Abstract
Self-transferable IncFI plasmid pIP1206, isolated from an Escherichia coli clinical isolate, carries two new resistance determinants: qepA, which confers resistance to hydrophylic fluoroquinolones by efflux, and rmtB, which specifies a 16S rRNA methylase conferring high-level aminoglycoside resistance. Analysis of the 168,113-bp sequence (51% G+C) revealed that pIP1206 was composed of several subregions separated by copies of insertion sequences. Of 151 open reading frames, 56 (37%) were also present in pRSB107, isolated from a bacterium in a sewage treatment plant. pIP1206 contained four replication regions (RepFIA, RepFIB, and two partial RepFII regions) and a transfer region 91% identical with that of pAPEC-O1-ColBM, a plasmid isolated from an avian pathogenic E. coli. A putative oriT region was found upstream from the transfer region. The antibiotic resistance genes tet(A), catA1, blaTEM-1, rmtB, and qepA were clustered in a 33.5-kb fragment delineated by two IS26 elements that also carried a class 1 integron, including the sulI, qacEΔ1, aad4, and dfrA17 genes and Tn10, Tn21, and Tn3-like transposons. The plasmid also possessed a raffinose operon, an arginine deiminase pathway, a putative iron acquisition gene cluster, an S-methylmethionine metabolism operon, two virulence-associated genes, and a type I DNA restriction-modification (R-M) system. Three toxin/antitoxin systems and the R-M system ensured stabilization of the plasmid in the host bacteria. These data suggest that the mosaic structure of pIP1206 could have resulted from recombination between pRSB107 and a pAPEC-O1-ColBM-like plasmid, combined with structural rearrangements associated with acquisition of additional DNA by recombination and of mobile genetic elements by transposition.
Plasmids are widespread in bacteria, where they represent an important part of the genome. Among these, conjugative plasmids are essential contributors to horizontal transfer of genetic information and to bacterial genome evolution. In addition, exchange of genetic material is enhanced by mobile genetic elements, such as transposons and insertion sequences (IS). Plasmids have been classified into incompatibility (Inc) groups on the basis of the impossibility for two plasmids to coexist stably within the same host (11, 14). Plasmids of the IncF group are considered of narrow host range. F-like plasmids are often (i) large molecules, from 40 to 200 kb that can harbor two or three sites for DNA replication initiation, (ii) conjugative, and (iii) composed of genes implicated in antibiotic resistance, virulence, and metabolic biochemical pathways. F-like plasmids are common in nature and have been found in 15% of an Escherichia coli collection containing 72 strains (10). IncFI self-transferable plasmid pIP1206 was detected in 2006 in E. coli 1540 during the screening of high-level aminoglycoside-resistant enterobacteria (8). pIP1206 contains the catI, tet(A), dfrA17, sulI, qacEΔ1, aadA4, and blaTEM-1 resistance genes that confer, respectively, resistance to chloramphenicol, tetracycline, trimethoprim, sulfonamides, quaternary ammonium compounds, streptomycin/spectinomycin, and β-lactams. Furthermore, pIP1206 carries genes responsible for two antibiotic resistance mechanisms recently detected in human pathogens: rmtB, which confers high-level resistance to 4,6-disubstituted deoxystreptamines by methylation at N-7 of residue G1405 of 16S rRNA (43), and qepA, which is responsible for diminished susceptibility to hydrophilic fluoroquinolones by synthesis of an efflux pump belonging to the major facilitator superfamily (MFS) (43).
Association of these two new genes with other antibiotic resistance determinants on the same plasmid is of concern, in particular because of the risk of dissemination due to coselection by various drugs. The aim of this work was to determine the genomic structure of pIP1206. This large plasmid presents similarities to pRSB107, originating from a bacterium in a sewage treatment plant (47), and pAPEC-O1-ColBM (24), isolated from an avian pathogenic E. coli, and it appears to be composed of DNA that has been acquired from various sources, probably following homologous recombination and transposition events.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strain 1540 was isolated from a patient in a Belgian hospital during the screening for aminoglycoside resistance of clinical isolates of Enterobacteriaceae (8). E. coli C600Rif, a rifampin-resistant derivative of C600, Acinetobacter sp. strain R1 (20), and Pseudomonas aeruginosa PAO38 (21) were used as recipients in conjugation experiments. The strains were grown in brain heart infusion broth or on agar at 37°C.
Filter mating.
Conjugation was performed by the solid mating-out assay with selection on rifampin (250 μg/ml) and kanamycin (40 μg/ml) for E. coli C600Rif and Acinetobacter sp. strain R1 or on rifampin (32 μg/ml) and kanamycin (250 μg/ml) for P. aeruginosa PAO38.
DNA sequencing.
Plasmid pIP1206 DNA was prepared by the alkaline lysis procedure (7). The sequence was determined by the shotgun cloning method (MWG Biotech, Champlan, France). The pGEM-Teasy vector was electroporated into E. coli DH10B, which is T1 phage resistant. Primers used for DNA sequencing were easyR, 5′-CAGGCGGCCGCGAATTCAC-3′, and easyF, 5′-TGGCGGCCGCGGGAATCCG-3′.
Sequence analysis.
Contig assembly and annotation were performed using the Contigs-Assembly and Annotation Tool-Box (CAAT-Box) software (16) developed at the Genopole of Institut Pasteur for the computational analysis of sequences obtained by the shotgun method, such as prediction of links between contigs as well as the annotation of the sequence. CAAT-Box was also used to create the EMBL file.
Nucleotide sequence accession number.
The sequence of pIP1206 was submitted to the EMBL database and assigned accession number AM886293.
RESULTS AND DISCUSSION
General structure of pIP1206.
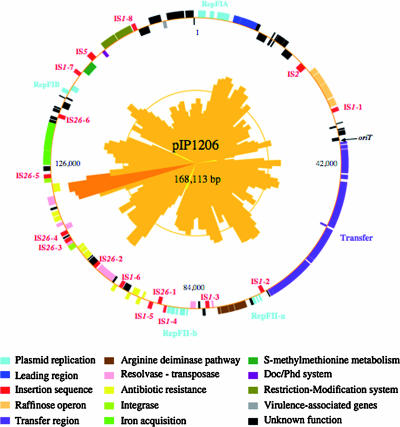
The sequence of pIP1206, isolated from E. coli 1540, was determined by a shotgun approach. The plasmid was found to be a circular molecule of 168,113 bp (Fig. 1) with a G+C content of 51%, ranging from 30% to 72%. Annotation of the data using the CAAT-Box tool (16) resulted in the detection of 176 coding sequences. Among these, 107 were assigned to six functions: replication, transfer, antibiotic resistance, biochemical pathways, restriction-modification (R-M), and virulence, whereas 44 encoded hypothetical proteins (Fig. 1 and Table 1). Of these 151 open reading frames (ORFs), 56 (37%) were also present in pRSB107, which was isolated from a bacterium in a sewage treatment plant (47), 44 (29%) in pAPEC-O1-ColBM (24), which was isolated from an avian pathogenic E. coli, 11 (7%) in both plasmids, and 40 (27%) from various sources. Furthermore, 25 coding sequences as part of 16 ISs, in particular IS26 and IS1, were found throughout the plasmid. It is noteworthy that (i) some sets of genes in pRSB107 are not present in pIP1206 and (ii) an inversion occurred that resulted in the presence of a Tn21-like sequence in both pRSB107 and pIP1206 but in the opposite orientation. Furthermore, it is likely that the IS copies also are responsible for inversion events. It thus appears that pIP1206 could result from acquisition by a pRSB107-like plasmid, which differs from prototype pRSB107 following deletions and inversions of several determinants by transpositions mediated by insertion sequences or transposons and of part of pAPEC-O1-ColBM-like plasmid, such as the entire transfer region, by homologous recombination.
