Abstract
In a phase I/II evaluation of the CXCR4 antagonist AMD3100, human immunodeficiency virus RNA levels were significantly reduced in a single study subject who harbored CXCR4 (X4)-tropic virus, but not in subjects who harbored either dual/mixed (DM)-tropic or CCR5 (R5)-tropic virus (C. W. Hendrix et al., J. Acquir. Immune Defic. Syndr. 37:1253-1262, 2004). In this study, we analyzed the envelope clones of DM-tropic virus in baseline and treated virus populations from 14 subjects. Ten subjects exhibited significant reductions in CXCR4-mediated infectivity after 10 days of AMD3100 therapy relative to baseline (X4 suppressor group), while four subjects had no reduction of CXCR4-mediated infectivity (X4 nonsuppressor group). The baseline viruses of the X4 suppressor group infected CXCR4-expressing cells less efficiently than those of the X4 nonsuppressor group. Clonal analysis indicated that the baseline viruses from the X4 suppressor group contained a higher proportion of R5-tropic variants mixed with CXCR4-using variants, while the X4 nonsuppressor group was enriched for CXCR4-using variants. AMD3100 suppressed X4-tropic variants in all subjects studied, but not all dualtropic variants. Furthermore, dualtropic variants that used CXCR4 efficiently were suppressed by AMD3100, while dualtropic variants that used CXCR4 poorly were not. This study demonstrated that AMD3100 has the ability to suppress both X4-tropic and certain dualtropic variants in vivo. The suppression of CXCR4-using variants by AMD3100 is dependent on both the tropism composition of the virus population and the efficiency of CXCR4 usage of individual variants.
Human immunodeficiency virus type 1 (HIV-1) envelope (env)-mediated entry is a multistep process, which requires a chemokine coreceptor (predominantly CCR5 or CXCR4) in addition to CD4 (3, 6). HIV-1 gp120 binds to CD4 on the cell surface, promoting a conformational change that allows gp120 to engage the coreceptor, which in turn triggers conformational changes in gp41 that lead to fusion of the viral and host membranes (1). Some viruses utilize both coreceptors and are characterized as dualtropic, while others use either CCR5 (R5 tropic) or CXCR4 (X4 tropic) exclusively. R5-tropic viruses are predominant at transmission and at early stages of infection, while most CXCR4-using (dualtropic and X4-tropic) viruses are present at later stages of disease (18, 21).
The switch from R5-tropic to CXCR4-using virus, first characterized as a switch from a non-syncytium-inducing to a syncytium-inducing phenotype in MT2 cells, occurs in approximately 50% of untreated HIV-1-infected individuals and is associated with decreased CD4+ T-cell counts and rapid disease progression (12, 13, 20). A number of recent studies have consistently demonstrated that the frequency of CXCR4-using viruses is approximately 20% in early infection (17) and increases to approximately 50% in late-stage disease (16; E. Coakley et al., presented at the 2nd International Workshop on Targeting HIV Entry, Boston, MA, 20 to 21 October 2006). The prevalence of CXCR4 use in antiretroviral treatment-experienced subjects is 50%, or slightly higher, depending on the cohort studied (17; E. Coakley et al., presented at the 2nd International Workshop on Targeting HIV Entry; J. Demarest et al., presented at the 44th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 30 October to 2 November 2004). Consequently, there is an interest in the development of drugs that block CXCR4-mediated HIV-1 infection in addition to CCR5 inhibitors.
Agents that inhibit HIV-1 entry are widely studied. Enfuvirtide targets virus-cell membrane fusion and is approved for use in North America and Europe. Compounds that block the interaction of HIV-1 gp120 with either CD4 (11, 15) or the CCR5 or CXCR4 chemokine coreceptor (5, 9, 22) are being evaluated in several clinical trials. The phase III trial of maraviroc, a CCR5 antagonist, provides compelling evidence that this class of compounds can efficiently block R5-tropic HIV-1 infection (J. Lalezari et al., presented at the 14th Conference on Retroviruses and Opportunistic Infections, Los Angeles, CA, 2007 [abstr. 104bLB]; M. Nelson et al., presented at the 14th Conference on Retroviruses and Opportunistic Infections, Los Angeles, CA, 2007 [abstr. 104aLB]).
One of the most-well-characterized CXCR4 antagonists is the bicyclam molecule AMD3100 (2, 4, 7, 8). Further studies have shown that AMD3100 also inhibits dualtropic virus in vitro (19). In a phase I/II clinical trial, AMD3100 was shown to significantly reduce HIV RNA levels (0.9-log10-unit reduction) in a single study subject who harbored pure X4-tropic virus but not in the remaining subjects who harbored either dual/mixed (DM)-tropic or R5-tropic viruses (9). In addition, AMD11070, another CXCR4 antagonist, was shown to reduce the fraction of CXCR4-using viruses in a phase I/II study (G. Moyle et al., presented at the 14th Conference on Retroviruses and Opportunistic Infections, Los Angeles, CA, 2007 [abstr. 511]).
