baiker, J. Bode, F. O. Fackelmayer, and H. J. Lipps. 2004. Nuclear scaffold/matrix attached region modules linked to a transcription unit are sufficient for replication and maintenance of a mammalian episome. Proc. Natl. Acad. Sci. USA 101:11322-11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipps, H. J., A. C. Jenke, K. Nehlsen, M. F. Scinteie, I. M. Stehle, and J. Bode. 2003. Chromosome-based vectors for gene therapy. Gene 304:23-33. [DOI] [PubMed] [Google Scholar]
- 9.Livingston, D. M., and D. M. Kupfer. 1977. Control of Saccharomyces cerevisiae 2μ DNA replication by cell division cycle genes that control nuclear DNA replication. J. Mol. Biol. 116:249-260. [DOI] [PubMed] [Google Scholar]
- 10.Manzini, S., A. Variolu, I. M. Stehle, M. L. Bacci, M. G. Cerrito, R. Giovannoni, A. Zannoni, M. R. Bianco, M. Forni, P. Donini, M. Papa, H. J. Lipps, and M. Lavitrano. 2006. Genetically modified pigs produced with a nonviral episomal vector. Proc. Natl. Acad. Sci. USA 103:17672-17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCaffrey, A. P., H. Nakai, K. Pandey, Z. Huang, F. H. Salazar, H. Xu, S. F. Wieland, P. L. Marion, and M. A. Kay. 2003. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 21:639-644. [DOI] [PubMed] [Google Scholar]
- 12.Mehta, A., X. Lu, T. M. Block, B. S. Blumberg, and R. A. Dwek. 1997. Hepatitis B virus (HBV) envelope glycoproteins vary drastically in their sensitivity to glycan processing: evidence that alteration of a single N-linked glycosylation site can regulate HBV secretion. Proc. Natl. Acad. Sci. USA 94:1822-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng, J., Y. Zhao, J. Mai, W. K. Pang, X. Wei, P. Zhang, and Y. Xu. 2005. Inhibition of hepatitis B virus replication by various RNAi constructs and their pharmacodynamic properties. J. Gen. Virol. 86:3227-3234. [DOI] [PubMed] [Google Scholar]
- 14.Piechaczek, C., C. Fetzer, A. Baiker, J. Bode, and H. J. Lipps. 1999. A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells. Nucleic Acids Res. 27:426-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Protzer, U., S. Seyfried, M. Quasdorff, G. Sass, M. Svorcova, D. Webb, F. Bohne, M. Hösel, P. Schirmacher, and G. Tiegs. 2007. Antiviral activity and hepatoprotection by heme oxygenase-1 in hepatitis B virus infection. Gastroenterology 133:1156-1165. [DOI] [PubMed] [Google Scholar]
- 16.Quasdorff, M., M. Hosel, M. Odenthal, U. Zedler, F. Bohne, P. Gripon, H. P. Dienes, U. Drebber, D. Stippel, T. Goeser, and U. Protzer. 26 March 2008, posting date. A concerted action of HNF4α and HNF1α links hepatitis B virus replication to hepatocyte differentiation. Cell. Microbiol. doi: 10.1111/j.1462-5822.2008.01141.x. [DOI] [PubMed]
- 17.Rehermann, B., and F. V. Chisari. 1995. The immunology of chronic hepatitis B, p. 85-117. In R. Koshy and W. H. Caselmann (ed.), Hepatitis B virus: molecular mechanisms in disease and novel strategies for therapy. Imperial College Press, London, United Kingdom.
- 18.Schaarschmidt, D., J. Baltin, I. M. Stehle, H. J. Lipps, and R. Knippers. 2004. An episomal mammalian replicon: sequence-independent binding of the origin recognition complex. EMBO J. 23:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scherer, L. J., and J. J. Rossi. 2003. Approaches for the sequence-specific knockdown of mRNA. Nat. Biotechnol. 21:1457-1465. [DOI] [PubMed] [Google Scholar]
- 20.Sells, M. A., M. L. Chen, and G. Acs. 1987. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA 84:1005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shlomai, A., and Y. Shaul. 2003. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology 37:764-770. [DOI] [PubMed] [Google Scholar]
- 22.Stehle, I. M., J. Postberg, S. Rupprecht, T. Cremer, D. A. Jackson, and H. J. Lipps. 2007. Establishment and mitotic stability of an extra-chromosomal mammalian replicon. BMC Cell Biol. 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stehle, I. M., M. F. Scinteie, A. Baiker, A. C. Jenke, and H. J. Lipps. 2003. Exploiting a minimal system to study the epigenetic control of DNA replication: the interplay between transcription and replication. Chromosome Res. 11:413-421. [DOI] [PubMed] [Google Scholar]
- 24.Uprichard, S. L., B. Boyd, A. Althage, and F. V. Chisari. 2005. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc. Natl. Acad. Sci. USA 102:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, Y., A. L. Huang, N. Tang, B. Q. Zhang, and N. F. Lu. 2005. Specific anti-viral effects of RNA interference on replication and expression of hepatitis B virus in mice. Chin. Med. J. 118:1351-1356. [PubMed] [Google Scholar]
- 26.Yotsuyanagi, H., and K. Koike. 2007. Drug resistance in antiviral treatment for infections with hepatitis B and C viruses. J. Gastroenterol. 42:329-335. [DOI] [PubMed] [Google Scholar]