FIG. 1.
Graphical map of pIP1206. The color code for the various gene functions is indicated below the map. The origin of transfer (oriT) is marked by a black arrow. The G+C plot is indicated on the inner circle (mean, 51%). G+C contents that were >2 standard deviations above and below the mean are indicated in dark orange and in yellow, respectively.
TABLE 1.
ORFs identified in pIP1206
| ORF | Gene | G+C (%) | Function | Identity (%) | Pfama | Accession no. | Organism (plasmid) |
|---|---|---|---|---|---|---|---|
| oriV-1 region | 86 | AJ851089 | Uncult.b (pRSB107) | ||||
| 1 | ccdA | 51 | Antitoxin | 100 | 07362 | AJ851089 | Uncult. (pRSB107) |
| 2 | ccdB | 50 | Toxin | 100 | 01845 | AJ851089 | Uncult. (pRSB107) |
| 3 | resD | 57 | Resolvase | 100 | 00589 | AJ851089 | Uncult. (pRSB107) |
| 4 | repE | 47 | Replication protein (RepE) | 100 | 01051 | AJ851089 | Uncult. (pRSB107) |
| 5 | sopA | 47 | Partitioning protein | 100 | 01656 | AJ851089 | Uncult. (pRSB107) |
| 6 | sopB | 45 | Partitioning protein | 100 | 02195 | AJ851089 | Uncult. (pRSB107) |
| 7 | orf7 | 55 | Hypothetical protein | 100 | 06924 | AJ851089 | Uncult. (pRSB107) |
| 8 | orf8 | 63 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 9 | orf9 | 56 | DNA methylase | 100 | 01555 | AJ851089 | Uncult. (pRSB107) |
| 10 | orf10 | 53 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 11 | orf11 | 57 | Hypothetical protein | 100 | 07128 | AJ851089 | Uncult. (pRSB107) |
| 12 | orf12 | 60 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 13 | orf13 | 63 | Hypothetical protein | 98 | 07339 | NC_002134 | E. coli (pR100) |
| 14 | klcA | 55 | Hypothetical antirestriction | 100 | 03230 | AJ851089 | Uncult. (pRSB107) |
| 15 | orf15 | 56 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 16 | orf16 | 51 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 17 | orf17 | 49 | RNA-directed DNA polymerase | 62 | 08388 | NC_008322 | Shewanella sp. |
| 18 | ydaA | 68 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 19 | orf19 | 61 | Hypothetical protein | 97 | NC_006671 | E. coli (pAPEC-O2-R) | |
| 20 | orf20 | 58 | Hypothetical protein | 100 | NC_002134 | E. coli (pR100) | |
| 21 | orf21 | 61 | Hypothetical protein | 98 | NZ_AAJX01000096 | E. coli | |
| 22 | ydbA | 56 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 23 | orf23 | 60 | Hypothetical DNA methylase | 96 | NC_006671 | E. coli (pAPEC-O2-R) | |
| 24 | orf24 | 61 | Hypothetical protein | 100 | NC_006671 | E. coli (pAPEC-O2-R) | |
| 25 | orf25 | 49 | RNA-directed DNA polymerization | 62 | 08388 | NC_008322 | Shewanella sp. |
| 26 | IS2-orf2 | 54 | Transposase | 98 | 00665 | AC_000091 | E. coli |
| 27 | IS2-orf1 | 54 | Transposase | 99 | 01527 | AC_000091 | E. coli |
| 28 | rafR | 48 | Transcriptional regulator | 89 | 00356 | M29849 | E. coli (pRSD2) |
| 29 | rafA | 51 | α-Galactosidase | 97 | 02065 | M27273 | E. coli (pRSD2) |
| 30 | rafB | 46 | Raffinose permease | 97 | 01306 | M27273 | E. coli (pRSD2) |
| 31 | rafD | 51 | Raffinose invertase | 96 | 08244 | M27273 | E. coli (pRSD2) |
| 32 | rafY | 39 | Putative glycoporine | 88 | U82290 | E. coli (pRSD2) | |
| 33 | IS1-insA | 52 | Transposase | 97 | 03811 | NC_006816 | Salmonella enterica serovar Typhimurium (pU302L) |
| 34 | IS1-insB | 52 | Transposase | 99 | 03400 | NC_006816 | Serovar Typhimurium (pU302L) |
| 35 | orf35 | 47 | Stable inheritance protein | 30 | 07885 | NC_002483 | E. coli (plasmid F) |
| 36 | yubN | 61 | Hypothetical protein | 90 | NC_002483 | E. coli (plasmid F) | |
| 37 | yubO | 52 | Hypothetical protein | 95 | NC_002483 | E. coli (plasmid F) | |
| 38 | yubP | 44 | Hypothetical protein | 99 | 06067 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 39 | orf39 | 43 | Transglycosylation | 95 | 01464 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 40 | traM | 38 | Mating signal | 99 | 05261 | NC_009602 | E. coli (pSFO157) |
| 41 | traJ | 33 | Regulation | 96 | M62986 | E. coli (IncFI p307) | |
| 42 | traY | 38 | oriT nicking | 95 | M62986 | E. coli (IncFI p307) | |
| 43 | traA | 52 | F pilin subunit | 98 | 05513 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 44 | traL | 47 | F pilus assembly | 100 | 07178 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 45 | traE | 47 | F pilus assembly | 99 | 05309 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 46 | traK | 57 | F pilus assembly | 98 | 06586 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 47 | traB | 57 | F pilus assembly | 99 | 06447 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 48 | traP | 49 | Conjugal transfer protein | 99 | 07296 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 49 | trbG | 42 | 100 | DQ381420 | E. coli (pAPEC-O1-ColBM) | ||
| 50 | traV | 52 | F pilus assembly | 97 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 51 | traC | 53 | F pilus assembly | 98 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 52 | trbI | 58 | 98 | DQ381420 | E. coli (pAPEC-O1-ColBM) | ||
| 53 | traW | 60 | F pilus assembly | 98 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 54 | traU | 55 | F pilus assembly | 97 | 06834 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 55 | trbC | 55 | F pilus assembly | 98 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 56 | traN | 52 | Aggregate stability | 99 | 06986 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 57 | trbE | 41 | 100 | DQ381420 | E. coli (pAPEC-O1-ColBM) | ||
| 58 | traF | 50 | F pilus assembly | 99 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 59 | trbA | 38 | Conjugal transfer protein | 95 | NC_007941 | E. coli (pUTI89) | |