DM-tropic virus populations can comprise either pure dualtropic variants or mixtures of R5-tropic, X4-tropic, and/or dualtropic variants. The effect of AMD3100 on X4-tropic and dualtropic variants in a DM-tropic virus population in vivo is not known. In this study, we evaluated the targeted antiviral activity of AMD3100 by characterizing the env clones of the baseline and treated virus populations in subjects with DM-tropic viruses at baseline. Both the tropism composition of the virus population (the relative proportions of R5-tropic, X4-tropic, and dualtropic clones) and the efficiency of CXCR4-mediated entry of individual variants were found to be associated with the ability of AMD3100 to suppress CXCR4-using variants in vivo.
MATERIALS AND METHODS
Study cohort.
Forty HIV-1-positive individuals were enrolled in a phase I/II multicenter, open-label, dose-escalating study of AMD3100 administered as a 10-day intravenous infusion (9). Subjects were randomized across a range of doses (2.5, 5, 10, 20, 40, 80, and 160 μg/kg of body weight/h). The protocol was approved by the local institutional review board, and all subjects gave written informed consent prior to their participation in the study. The coreceptor tropism of baseline plasma viruses (drug screen or treatment initiation) and all available day 11 plasma viruses (completion of 10 days of AMD3100) was determined. Of the 40 subjects, 14 subjects who harbored DM-tropic viruses at baseline, and for whom paired samples taken at day 11 were available, were selected for this follow up study. Viral loads at baseline and at day 11 of treatment were measured using the Roche HIV-1 RNA Amplicor monitor assay.
Determination of HIV-1 coreceptor phenotype.
Coreceptor tropism was measured using the Trofile assay (Monogram Biosciences) (24). Specifically, a replication-defective retroviral vector containing a luciferase gene was used to cotransfect human embryonic kidney 293 cell cultures (AIDS Research and Reference Reagent Program, NIH) along with env expression vectors containing patient-derived viral envelope sequences (24). Pseudotyped viruses were harvested 2 days after transfection and were assessed for their ability to infect U87 cells expressing CD4 and either CCR5 or CXCR4 (provided by Nathaniel Landau) (24). Viruses were classified as R5 tropic, X4 tropic, or DM tropic based on two criteria: (i) the production of luciferase activity (expressed in relative light units [RLU]) in U87 CD4 CCR5 and U87 CD4 CXCR4 cells and (ii) the specific inhibition of luciferase activity by a CCR5 antagonist [a member of the 4-(piperidin-1-yl)butane family, provided by Merck] or a CXCR4 antagonist (AMD3100, provided by AnorMED) (24).
Clonal analysis of viral populations.
Forty-eight env clones were isolated from each viral population and screened for their ability to mediate pseudovirion infection of U87 cells expressing CD4 and either CCR5 or CXCR4 (23). The coreceptor tropism of a subset of viable clones from selected subjects was confirmed using the standard Trofile assay (24). The sequences of the gp160 env regions of these clones were determined using standard dye-deoxy chain terminator chemistry (ABI, Foster City, CA).
Phylogenetic analysis of env clones.
Two subjects (subjects 33 and 35) had baseline (day 0), on-treatment (day 11), and posttreatment (days 18 and 39) samples available. Ten to 15 clones from each time point were sequenced, and phylogenetic analysis of gp160 env nucleotide sequences was performed using neighbor-joining methods (14) and bootstrap resampling (1,000 replicates). For all phylogenetic trees shown in the study, coreceptor tropism designations of env clones were assigned based on the results of the Trofile assay (24). The amino acid sequences of the V3 loop regions of these clones and their correlation with coreceptor tropism before and after AMD3100 treatment were also compared.
Statistical analyses.
The Wilcoxon signed-rank test was used to compare viral infectivities (measured as RLU) in the CXCR4+ and CCR5+ cells of X4 suppressor and X4 nonsuppressor groups at baseline and day 11. All analyses were performed using Prism, version 4.03 (GraphPad, San Diego CA).
Nucleotide sequence accession numbers.
The gp160 env nucleotide sequences obtained in this study have been submitted to GenBank under accession numbers EU604549 to EU604642.
RESULTS
Reduction of viral infectivity by AMD3100 is correlated with CXCR4 usage of DM-tropic viruses.
The coreceptor tropism of viruses isolated from 14 subjects at baseline and after 10 days of AMD3100 treatment (day 11) was determined using the Trofile assay (24). Under AMD3100 drug pressure, DM-tropic virus populations in some subjects converted to R5 tropism while others remained DM tropic (Table 1). The baseline viruses displayed a broader range of infectivity in CXCR4-expressing cells than in CCR5-expressing cells, as measured by luciferase activity (RLU). Based on infectivity in CXCR4-expressing cells before and after AMD3100 treatment, we were able to classify the 14 subjects into two groups: X4 suppressors (10 subjects) and X4 nonsuppressors (4 subjects). These two groups differed in the ability of viruses to infect CXCR4-expressing cells at baseline. The infectivity of baseline viruses in CXCR4-expressing cells ranged from 306 to 48,606 RLU for X4 suppressors, compared to 216,896 to 1,647,981 RLU for X4 nonsuppressors. In contrast, the infectivity of baseline viruses in CCR5-expressing cells was similar for X4 suppressors (range, 34,494 to 813,948 RLU) and X4 nonsuppressors (range, 35,731 to 720,638 RLU). Following 10 days of AMD3100 therapy, 8 of 10 subjects classified as X4 suppressors had virus populations that became R5 tropic, and the other 2 remained DM tropic but displayed a >50-fold decrease in infectivity in CXCR4-expressing cells. In contrast, none of the four subjects classified as X4 nonsuppressors had virus populations showing reductions in infectivity in CXCR4-expressing cells after 10 days of AMD3100 therapy. Comparison of luciferase activities generated from baseline and day 11 virus samples showed reductions in viral infectivity on CXCR4 target cells for the 10 subjects classified as X4 suppressors (P = 0.002 by the Wilcoxon signed-rank test) but not for X4 nonsuppressors (P = 0.625). Viral infectivity in CCR5 target cells did not differ between the two groups (P > 0.08). No dose-related difference between the two groups was observed, since subjects on low and high doses were found in both groups (Table 1).