| 60 | artA | 33 | 96 | NC_007941 | E. coli (pUTI89) | ||
| 61 | traQ | 54 | F pilin synthesis | 95 | NC_007941 | E. coli (pUTI89) | |
| 62 | trbB | 57 | 96 | DQ381420 | E. coli (pAPEC-O1-ColBM) | ||
| 63 | trbJ | 47 | Conjugal transfer protein | 69 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 64 | trbF | 38 | 60 | NC_007941 | E. coli (pUTI89) | ||
| 65 | traH | 51 | F pilus assembly | 99 | 06122 | NC_006671 | E. coli (pAPEC-O2-R) |
| 66 | traG | 50 | F pilus assembly | 92 | 07916 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 67 | traS | 30 | Surface exclusion | 54 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 68 | traT | 50 | Surface exclusion | 99 | 05818 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 69 | traD | 52 | DNA transport | 96 | 02534 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 70 | traI | 59 | DNA helicase | 96 | 07057 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 71 | traX | 57 | F pilin acetylation | 97 | 05857 | AJ851089 | Uncult. bact. (pRSB107) |
| 72 | finO | 55 | Fertility inhibition protein | 97 | 05286 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 73 | orf73 | 61 | Hypothetical protein | 92 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 74 | IS1-insB | 52 | Transposase | 99 | 03400 | NC_006816 | Serovar Typhimurium (pU302L) |
| 75 | IS1-insA | 52 | Transposase | 97 | 03811 | NC_006816 | Serovar Typhimurium (pU302L) |
| 76 | srnB | 49 | Postsegregation killing | 96 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 77 | repA2 | 46 | Regulation of RepA1 expression | 84 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 78 | repA6 | 49 | Regulation of RepA1 expression | 83 | AJ851089 | Uncult. (pRSB107) | |
| 79 | ΔrepA1 | 56 | Replication initiation (truncated) | 50 | 02387 | AJ851089 | Uncult. (pRSB107) |
| 80 | orf80 | 50 | Hypothetical protein | ||||
| 81 | arcA | 53 | Arginine deiminase | 95 | 02274 | CP000653 | Enterobacter sp. strain 638 |
| 82 | arcC | 57 | Carbamate kinase | 88 | 00696 | CP000653 | Enterobacter sp. strain 638 |
| 83 | arcB | 53 | Ornithine carbamoyl transferase | 91 | 00185 | CP000653 | Enterobacter sp. strain 638 |
| 84 | arcD | 54 | C-4 dicarboxylate anaerobic carrier | 91 | 03606 | CP000653 | Enterobacter sp. strain 638 |
| 85 | argR | 48 | Arginine repressor | 84 | 01316 | CP000653 | Enterobacter sp. strain 638 |
| 86 | orf86 | 52 | Putative transposase | 98 | 05717 | NC_002655 | E. coli |
| 87 | IS1-insB | 52 | Transposase | 99 | 03400 | NC_006816 | Serovar Typhimurium (pU302L) |
| 88 | IS1-insA | 52 | Transposase | 97 | 03811 | NC_006816 | Serovar Typhimurium (pU302L) |
| 89 | orf89 | 52 | Truncated hypothetical protein | 84 | AY545598 | E. coli (pAPEC-O2-ColV) | |
| 90 | orf90 | 43 | Hypothetical protein | 69 | NC_008253 | E. coli | |
| 91 | orf91 | 49 | Transposase | 95 | 02371 | AF389912 | S. marcescens |
| 92 | repA2 | 47 | Regulation of RepA1 expression | 97 | NC_007635 | E. coli (pCoo) | |
| 93 | repA6 | 49 | Regulation of RepA1 expression | 83 | AJ851089 | Uncult. (pRSB107) | |
| 94 | repA1 | 56 | Replication initiation | 96 | 02387 | AJ851089 | Uncult. (pRSB107) |
| 95 | repA4 | 58 | Replication protein | 99 | V00351 | E. coli (pRSC13) | |
| 96 | tir | 52 | Transfer inhibition | 100 | 02517 | AJ851089 | Uncult. (pRSB107) |
| 97 | pemI | 53 | Plasmid stable inheritance | 100 | 04014 | AJ851089 | Uncult. (pRSB107) |
| 98 | pemK | 55 | Plasmid stable inheritance | 100 | 02452 | AJ851089 | Uncult. (pRSB107) |
| 99 | IS1-insA | 52 | Transposase | 97 | 03811 | NC_006816 | Serovar Typhimurium (pU302L) |
| 100 | ΔIS1-insB | 52 | Transposase (truncated) | 64 | 03400 | NC_006816 | Serovar Typhimurium (pU302L) |
| 101 | IS26-tnpA | 52 | Transposase | 100 | 00665 | NC_006856 | S. enterica |
| 102 | IS1-insA | 52 | Transposase | 97 | 03811 | NC_006816 | Serovar Typhimurium (pU302L) |
| 103 | IS1-insB | 52 | Transposase | 99 | 03400 | NC_006816 | Serovar Typhimurium (pU302L) |
| 104 | tetD | 35 | Tetracycline resistance | 100 | 00165 | AJ851089 | Uncult. (pRSB107) |
| 105 | tetC | 32 | Tetracycline transcriptional regulator | 100 | 00440 | AJ851089 | Uncult. (pRSB107) |
| 106 | tetA | 43 | MFS efflux pump | 99 | 07690 | AJ851089 | Uncult. (pRSB107) |
| 107 | tetR | 40 | Tetracycline repressor protein | 100 | 02909 | AJ851089 | Uncult. (pRSB107) |
| 108 | jemC | 41 | Transcriptional regulator | 100 | 01022 | AJ851089 | Uncult. (pRSB107) |
| 109 | orf109 | 37 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 110 | IS1-insB | 52 | Transposase | 99 | 03400 | NC_006816 | Serovar Typhimurium (pU302L) |
| 111 | IS1-insA | 52 | Transposase | 97 | 03811 | NC_006816 | Serovar Typhimurium (pU302L) |
| 112 | catA | 44 | Chloramphenicol resistance | 100 | 00302 | AP000342 | E. coli (pR100) |
| 113 | ybjA | 48 | Acetyltransferase | 100 | 00583 | AP000342 | E. coli (pR100) |
| 114 | tnpA-Tn21 | 63 | Transposase | 100 | 01526 | AP000342 | E. coli (pR100) |
| 115 | tnpR-Tn21 | 57 | Resolvase | 100 | 02796 | AP000342 | E. coli (pR100) |
| 116 | IS26-tnpA | 52 | Transposase | 100 | 00665 | NC_006856 | S. enterica |
| 117 | ΔchrA | 58 | Chromate ion transport (truncated) | 100 | 02417 | AJ698325 | Uncult. (pRSB101) |
| 118 | orf118 | 62 | Hypothetical protein | 100 | DQ390454 | E. coli (pLEW517) | |
| 119 | sulI | 62 | Sulfonamide resistance | 99 | 00809 | AP000342 | E. coli (pR100) |
| 120 | qacEΔ1 | 50 | Quaternary ammonium compound resistance | 100 | 00893 | AP000342 | E. coli (pR100) |
| 121 | aadA4 | 58 | Streptomycin-spectinomycin resistance | 100 | 01909 | AY214164 | E. coli (pAPEC-O2-R) |
| 122 | dfrA17 | 36 | Trimethoprim resistance | 100 | 00186 | AY828551 | E. coli |
| 123 | int | 61 | Integrase | 100 | 02899 | AP000342 | E. coli (pR100) |
| 124 | ΔtnpM | 63 | Transposase (truncated) | 81 | 00563 | AJ851089 | Uncult. (pRSB107) |
| 125 | IS26-tnpA | 52 | Transposase | 100 | 00665 | NC_006856 | S. enterica |
| 126 | orf126 | 55 | Hypothetical protein | 100 | 03551 | AJ698325 | Uncult. (pRSB101) |
| 127 | IS26-tnpA | 52 | Transposase | 100 | 00665 | NC_006856 | S. enterica |
| 128 | ΔtnpA-Tn3 | 51 | Transposase (truncated) | 99 | 01526 | CP000057 | H. influenzae |
| 129 | tnpR-Tn3 | 51 | Resolvase | 99 | 02796 | CP000057 | H. influenzae |
| 130 | blaTEM-1 | 49 | β-Lactam resistance | 100 | 00144 | AB070224 | S. marcescens |
| 131 | rmtB | 56 | Aminoglycoside resistance | 100 | 07091 | AB103506 | S. marcescens |
| 132 | orf132 | 59 | Hypothetical protein | ||||
| 133 | orf133 | 69 | Transposase | 99 | 04986 | CT025832 | A. baumannii |
| 134 | qepA | 72 | Fluoroquinolone resistance | 56 | 07690 | CP000316 | Polaromonas sp. |
| 135 | Δint | 61 | Integrase (truncated) | 100 | 02899 | DQ390454 | E. coli (pLEW517) |
| 136 | IS26-tnpA | 52 | Transposase | 100 | 00665 | NC_006856 | S. enterica |
| 137 | nqrc | 47 | NADH-ubiquinone oxidoreductase | 100 | 04205 | AJ851089 | Uncult. (pRSB107) |
| 138 | orf138 | 55 | Hypothetical protein | 99 | 03239 | AJ851089 | Uncult. (pRSB107) |
| 139 | orf139 | 53 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 140 | orf140 | 57 | Hypothetical protein | 100 | 04945 | AJ851089 | Uncult. (pRSB107) |
| 141 | orf141 | 59 | Hypothetical protein | 100 | 02687 | AJ851089 | Uncult. (pRSB107) |
| 142 | orf142 | 58 | Hypothetical protein | 100 | 02687 | AJ851089 | Uncult. (pRSB107) |
| 143 | orf143 | 57 | ABC transporter ATP binding protein | 100 | 00005 | AJ851089 | Uncult. (pRSB107) |
| 144 | orf144 | 58 | Hypothetical protein | 100 | 08534 | AJ851089 | Uncult. (pRSB107) |
| 145 | orf145 | 49 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 146 | orf146 | 57 | Glucose-1-phosphatase | 100 | 00328 | AJ851089 | Uncult. (pRSB107) |
| 147 | IS26-tnpA | 52 | Transposase | 100 | 00665 | CT025832 | A. baumannii |
| 148 | orf148 | 54 | Hypothetical protein | 100 | 01402 | AJ851089 | Uncult. (pRSB107) |
| 149 | orf149 | 48 | Plasmid stabilization system | 100 | 05016 | AJ851089 | Uncult. (pRSB107) |
| 150 | orf150 | 38 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 151 | orf151 | 33 | Hypothetical protein | 100 | AJ851089 | Uncult. (pRSB107) | |
| 152 | orf152 | 49 | Hypothetical protein | 100 | NC_006816 | Serovar Typhimurium (pU302L) | |
| 153 | orf153 | 45 | Hypothetical protein | ||||
| 154 | int | 57 | Integrase | 99 | 00589 | AJ851089 | Uncult. bact. (pRSB107) |
| 155 | repA | 51 | Replication | 99 | 01051 | AJ851089 | Uncult. bact. (pRSB107) |
| 156 | IS1-insA | 52 | Transposase | 97 | 03811 | NC_006816 | Serovar Typhimurium (pU302L) |
| 157 | IS1-insB | 52 | Transposase | 99 | 03400 | NC_006816 | Serovar Typhimurium (pU302L) |
| 158 | mmuM | 60 | Homocysteine S-methyltransferase | 100 | 02574 | U70214 | E. coli |
| 159 | mmuP | 54 | S-methylmethionine permease | 99 | 00324 | U70214 | E. coli |
| 160 | IS5-tnpA | 55 | Transposase | 99 | 01609 | AJ001620 | E. coli |
| 161 | doc | 52 | Death on curing protein | 99 | 05012 | AF234173 | Bacteriophage P1 |
| 162 | phd | 51 | Prevent host death protein | 100 | 02604 | AF234173 | Bacteriophage P1 |
| 163 | hsdM | 45 | Type I restriction enzyme, methylase | 99 | 02384 | X13145 | E. coli (pR124/3) |
| 164 | hsdS | 39 | Type I restriction enzyme, specificity | 57 | 01420 | CP000447 | S. frigidimarina |
| 165 | hsdR | 43 | Type I restriction enzyme, nuclease | 97 | 04313 | X13145 | E. coli (pR124/3) |
| 166 | IS1-insA | 52 | Transposase | 97 | 03811 | NC_006816 | Serovar Typhimurium (pU302L) |
| 167 | IS1-insB | 52 | Transposase | 99 | 03400 | NC_006816 | Serovar Typhimurium (pU302L) |
| 168 | orf168 | 48 | Hypothetical protein | 23 | AAMR01000022 | V. splendidus | |
| 169 | orf169 | 49 | Hypothetical protein | 41 | AAMR01000022 | V. splendidus | |
| 170 | orf170 | 49 | Hypothetical protein | 100 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 171 | vagD | 59 | Virulence-associated protein | 99 | 01850 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 172 | vagC | 54 | Virulence-associated protein | 100 | 04014 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
| 173 | orf173 | 44 | Hypothetical protein | 89 | DQ449578 | Klebsiella pneumoniae (pK245) | |
| 174 | orf174 | 50 | Hypothetical protein | 100 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 175 | korC | 57 | Hypothetical protein | 100 | DQ381420 | E. coli (pAPEC-O1-ColBM) | |
| 176 | orf176 | 45 | Hypothetical protein | 100 | DQ381420 | E. coli (pAPEC-O1-ColBM) |
Pfam is a large collection of multiple sequence alignments and hidden Markov models covering many common protein domains and families.
Uncult., uncultured bacterium.
Replication regions.
Plasmid pIP1206 contained four replication regions, RepFIA, RepFIB (Fig. 1), RepFII-a, and RepFII-b (Fig. 1 and 2). RepFIA, a 6,498-bp fragment (ORFs 1 to 6; G+C content, 47%) contained the oriV-1 origin of replication and the genes necessary for maintenance of the plasmid, such as DNA replication (repE), partition (the ccdA/ccdB and sopABC operons), multimer resolution (resD), and incompatibility (incC), and was 99% identical to the RepFIA primary replication region of pRSB107 (47).