TABLE 1.
Coreceptor tropism of virus population from DM subjects at baseline and day 11
| Groupa and subject | AMD3100 dose (μg/kg/h) | Tropism, day 11 | RLU produced in:
|
|||
|---|---|---|---|---|---|---|
| CCR5+ cells
|
CXCR4+ cells
|
|||||
| Baseline | Day 11 | Baseline | Day 11 | |||
| X4 suppressor | ||||||
| 74 | 80 | R5 | 52,546 | 87,159 | 308 | 56 |
| 31 | 20 | R5 | 114,206 | 316,970 | 575 | 107 |
| 9 | 5 | R5 | 39,361 | 50,543 | 998 | 112 |
| 32 | 40 | R5 | 46,180 | 140,449 | 1,904 | 107 |
| 28 | 20 | R5 | 409,212 | 384,574 | 2,046 | 81 |
| 35 | 40 | R5 | 34,494 | 153,409 | 2,443 | 56 |
| 73 | 80 | R5 | 143,194 | 134,000 | 3,021 | 97 |
| 11 | 5 | DM | 224,031 | 256,901 | 13,710 | 199 |
| 10 | 5 | R5 | 813,948 | 932,650 | 32,597 | 77 |
| 33 | 40 | DM | 241,910 | 170,994 | 48,606 | 505 |
| Median (range) | 128,670 (34,494-813,948) | 162,201 (50,543-932,650) | 2,244 (308-48,606) | 102 (56-505) | ||
| X4 nonsuppressor | ||||||
| 70 | 40 | DM | 124,598 | 157,147 | 216,896 | 256,772 |
| 2 | 2.5 | DM | 272,689 | 346,774 | 342,258 | 496,095 |
| 21 | 10 | DM | 35,731 | 223,413 | 389,548 | 209,882 |
| 71 | 40 | DM | 720,638 | 851,150 | 1,647,981 | 1,130,617 |
| Median (range) | 198,644 (35,731-720,638) | 285,093 (15,714-851,149) | 365,903 (216,896-1,647,980) | 376,433 (209,881-1,130,616) | ||
Defined by changes in viral infectivity in CXCR4+ cells after AMD3100 treatment.
The composition of the DM-tropic virus population affects the antiviral activity of AMD3100 in vivo.
To further explore whether the difference in the responses of DM-tropic viruses to AMD3100 treatment was due to different types of CXCR4-using variants (for example, X4 tropic or dualtropic) in the DM-tropic virus populations, individual env clones were isolated from baseline and treated samples of all 14 DM subjects, and the ability of individual clones to infect CCR5 or CXCR4 target cells was assessed. For each subject, approximately 20 to 38 functional clones were analyzed, and the percentage of env clones exhibiting R5, X4, or dual tropism in the virus population was determined (Table 2). All baseline samples from the 10 subjects in the X4 suppressor group contained R5-tropic clones mixed with X4-tropic and/or dualtropic clones. In contrast, only one of four baseline samples from the X4 nonsuppressor group had a mixture of R5-tropic and dualtropic clones. The remaining baseline X4 nonsuppressor samples contained only dualtropic clones (n = 2) or a mixture of dualtropic and X4-tropic clones (n = 1). Baseline virus populations in the X4 suppressor group tended to have more R5-tropic clones (range, 52 to 95%) and fewer CXCR4-using clones (range, 5 to 48%). In contrast, those in the X4 nonsuppressor group had markedly fewer R5-tropic clones (range, 0 to 8%) and more CXCR4-using clones (range, 92 to 100%). Thus, the proportion of CXCR4-using clones isolated from the virus populations at baseline differed greatly between these two groups, with predominantly dualtropic variants in X4 nonsuppressors.