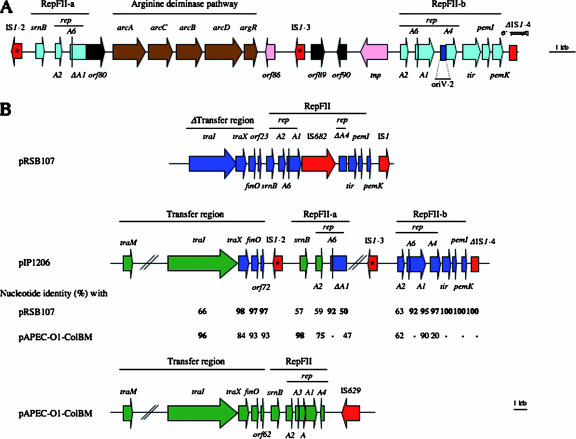
FIG. 2.
(A) Schematic representation of the 19-kb fragment of pIP1206 containing the RepFII replicons and the genes for the arginine deiminase pathway. The RepFII genes are shown in blue, those of the arginine pathway in brown, IS elements in red, transposase in pink, and ORFs for hypothetical proteins in black. The OriV-2 replication origin is shown in dark blue. (B) Comparison of the RepFII regions of pRSB107, pIP106, and pAPEC-01-ColBM. The genes in pIP1206 exhibiting the highest level of identity with pRSB107 are indicated in blue, and those with identify with pAPEC-01-ColBM are in green. Insertion sequences are in red. Asterisks indicate a 4-bp deletion in insB′ of IS1 sequences IS1-2 and IS1-3. Arrows represent coding sequences and indicate the direction of transcription. The Δ symbol denotes a deletion.
The ccdA-ccdB antitoxin-toxin operon (22) enforces stabilization of the plasmid. The CcdB toxin acts on the A subunit of the gyrase, an essential topoisomerase involved in DNA replication, recombination, and transcription, by catalyzing ATP-dependent negative supercoiling of DNA (35). The CcdB-gyrase complex is responsible for the relaxation of supercoiled DNA. The toxic effect of CcdB is antagonized by CcdA, but the half-life of the CcdA antitoxin is shorter than that of CcdB.
The resD gene encodes a recombinase that resolves multimers to monomers. Partition of the plasmid is ensured by the sopABC cluster, which is responsible for adequate subcellular positioning of plasmid DNA molecules and is thus necessary for stable maintenance of low-copy-number F-plasmids (40, 41). sopABC of pIP1206 was identical to that in pRSB107.
RepE binds specifically to four incB iterons, which are 19-bp directly repeated sequences in oriV-2, and causes bending of this region, which is a critical first step in replication of F-plasmids (42, 57). The incC region is implicated in incompatibility with plasmids of the same group and in the control of the plasmid copy number (49, 51).
The second replication region, RepFIB (ORFs 154 and 155; G+C content, 52%) is composed of a repA replication and an int site-specific integrase gene that share more than 99% identity with the RepFIB replicon from several IncF plasmids, including pRSB107 and pAPEC-O1-ColBM. Two genes (ORFs 148 and 149) encoding proteins putatively involved in the regulation of plasmid copy number and in plasmid stabilization were found upstream from the int gene.
Two RepFII replicons (FII-a, ORFs 76 to 79 and G+C content of 51%; FII-b, ORFs 92 to 98 and G+C content of 52%) were also present in pIP1206 (Fig. 2). FII-a was composed of genes encoding SrnB, involved in postsegregational killing of plasmid-free cells, and RepA2 and RepA6, responsible for negative and positive regulation of RepA1 transcription, respectively. RepA2 shared 84% and 48% identity with RepA2 of pAPEC-O1-ColBM and pRSB107, respectively. Downstream from these genes, there was a 3′-end truncated repA1 gene. FII-b, the second RepFII replicon, was composed of the repA2, repA6, repA1, and repA4 genes. The RepA2FII-b protein was 48%, 52%, and 61% identical with RepA2FII-a of pIP1206, pAPEC-O1-ColBM, and pRSB107, respectively. RepA6FII-b was identical with RepA6FII-a of pIP1206 and had 83% identity with RepA6pRSB107; there is no report of RepA6 in pAPEC-O1-ColBM. The repA4 region shared 97% identity with repA4pRSB107 and only 20% with that of repA4pAPECO1-ColBM. However, a mutation in the 5′ portion of repA4pRSB107 is responsible for a truncated RepA4 of only 4 amino acids. As in pRSB107, the tir, pemI, and pemK genes were present downstream from repA4. The tir gene encodes a putative protease, and pemI/pemK were similar to chpAI/chpAK (69% and 81% identity, respectively), which have properties of an addiction module, ChpAI, protecting the bacterium from the toxic effect of ChpAK (15, 38). As in pRSB107, an IS1 copy was located downstream from FII-b (47). In summary, these data indicate that the two RepFII replicons have distinct origins and suggest that pIP1206 could have resulted from a double homologous recombination event between pRSB107 and a pAPEC-O1-ColBM-like plasmid.
IncF plasmids frequently possess more than one replication region, and it is thought that this could provide the plasmid with a broader range of hosts. However, we could not obtain, in repeated attempts, transfer of pIP1206 from E. coli Top10/pIP1206 to Acinetobacter sp. strain R1 and to P. aeruginosa PAO38 by filter mating. These results indicate that, despite the presence of four replication regions, pIP1206 is, as shown for other plasmids of the IncF group, of narrow host range.
The pIP1206 leading region is highly similar to that of pRSB107.
Downstream from RepFIA, a 5,179-bp fragment (ORFs 7 to 16; G+C content, 58%) was identical to a portion in pRSB107. This region, highly similar to the F-plasmid leading region, is the first piece of DNA to enter the recipient cell during conjugative transfer (47). In pIP1206 and pRSB107, this region is composed of 10 genes encoding eight proteins with unknown functions, ORF9 for a DNA methylase, and ORF14 for a putative antirestriction protein (Fig. 1). The methylase is a member of the N6-methyltransferase group that has been proposed to protect the transferred DNA from endonucleases in the recipient cell (37). Surprisingly, another gene for a methyltransferase (ORF23), partially present in pRSB107, was located downstream from the leading region. This methylase, which is very distantly related to the previous one (only 15% identity), was 97% identical to an N6-methyltransferase encoded by pAPEC-O1-ColBM (accession number DQ381420). Of note, the ydbA gene, located just upstream from ORF23 in pIP1026, is also present in pRSB107 and pAPEC-O1-ColBM. Thus, these two ORFs could be implicated in a double homologous recombination event between pRSB107 and pAPEC-O1-ColBM.
Two copies (ORFs 17 and 25; 99.3% identity; 49% G+C content) of an additional gene for a putative reverse transcriptase were found in pIP1206 downstream from the plasmid leading region. These proteins exhibited 62% identity with an RNA-directed DNA polymerase found in Shewanella sp. (accession number CP00034).
Transfer region.
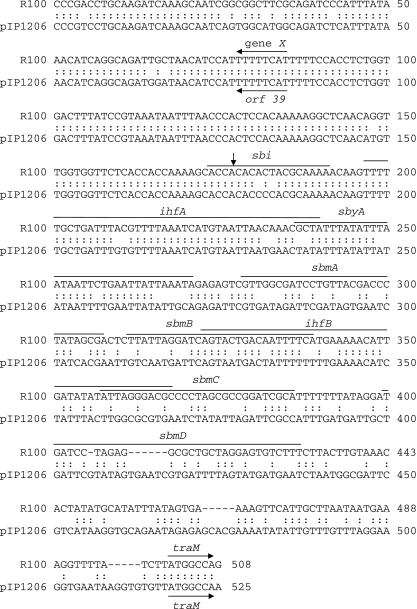
Upstream from RepFII-a, a 32,029-bp fragment (ORFs 40 to 72; G+C content, 52%) contained a complete transfer region (Fig. 1) with 91% identity to that of pAPEC-O1-ColBM. This region comprised 24 tra genes (traM, traJ, traY, traA, traL, traE, traK, traB, traP, traV, traR, traC, traW, traU, traN, traF, traQ, traH, traG, traS, traT, traD, traI, and traX), and 8 trb genes (trbG, trbI, trbC, trbE, trbA, trbB, trbF, and trbJ), artA, and finO. Twenty-three of the predicted proteins shared more than 95% identity with those encoded by pAPEC-O1-ColBM; the level of identity was lower for TraM (82%), TraJ (26%), TraY (25%), TrbA (58%), TrbF (69%), TrbJ (69%), TraG (92%), TraS (66%), TraX (88%), ArtA (74%), and FinO (94%). A frameshift mutation in the trbJ gene of pIP1206 resulted in a premature stop codon and thus to synthesis of a truncated protein. The function of the putative inner membrane protein TrbJ is unknown. However, mutations in trbJ do not significantly alter transfer efficiency (36). TraJ and TraY, implicated in the transcriptional regulation of tra gene expression (17), were highly similar to their counterparts in IncFI plasmid P307 (Table 1). TraD of pIP1206 contained a triplication of the 621-PQQ-623 motif compared to the sequence deduced from pAPEC-O1-ColBM. The number of repetitions of this motif in TraD seems to be polymorphic: 6 in pAPEC-O2-ColV (accession number NC_007675) and pAPEC-O2-R (accession number NC_006671), 7 in pSFO157 (accession number NC_009602), R1 (accession number AY684127), and pC15-1a (accession number NC_005327), 9 in R100 (accession number NC_002134), 10 in NR1 (accession number NC_009133), and 13 in pSS_046 (accession number NC_007385). Transfer by conjugation is initiated by site- and strand-specific nicking of plasmid DNA at the oriT transfer origin, which is located near the transfer region. Upstream from traMpIP1206, a 432-bp fragment exhibited 61% identity with the oriT region of plasmid R100 (Fig. 3) (1). Eight recognition and binding sites, sbi, ihfA, sbyA, sbmA, sbmB, ihfB, sbmC, and sbmD, have been shown to be involved in DNA transfer (Fig. 3). In pIP1206, these eight sequences were present and shared 89%, 86%, 71%, 53%, 61%, 61%, 38%, and 47% identity, respectively, with their counterparts in R100. It has been suggested that another putative functional site, located between gene X and the sbi site in R100 (Fig. 3), is implicated in efficient DNA transfer. In pIP1206, a 99% identical fragment was found (Fig. 3). Furthermore, the X gene of R100 and orf39 of pIP1206, which are located upstream from traM, exhibited 92% identity. These observations strongly suggest that pIP1206 possesses a functional oriT. Transfer of plasmid pIP1206 from E. coli Top10/pIP1206 to E. coli C600Rif was obtained at a frequency of 10−4 per donor. This result confirmed that, despite the presence of a mutated trbJ and of trbF and traS exhibiting unusual sequences, the transfer region of pIP1206 is functional.
FIG. 3.
Sequence comparison of the oriT regions of plasmids R100 and pIP1206. The TraI recognition site (sbi) and the TraY (sbyA), TraM (sbmA, -B, -C, and -D), and integration host factor (ihfA and -B) binding sites are indicated and are as defined previously (1). The vertical arrow indicates the oriT origin of DNA transfer.
Plasmid pRSB107 contains an incomplete transfer region composed of part of traI, traX, and finO. Deduced amino acid sequences revealed that TraI of pIP1206 shared a higher level of identity with TraI of pAPEC-O1-ColBM than that of pRSB107, whereas TraX and FinO presented a higher identity with the corresponding genes in pRSB107 (Fig. 2B). Thus, the traI traX finO region could be implicated in the homologous recombination that took place between pRSB107 and a pAPEC-O1-ColBM-like plasmid to generate pIP1206.
Raffinose operon and cluster for an arginine deiminase pathway.
A raffinose operon (ORFs 28 to 32; G+C content, 47%) was detected in pIP1206 between the replication and the transfer regions (Fig. 1). It was composed of five genes encoding a repressor (RafR), an α-d-galactosidase (RafA), a permease (RafB), a sucrose hydrolase (RafD), and a porin (RafY) (2, 4, 52). These proteins were 89%, 97%, 97%, 96%, and 88% identical to those encoded by the raffinose operon in plasmid pRSD2, which allows growth of E. coli on raffinose (4). Between the rafR and rafA genes were two symmetrical 18-bp sequences form the rafO operator (5). The sequence of rafO from pIP1206 exhibited 90% identity with that from pRSD2.
An arcACBD gene cluster was located downstream from the transfer region (ORFs 81 to 85; G+C content, 53%) (Fig. 2A) encoding proteins involved in the arginine deiminase pathway, one of the four arginine catabolic pathways (for a review see reference 30). The pIP1206 gene set arcA, arcB, and arcC encoded, respectively, an arginine deiminase, an ornithine transcarbamylase, and a carbamate kinase. These proteins exhibited 95%, 91%, and 88% identity with their counterparts in Enterobacter sp. strain 638 (Table 1). The deduced amino acid sequence of the additional ORF84 (arcD) downstream from arcB for ornithine transcarbamylase shared, respectively, 92% and 82% identity with a protein previously described in GenBank as a C-4 dicarboxylate anaerobic carrier (CduC) from Enterobacter sp. strain 638 (accession number CP000653) and as an arginine-ornithine antiporter in Aeromonas salmonicida subsp. salmonicida (accession number CP000462). In these organisms, the protein is also specified by a gene downstream from arcB. The arginine biosynthetic pathway is regulated by a repressor (ArgR) (for a review, see reference 31). ArgR of pIP1206 had 84% identity with ArgR of Enterobacter sp. strain 638. arc operons have been detected in various species, but the gene order differs: arcDABC in Pseudomonas aeruginosa (9, 53), arcABCRD in Enterococcus faecalis (6), and arcABDCR in Bacillus licheniformis (33). In pIP1206, the gene organization (arcACBDR) was identical to that in Enterobacter sp. strain 638.
Mobile elements in pIP1206.
Insertion sequences can be responsible for DNA rearrangements (deletions, duplications, or inversions) that are associated with transposition (34). Class I transposons consist of a DNA segment flanked by two copies of an IS, in direct or in opposite orientation.
Analysis of pIP1206 revealed the presence of 16 copies of IS elements: six IS26, four IS1, three mutated IS1, one IS1 truncated at the 3′ end, one IS2, and one IS5 (Fig. 1). The three mutated IS1 elements had the same deletion of four bases (351-CAGT-354) in the insA-insB intergenic region, precisely within the insB′ ORF, which is a 5′ extension of insB. It has been demonstrated that insA and insB, but not insB′, are essential for cointegrate formation. A mutation in the latter ORF is responsible for a reduction in cointegration frequency (23, 32). Thus, these three IS1 copies are potentially functional. Three transposons, Tn3, Tn10 and a Tn21-like transposon, were identified, but none of them was intact. Three genes (ORFs 86, 91, and 133) encoding putative transposases were also found throughout pIP1206.