TABLE 2.
env clonal composition of DM subjects at baseline and day 11
| Groupa and subject | Tropism (env pools), day 11 | % of clones that were:
|
|||||
|---|---|---|---|---|---|---|---|
| R5 tropic
|
X4 tropic
|
Dualtropic
|
|||||
| Baseline | Day 11 | Baseline | Day 11 | Baseline | Day 11 | ||
| X4 suppressor | |||||||
| 74 | R5 | 95 | 100 | 0 | 0 | 5 | 0 |
| 31 | R5 | 92 | 100 | 4 | 0 | 4 | 0 |
| 9 | R5 | 93 | 98 | 0 | 0 | 7 | 2 |
| 32 | R5 | 80 | 94 | 0 | 0 | 20 | 6 |
| 28 | R5 | 72 | 100 | 14 | 0 | 14 | 0 |
| 35 | R5 | 61 | 100 | 39 | 0 | 0 | 0 |
| 73 | R5 | 82 | 100 | 3 | 0 | 15 | 0 |
| 11 | DM | 52 | 97 | 0 | 0 | 48 | 3 |
| 10 | R5 | 65 | 98 | 15 | 0 | 20 | 2 |
| 33 | DM | 57 | 85 | 8 | 0 | 35 | 15 |
| X4 nonsuppressor | |||||||
| 70 | DM | 0 | 0 | 0 | 0 | 100 | 100 |
| 2 | DM | 8 | 10 | 0 | 0 | 92 | 90 |
| 21 | DM | 0 | 0 | 58 | 0 | 42 | 100 |
| 71 | DM | 0 | 0 | 0 | 0 | 100 | 100 |
Defined by changes in viral infectivity in CXCR4+ cells after AMD3100 treatment.
Following 10 days of therapy with AMD3100, CXCR4-using clones in the virus populations from the X4 suppressor group at baseline were either undetectable or their numbers were greatly reduced, and there was a concomitant increase in the proportion of R5-tropic clones. In contrast, the relative proportions of CXCR4-using clones (predominantly dualtropic clones) and R5-tropic clones in X4 nonsuppressors showed no change from those at baseline. Interestingly, the X4-tropic clones identified in the baseline samples of seven subjects were suppressed by AMD3100, regardless of whether they were present in X4 suppressors or X4 nonsuppressors. However, the ability of AMD3100 to suppress dualtropic clones differed between the two groups. For all 10 subjects in the X4 suppressor group, disappearance of dualtropic clones or a reduction in their proportion was observed at day 11. Conversely, for all four subjects in the X4 nonsuppressor group, no reduction in the relative proportion of dualtropic clones at day 11 was found. Furthermore, only X4-tropic clones, not dualtropic clones, were suppressed at day 11 in a single subject, subject 21, who had a mixture of X4-tropic and dualtropic clones at baseline.
Taken together, these data show that AMD3100 can suppress X4-tropic variants in all subjects tested here and that the ability to suppress dualtropic variants in virus populations was associated with the relative proportions of R5-tropic and dualtropic variants present in baseline virus populations.
Reemergence of CXCR4-using viruses after AMD3100 treatment.
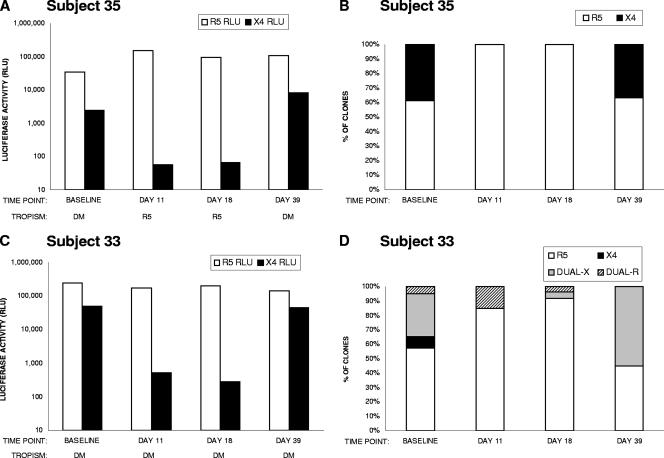
Two subjects (subjects 33 and 35) in the X4 suppressor group who received 40 μg/kg/h of AMD3100 had additional posttreatment plasma samples available (days 18 and 39) for coreceptor tropism analysis of env populations and clones. The virus population of subject 35 comprised R5-tropic (39%) and X4-tropic (61%) clones at baseline, R5-tropic clones only at days 11 and 18, and a mixture of R5-tropic (63%) and X4-tropic (37%) clones at day 39 (Fig. 1A and B). The virus population of subject 33 comprised R5-tropic (57%), X4-tropic (8%), and dualtropic (35%) clones at baseline. On days 11 and 18, the infectivity of the virus population in CXCR4-expressing cells was reduced more than 50-fold from that at baseline, and no X4-tropic clones were detected. The percentages of R5-tropic clones increased to 85% and 92%, and those of dualtropic clones decreased to 15% and 8%, respectively, at days 11 and 18. On day 39, the infectivity of the virus population in CXCR4-expressing cells was comparable to that of the baseline sample. The proportion of dualtropic clones increased to 55%, and that of R5-tropic clones decreased to 45% (Fig. 1C and D).
FIG. 1.
Analysis of tropism of viruses from subjects 35 and 33 before, during, and after treatment with AMD3100. (A and C) The coreceptor tropism of the virus population at baseline and on days 11, 18, and 39 was determined using Trofile. Infectivity in CD4+ CCR5+ (open bars) and CD4+ CXCR4+ (solid bars) U87 cells, as measured by luciferase activity (RLU) in a single measurement, is shown. (B and D) Proportions of clones with the indicated tropisms (R5, X4, dual-X, and dual-R), represented as percentages of the total number of functional clones for each time point, are shown. Open bars, R5-tropic clones; solid bars, X4-tropic clones; shaded bars, dual-X-tropic clones; hatched bars, dual-R-tropic clones.