The movement of transposons or insertion sequences is usually associated with a duplication of target DNA at the site of insertion, leading to short, directly repeated sequences at the ends of the element. In pIP1206, only a single perfect duplication was found that flanked IS1-5. Thus, as previously proposed for the mosaic IncF p1658/97 plasmid (56), some of the mobile genetic elements have probably been acquired by recombination.
Antibiotic resistance genes.
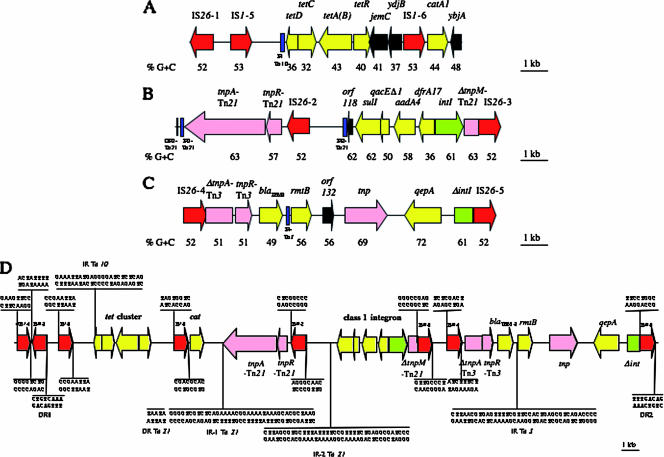
A 10-kb segment (G+C content, 45%), identical to a portion of pRSB107, contained genes responsible for tetracycline [tet(A)] and chloramphenicol (catA) resistance (Fig. 4A). As in pRSB107, the tetracycline resistance determinant (ORFs 104 to 107) was borne by a truncated derivative of Tn10 bordered by two IS1 sequences (IS1-5 and IS1-6). The tet cluster is composed of four genes encoding a repressor (TetR), a tetracycline efflux antiporter [TetA(B)] belonging to the major facilitator superfamily, a transcriptional regulator (TetC), and a putative transcriptional regulator (TetD). Upstream from this cluster was jemC (encoding a putative repressor of arsenic and mercury resistance operons) and an ORF for a hypothetical protein (named YdjB in plasmid R100). In pIP1206, the jemA/jemB genes were not present, as opposed to Tn10 (12). The catA gene (ORF 112) was located immediately after this truncated transposon. ybjA (ORF 113), which codes for an N-acetyltransferase, was downstream from catA.
FIG. 4.
Schematic representation of antibiotic resistance regions of pIP1206 to tetracycline and chloramphenicol (A), to sulfonamides, quaternary ammonium, streptomycin/spectinomycin, and trimethoprim (B), and to β-lactams, aminoglycosides, and fluoroquinolones (C). (D) Nucleotide sequences flanking the mobile elements. DR1 and DR2 flank at one end IS26-1 and IS26-5. Genes for resistance are indicated in yellow, those for transposases/resolvases are in pink, integrases are in green, IS are in red, IRs and DRs are in blue, and ORFs for hypothetical proteins are in black. Arrows represent coding sequences and indicate the direction of transcription. The percent G+C content is indicated under the arrows. Truncated genes are indicated by a Δ symbol.
Downstream from the previous fragment, a remodelled Tn21-like transposon (ORFs 114 to 124; G+C content, 58%) was identified (Fig. 4B) (for a review, see reference 29). A Tn21 derivative is also present in the pRSB107 antibiotic resistance region (47). The Tn21-like structure in pIP1206 was characterized by the presence of transposition genes (a truncated tnpM, tnpR, and tnpA) of a class I integron and by the absence of a set of genes encoding mercury resistance. The global structure of the transposon also differed from that in pRSB107. In pIP1206, direct repeat 1 (DR1), inverted repeat 1 (IR1), and the tnpA-tnpR genes were at the left end of the element. A 7.6-kb fragment, delineated by IS26-2 and IS26-3 and containing a second 38-bp IR (IR2), a class I integron, and the 3′-truncated regulator tnpM, constituted the remaining of the Tn21-like transposon. Curiously, this fragment was inverted relative to typical Tn21, tnpM being in the opposite orientation and at the right end of the transposon and IR2 being located between tnpA and tnpM (Fig. 4B). Analysis of the sequences flanking the IS26 copies revealed that the 8-bp sequence downstream from IS26-2 was identical to the 8 bp upstream from IS26-3 and that the 8-bp sequence upstream from IS26-2 was identical to the 8 bp downstream from IS26-3 (Fig. 4D). Thus, recombination could have occurred between the two copies of IS26, leading to the final structure of the Tn21-like transposon in pIP1206. In summary, the evolution of this transposon could have occurred as follows: acquisition of IS26-2 downstream from the Tn21-like transposon and of IS26-3 within tnpM, followed by recombination between the two IS elements.
The class I integron (ORFs 119 to 123) is composed of intI1 and four resistance genes: sulI (resistance to sulfonamides), qacEΔ1 (to ammonium antiseptics), aadA4 (to streptomycin and spectinomycin), and dfrA17 (to trimethoprim) (Fig. 4B). The integron was identical to one located in the pEC1072 plasmid of E. coli EC107 and to another carried by a plasmid in E. coli 9516014-1 (13, 46) and similar to that of plasmid pAPEC-O2-R (95% identity at the nucleotide level), except for the presence of a catB3 gene in pAPEC-O2-R.
Three additional resistance genes for a class A β-lactamase (blaTEM-1), a methylase (rmtB), and an efflux pump (qepA) were located on an 11.3-kb fragment (ORFs 130, 131, and 134; G+C content, 59%) delineated by IS26-4 and IS26-5 (Fig. 4C). RmtB is a 16S rRNA m7G methyltransferase that modifies the N-7 position of nucleotide G1405 located in the A site of 16S rRNA and confers high-level resistance to all available aminoglycosides used for therapy, except streptomycin (43).
The qepA gene confers resistance to hydrophilic fluoroquinolones. The deduced sequence of QepA is 45% to 56% identical with various 14-transmembrane segment proton-dependent efflux pumps belonging to the MFS superfamily. The G+C content of qepA (72%) significantly differs from that of the chromosome of Enterobacteriaceae (50%), suggesting that qepA could have originated from actinomycetes (43, 54).
ORF133, having 99% identity with a putative transposase (accession number CT025832), was detected between the rmtB and qepA genes. Upstream from qepA, a gene for a class I integron integrase was disrupted by insertion of IS26-5 (Fig. 4C). A Tn3-like transposon, containing the blaTEM-1 gene, was present upstream from rmtB (Fig. 4C). The structural gene for the Tn3 transposase was truncated due to insertion of IS26-4. A 38-bp sequence corresponding to an inverted terminal repeat typical of Tn3 elements (accession number V00613) was found between blaTEM-1 and rmtB (Fig. 4D). A similar fragment containing blaTEM-1, rmtB, and qepA has been reported in plasmid pHPA from an E. coli strain in Japan (54), suggesting that the association of these three resistance genes has a worldwide distribution.