The suppression of dualtropic variants in a population is associated with the infectivity of individual variants in CXCR4 target cells.
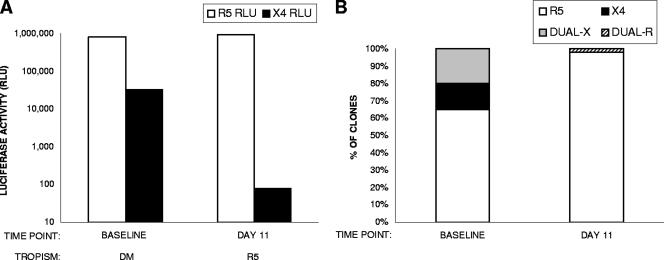
It should be noted that in subject 33, all dualtropic clones detected at day 11 were phenotypically distinct from the majority of dualtropic clones detected in the paired baseline sample. We have previously described the existence of dualtropic viruses that have differing abilities to use CXCR4 and CCR5. Dual-X-tropic viruses use CXCR4 efficiently and have V3 sequences distinct from those of R5-tropic viruses from the same patient (X4-like), while dual-R-tropic viruses use CXCR4 poorly and have V3 sequences that are similar or identical to those of R5-tropic viruses from the same patient (R5-like) (10). In addition to the presence of R5-tropic clones at all time points, subject 33 had a baseline virus population composed of X4-tropic (8%), dual-X-tropic (30%), and dual-R-tropic (5%) clones. Under drug pressure, only dual-R-tropic clones (15%), not X4-tropic or dual-X-tropic clones, were detected at day 11. The dual-X-tropic clones reemerged following cessation of AMD3100 treatment: 4% dual-X-tropic clones were detected at day 18, and this proportion increased to 55% by day 39 (Fig. 1D). Consistent with our previous observations, the average luciferase readout in CXCR4-expressing cells was 106 RLU for dual-X-tropic clones and 103 RLU for dual-R-tropic clones (Table 3). Dual-R-tropic clones were also observed in one other X4 suppressor, subject 10 (Fig. 2). In this subject, the baseline virus population comprised R5-tropic (65%), X4-tropic (15%), and dual-X-tropic (20%) clones. The virus population at day 11 was essentially R5 tropic, with 2% dual-R-tropic clones, but no dual-X-tropic or X4-tropic clones were identified. Although limited and preliminary, the data from these two cases suggest that AMD3100 can suppress X4-tropic and dual-X-tropic variants but not dual-R-tropic variants.
TABLE 3.
Coreceptor usage and V3 sequence analysis of env clones from subjects 33 and 35 at baseline and days 11, 18, and 39
| Subject and time point | No. of clones | Tropism | Avg RLU
|
V3 loop amino acid sequence | |
|---|---|---|---|---|---|
| R5 | X4 | ||||
| Subject 35 | |||||
| Baseline | 6 | R5 | 469,080 | 84 | CTRPSNNTRKSINMGPGRAFYTTGEIIGDIRQAHC |
| 1 | R5 | 366,742 | 113 | ............................N...... | |
| 4 | X4 | 82 | 355,190 | ......H...RVTL..S.VY......T....R... | |
| 1 | X4 | 71 | 388,674 | ....N.H...RVTL..S.VY......T....R... | |
| Day 11 | 10 | R5 | 201,554 | 72 | ................................... |
| Day 18 | 10 | R5 | 179,136 | 79 | ................................... |
| Day 39 | 5 | R5 | 248,231 | 85 | ................................... |
| 5 | X4 | 75 | 252,333 | ......H...RVTL..S.VY......T....R... | |
| Subject 33 | |||||
| Baseline | 7 | R5 | 1,016,370 | 104 | CTRPNNNTRKSINIGPGRAWYATGQIIGDIRQAYC |
| 2 | Dual-R | 1,858,036 | 307 | ................................... | |
| 5 | Dual-X | 305,294 | 1,730,180 | ..........RVTM...KV..T.........K... | |
| 1 | X4 | 111 | 1,679,250 | ..........RVTM...KV.........G...... | |
| Day 11 | 8 | R5 | 1,097,747 | 110 | ................................... |
| 4 | Dual-R | 1,310,893 | 1,368 | ................................... | |
| Day 18 | 8 | R5 | 1,856,059 | 172 | ................................... |
| 1 | Dual-R | 2,278,396 | 258 | ................................... | |
| 1 | Dual-X | 97,013 | 1,345,964 | ..........RVTM...KV..T.........K... | |
| Day 39 | 8 | R5 | 1,487,627 | 114 | ................................... |
| 6 | Dual-X | 302,686 | 1,899,393 | ..........RVTM...KV..T.........K... | |
| 1 | Dual-X | 739,163 | 170,982 | ..........GVTM...KV..T.........K... | |
FIG. 2.
Analysis of tropism of viruses from subject 10 before and after treatment with AMD3100. (A) The coreceptor tropism of the virus population at baseline and day 11 was determined using Trofile. Infectivity in CD4+ CCR5+ (open bars) and CD4+ CXCR4+ (solid bars) U87 cells, as measured by luciferase activity (RLU) in a single measurement, is shown. (B) Proportions of clones with the indicated tropisms (R5, X4, dual-X, and dual-R), represented as percentages of the total number of functional clones isolated at each time point, are shown. Open bars, R5-tropic clones; solid bars, X4-tropic clones; shaded bars, dual-X-tropic clones; hatched bars, dual-R-tropic clones.
Phylogenetic analysis of gp160 and V3 sequences.
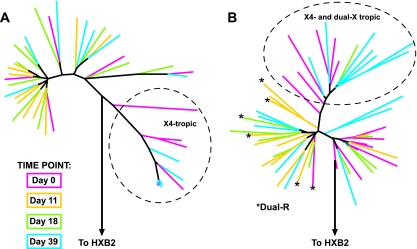
Ten to 15 gp160 env clones obtained from subjects 33 and 35 at each time point were sequenced, and phylogenetic analysis was performed (Fig. 3). X4-tropic and dual-X-tropic clones clustered together and were distinct from R5-tropic clones from the same subject, regardless of drug exposure. This suggests that X4-tropic and dual-X-tropic clones are genetically distinct from R5-tropic clones and that reemerging CXCR4-using clones, following treatment with AMD3100, were similar to CXCR4-using clones present at baseline. The dual-R-tropic clones that were not suppressed by AMD3100 in subject 33 clustered more closely with R5-tropic clones than with dual-X-tropic or X4-tropic clones. Furthermore, the V3 sequences of the dual-R-tropic clones were indistinguishable from those of R5-tropic clones (Table 3). In addition, the V3 loop amino acid sequences of X4-tropic or dual-X-tropic clones that reemerged posttreatment were the same as those isolated at baseline in both subjects, as were the V3 loop amino acid sequences of R5-tropic clones isolated pre- and post-drug exposure (Table 3).
FIG. 3.
Phylogenetic analysis of gp160 env clones isolated at baseline and on days 11, 18, and 39 for subjects 35 and 33. Phylogenetic trees show the association of the time point with the coreceptor tropism at baseline and on days 11, 18, and 39 for subjects 35 (A) and 33 (B). Colored lines represent env clones isolated at baseline (pink), day 11 (gold), day 18 (green), and day 39 (blue). Clones with X4 or dual-X tropism are shown within dashed circles. Asterisks indicate clones with dual-R tropism.
DISCUSSION
In this study, we retrospectively analyzed the coreceptor tropism of 14 DM-tropic subjects enrolled in a phase I/II study of the CXCR4 antagonist AMD3100. Other subjects enrolled in this phase I/II study harbored X4- or R5-tropic viruses at baseline and did not demonstrate tropism changes following AMD3100 treatment. Following 10 days of AMD3100 therapy, DM-tropic viruses from 10 subjects either became R5 tropic or exhibited a reduction of infectivity (>50-fold) in CXCR4-expressing cells (X4 suppressors), while the viruses from 4 subjects remained DM tropic and showed no reduction of infectivity in CXCR4-expressing cells (X4 nonsuppressors). The X4 suppressors also displayed lower levels of infectivity in CXCR4-expressing cells at baseline than the X4 nonsuppressors. env clonal analysis indicated that X4 suppressors had a much higher proportion of R5-tropic clones and a lower proportion of dualtropic clones than X4 nonsuppressors.
Following 10 days of AMD3100 treatment, all X4-tropic clones were suppressed in the virus populations studied, regardless of X4 suppressor or nonsuppressor classification. However, differences in the extent of suppression of dualtropic clones were observed. We found that AMD3100 suppresses dualtropic viruses when the virus population is predominantly R5 tropic. We hypothesize that in the absence of an R5-tropic virus population, dualtropic viruses escape AMD3100 suppression by using the CCR5 coreceptor (X4 nonsuppressors). Conversely, in mixed populations where R5-tropic viruses predominate, dualtropic viruses may compete less well than R5-tropic viruses for the CCR5 coreceptor and consequently are effectively suppressed by AMD3100 (X4 suppressors).
In addition to the composition of the virus population, the suppression of dualtropic variants is also dependent on the type of dualtropic variant present. We have previously described the existence of two distinct types of dualtropic viruses that differ in their ability to use the CXCR4 and CCR5 coreceptors in vitro (10). The preferential use of CCR5 (dual-R) or the ability to utilize both coreceptors efficiently (dual-X) could affect the efficacy of CCR5 and CXCR4 coreceptor inhibitors. The present study demonstrates that dual-X-tropic variants, which efficiently mediated infection in both CXCR4- and CCR5-expressing cells in vitro, could be suppressed by AMD3100 in X4 suppressors for whom R5-tropic variants predominated, while dual-R-tropic variants, which use CCR5 efficiently and CXCR4 inefficiently, could not. We hypothesize that dual-R-tropic viruses preferentially use CCR5, while dual-X-tropic viruses use both CXCR4 and CCR5 efficiently but are unable to compete effectively with R5-tropic viruses in the presence of CXCR4 drug pressure. Further studies are under way to investigate the relationship between coreceptor usage by different dualtropic variants and inhibition of these variants by CXCR4 and CCR5 inhibitors.
Using currently available technologies, it is impossible to define the composition of a virus population without performing clonal analysis. DM-tropic virus populations that exhibit low levels of infectivity in CXCR4 target cells may contain either a minor virus population that uses CXCR4 efficiently (X4 or dual-X tropic) or many dualtropic variants that use CXCR4 poorly (dual-R tropic). Thus, it cannot be assumed that virus populations with low levels of infectivity in CXCR4-expressing cells harbor CXCR4-using viruses that will respond to a CXCR4 antagonist.
From this study it appears that suppression of CXCR4-using variants may or may not be associated with an overall reduction in viral load. In this trial, a single subject with an X4-tropic virus did exhibit a 0.9-log10-unit reduction in viral load after 10 days of treatment with AMD3100 (9). However, no significant reductions in viral load were observed in any of the 14 subjects with DM-tropic virus, including subjects with undetectable levels of CXCR4-using variants at day 11. In X4 nonsuppressors, the majority of the virus populations comprised dualtropic variants that most likely were able to use CCR5 when under CXCR4 antagonist pressure, and thus, viral loads were not suppressed by AMD3100. In X4 suppressors, the suppression of CXCR4-using variants (dualtropic and/or X4 tropic) may have been accompanied by an increase in the proportion of R5-tropic variants, which could explain the lack of a measurable viral load reduction. However, if the population of R5-tropic variants remains unchanged, the lack of a measurable reduction in viral load may simply reflect the small proportion of CXCR4-using viruses in the population. For example, in order to realize a 0.3-log10-unit reduction in viral load, 50% of the virus population would need to be suppressed.
Regardless of whether or not viral load reductions are observed, successful suppression of CXCR4-using variants by a CXCR4 inhibitor may be an important determinant of clinical outcome. In addition, CXCR4 utilization may serve as a useful screening criterion for enrollment in clinical studies of CXCR4 inhibitors. One could envision enrolling subjects with baseline virus populations comprising CXCR4-using variants (X4 tropic or dualtropic) into trials involving an inhibitor of CXCR4, and using the reduction of CXCR4-using variants as a measure of response to inhibitor treatment (G. Moyle et al., presented at the 14th Conference on Retroviruses and Opportunistic Infections, Los Angeles, CA, 2007 [abstr. 511]). This approach could be used to determine whether CXCR4-using variants are effectively suppressed by treatment, independent of a reduction in viral load or the complete suppression of infectivity in CXCR4-expressing cells.
In this study, dual-R-tropic variants that use CCR5 efficiently and CXCR4 inefficiently were not suppressed by AMD3100. It is possible that subjects with this type of dualtropic variant may benefit from treatment with a CCR5 inhibitor. In this regard, a similar analysis of subjects with dualtropic viruses receiving a CCR5 antagonist would be highly informative (such as the phase 2b study of maraviroc for patients infected with non-CCR5-tropic HIV-1 [H. Mayer et al., presented at the XVI International AIDS Conference, Toronto, Ontario, Canada, 13 to 18 August 2006 {abstr. THLB0215}]). Further analyses of the coreceptor tropism of HIV-1 variants before and after treatment with coreceptor inhibitors are likely to provide valuable information that will advance our understanding of this important drug class and its clinical utility.
Acknowledgments
We are grateful to the Monogram Biosciences Clinical Reference Laboratory for performance of Trofile assays. We thank Cynthia Sedik for editorial assistance.
Trofile assay development was supported in part by NIH-NIAID SBIR grant R44-AI048990.
Footnotes
Published ahead of print on 28 April 2008.
REFERENCES
- 1.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 2.Bridger, G. J., R. T. Skerlj, S. Padmanabhan, S. A. Martellucci, G. W. Henson, S. Struyf, M. Witvrouw, D. Schols, and E. De Clercq. 1999. Synthesis and structure-activity relationships of phenylenebis(methylene)-linked bis-azamacrocycles that inhibit HIV-1 and HIV-2 replication by antagonism of the chemokine receptor CXCR4. J. Med. Chem. 42:3971-3981. [DOI] [PubMed] [Google Scholar]
- 3.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 4.Donzella, G. A., D. Schols, S. W. Lin, J. A. Este, K. A. Nagashima, P. J. Maddon, G. P. Allaway, T. P. Sakmar, G. Henson, E. De Clercq, and J. P. Moore. 1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 4:72-77. [DOI] [PubMed] [Google Scholar]
- 5.Dorr, P., M. Westby, S. Dobbs, P. Griffin, B. Irvine, M. Macartney, J. Mori, G. Rickett, C. Smith-Burchnell, C. Napier, R. Webster, D. Armour, D. Price, B. Stammen, A. Wood, and M. Perros. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49:4721-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 7.Gerlach, L. O., R. T. Skerlj, G. J. Bridger, and T. W. Schwartz. 2001. Molecular interactions of cyclam and bicyclam non-peptide antagonists with the CXCR4 chemokine receptor. J. Biol. Chem. 276:14153-14160. [DOI] [PubMed] [Google Scholar]
- 8.Hatse, S., K. Princen, L. O. Gerlach, G. Bridger, G. Henson, E. De Clercq, T. W. Schwartz, and D. Schols. 2001. Mutation of Asp(171) and Asp(262) of the chemokine receptor CXCR4 impairs its coreceptor function for human immunodeficiency virus-1 entry and abrogates the antagonistic activity of AMD3100. Mol. Pharmacol. 60:164-173. [DOI] [PubMed] [Google Scholar]
- 9.Hendrix, C. W., A. C. Collier, M. M. Lederman, D. Schols, R. B. Pollard, S. Brown, J. B. Jackson, R. W. Coombs, M. J. Glesby, C. W. Flexner, G. J. Bridger, K. Badel, R. T. MacFarland, G. W. Henson, and G. Calandra. 2004. Safety, pharmacokinetics, and antiviral activity of AMD3100, a selective CXCR4 receptor inhibitor, in HIV-1 infection. J. Acquir. Immune Defic. Syndr. 37:1253-1262. [DOI] [PubMed] [Google Scholar]
- 10.Huang, W., S. H. Eshleman, J. Toma, S. Fransen, E. Stawiski, E. E. Paxinos, J. M. Whitcomb, A. M. Young, D. Donnell, F. Mmiro, P. Musoke, L. A. Guay, J. B. Jackson, N. T. Parkin, and C. J. Petropoulos. 2007. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J. Virol. 81:7885-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobson, J. M., R. J. Israel, I. Lowy, N. A. Ostrow, L. S. Vassilatos, M. Barish, D. N. Tran, B. M. Sullivan, T. J. Ketas, T. J. O'Neill, K. A. Nagashima, W. Huang, C. J. Petropoulos, J. P. Moore, P. J. Maddon, and W. C. Olson. 2004. Treatment of advanced human immunodeficiency virus type 1 disease with the viral entry inhibitor PRO 542. Antimicrob. Agents Chemother. 48:423-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jekle, A., O. T. Keppler, E. De Clercq, D. Schols, M. Weinstein, and M. A. Goldsmith. 2003. In vivo evolution of human immunodeficiency virus type 1 toward increased pathogenicity through CXCR4-mediated killing of uninfected CD4 T cells. J. Virol. 77:5846-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koot, M., I. P. Keet, A. H. Vos, R. E. de Goede, M. T. Roos, R. A. Coutinho, F. Miedema, P. T. Schellekens, and M. Tersmette. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118:681-688. [DOI] [PubMed] [Google Scholar]
- 14.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 15.Kuritzkes, D. R., J. Jacobson, W. G. Powderly, E. Godofsky, E. DeJesus, F. Haas, K. A. Reimann, J. L. Larson, P. O. Yarbough, V. Curt, and W. R. Shanahan, Jr. 2004. Antiretroviral activity of the anti-CD4 monoclonal antibody TNX-355 in patients infected with HIV type 1. J. Infect. Dis. 189:286-291. [DOI] [PubMed] [Google Scholar]
- 16.Melby, T., M. Despirito, R. Demasi, G. Heilek-Snyder, M. L. Greenberg, and N. Graham. 2006. HIV-1 coreceptor use in triple-class treatment-experienced patients: baseline prevalence, correlates, and relationship to enfuvirtide response. J. Infect. Dis. 194:238-246. [DOI] [PubMed] [Google Scholar]
- 17.Moyle, G. J., A. Wildfire, S. Mandalia, H. Mayer, J. Goodrich, J. Whitcomb, and B. G. Gazzard. 2005. Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J. Infect. Dis. 191:866-872. [DOI] [PubMed] [Google Scholar]
- 18.Philpott, S. M. 2003. HIV-1 coreceptor usage, transmission, and disease progression. Curr. HIV Res. 1:217-227. [DOI] [PubMed] [Google Scholar]
- 19.Princen, K., S. Hatse, K. Vermeire, E. De Clercq, and D. Schols. 2004. Establishment of a novel CCR5 and CXCR4 expressing CD4+ cell line which is highly sensitive to HIV and suitable for high-throughput evaluation of CCR5 and CXCR4 antagonists. Retrovirology 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuitemaker, H., N. A. Kootstra, R. E. de Goede, F. de Wolf, F. Miedema, and M. Tersmette. 1991. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J. Virol. 65:356-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strizki, J. M., S. Xu, N. E. Wagner, L. Wojcik, J. Liu, Y. Hou, M. Endres, A. Palani, S. Shapiro, J. W. Clader, W. J. Greenlee, J. R. Tagat, S. McCombie, K. Cox, A. B. Fawzi, C. C. Chou, C. Pugliese-Sivo, L. Davies, M. E. Moreno, D. D. Ho, A. Trkola, C. A. Stoddart, J. P. Moore, G. R. Reyes, and B. M. Baroudy. 2001. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. USA 98:12718-12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westby, M., M. Lewis, J. Whitcomb, M. Youle, A. L. Pozniak, I. T. James, T. M. Jenkins, M. Perros, and E. van der Ryst. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80:4909-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitcomb, J. M., W. Huang, S. Fransen, K. Limoli, J. Toma, T. Wrin, C. Chappey, L. D. Kiss, E. E. Paxinos, and C. J. Petropoulos. 2007. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob. Agents Chemother. 51:566-575. [DOI] [PMC free article] [PubMed] [Google Scholar]