In summary, all the antibiotic resistance genes were located within a 33.5-kb fragment delineated by IS26-1 and IS26-5 (Fig. 4D). IS26 generates 8-bp DRs of target DNA upon transposition. Examination of the flanking sequences of each IS26 copy revealed two identical DR sequences, one upstream from IS26-1 (DR1) and the other downstream from IS26-5 (DR2). However, the two IS26 sequences were in opposite orientations (Fig. 4D), so that DR1 was located inside the 33.5-kb fragment. Of note, two IS1 sequences (the 3′-truncated IS1-4 and IS1-5) flanked IS26-1. One can thus propose that inversion of the IS1-5-DR1-IS26-1-ΔIS1-4 fragment led to the sequence ΔIS1-4-IS26-1-DR1-IS1-5. Thus, this 33.5-kb fragment could (i) have been acquired from another replicon (plasmid or chromosome) by transposition or (ii) have resulted from successive integrations of mobile elements in a smaller fragment flanked by IS26-1 and IS26-5.
Putative iron acquisition gene cluster.
A fragment of 12 kb (ORFs 137 to 146; G+C content, 55%) flanked by IS26-5 and IS26-6 and identical to a segment of pRSB107 contained a putative iron acquisition system (47). This region is composed of genes for an iron permease, two integral membrane proteins, an ABC transporter, and a protein of the thioredoxin family. It has been suggested that pathogenic bacteria harboring plasmids with iron acquisition systems are more virulent (47).
S-Methylmethionine metabolism operon.
S-methylmethionine is an abundant plant product that can be used for methionine biosynthesis. An operon composed of mmuP and mmuM (ORFs 158 and 159; G+C content, 57%), two members of the methionine regulon, was present in pIP1206. MmuP is a permease involved in the uptake of S-methylmethionine, and MmuM is an S-methylmethionine:homocysteine methyltransferase that transfers a methyl group from S-methylmethionine to homocysteine to form two molecules of methionine (48). It has been proposed that expression of methyltransferase genes increases selenium tolerance and reduces nonspecific selenium incorporation into proteins (39).
Type I DNA restriction and modification system.
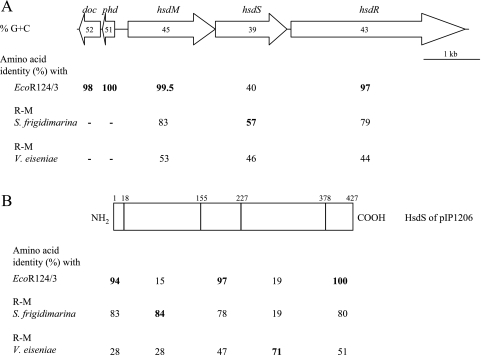
R-M systems (i) protect the host genome from restriction by adding a methyl group on residues within specific target DNA sequences and (ii) restrict unmodified foreign DNA that penetrates into the cell (for a review, see reference 55). R-M enzymes have been classified into three types. The type I system, which recognizes two specific DNA sequences separated by a nonspecific spacer, contains genes coding for three subunits: HsdM, implicated in DNA methylation, HsdS, which determines DNA specificity, and HsdR, which is responsible for restriction. HsdM, HsdS, and HsdR (ORFs 163 to 165; G+C content, 44%) (Fig. 4) in pIP1206 exhibited, respectively, 99%, 43%, and 97% identity with the EcoR124/3-type IC R-M system (44) (Fig. 5A). DNA sequence specificity of the R-M endonuclease is ensured by HsdS, which is responsible for binding to the enzyme's DNA recognition site (18, 19, 44). This protein shared only a low degree of identity with known HsdS proteins, the highest level (57%) being with the R-M system of Shewanella frigidimarina (Table 1; Fig. 5A). It has been hypothesized that the type I HsdS consists of two specific domains, corresponding to the central and distal conserved regions, which recognize the two components of the target sequence (3). The sequence of HsdS of pIP1206 could be divided in five portions (Fig. 5B): part 1 (amino acids 1 to 18), part 3 (amino acids 156 to 227), and part 5 (amino acids 378 to 427) had a high degree of identity with HsdS of EcoR124/3 (from 94% to 100%), whereas part 2 (amino acids 19 to 155) and part 4 (amino acids 228 to 377) exhibited a higher level of identity with the R-M systems of S. frigidimarina and Verminephrobacter eiseniae (accession number CP000542), respectively. Amino acids 156 to 227 and 378 to 427 of HsdS of pIP1206 could represent the two specific domains. From these observations, we suggest that the specificity of the R-M system of pIP1206 could be a hybrid between those of S. frigidimarina and V. eiseniae and that the HsdS protein of pIP1206 has a common ancestry with that of EcoR124/3.
FIG. 5.
Schematic representation of the 7-kb fragment containing the Hsd restriction-modification system (A) and HsdS of pIP1206 (B). The percent G+C content is indicated within the arrows. The highest amino acid identity is indicated in bold. A dash indicates absence of the gene.
It has been argued that R-M systems can (i) be considered as selfish systems (25), (ii) mediate plasmid maintenance (27), and (iii) be part of mobile genetic elements (26). Thus, the R-MpIP1206 system could also be implicated in stabilization of pIP1206 in the host strain. It is flanked by two IS fragments, which makes its mobility likely.
It has been shown that the sequence upstream from the hsd coding region of EcoR124/3 and EcoprrI, another type I R-M system, is conserved and composed of doc and phd, which are also present in the genome of bacteriophage P1 (50). The phd and doc genes encode an addiction system that stabilizes the P1 prophage:Doc, which is toxic to the host, with Phd as the antidote (28). The two genes (ORFs 161 and 162) also were present in pIP1206 upstream from hsdM. The deduced sequences shared 99% and 100% identity with Doc and Phd of P1, respectively. These genes are not present in S. frigidimarina and V. eiseniae.
Virulence-associated genes.
A fragment of 8.6 kb (ORFs 171 and 172; G+C content, 47.4%) contained vagC and vagD (virulence-associated genes). This portion was identical to a region of pAPEC-O1-ColBM, except for the presence in pIP1206 of an additional gene (ORF173) encoding a hypothetical protein. The vagC/vagD locus is required for plasmid maintenance by coupling plasmid replication with cell division; VagD exerts the biological effect, whereas VagC regulates vagD expression (45). Upstream from this fragment, a segment of 2.3 kb (G+C content, 48%) contained ORFs 168 and 169, which encode two hypothetical proteins having 21% and 41% identity with proteins in Vibrio splendidus.
Concluding remarks.
The entire sequence of pIP1206, isolated from an E. coli clinical strain, was determined because this plasmid carries two new genes for resistance to antibiotics: qepA, which confers resistance to hydrophilic fluoroquinolones by efflux, and rmtB, which directs production of a 16S rRNA methylase that confers high-level resistance to aminoglycosides (43). Analysis of the sequence revealed that pIP1206 contained a number of discrete functional elements. These subregions were delineated by insertion sequences, indicating that mobile elements play a major role in the mosaic structure of the replicon. (i) The main part of pIP1026 was probably derived from pRSB107, isolated from a bacterium in a wastewater treatment plant. (ii) Two IS1 sequences flank the transfer region. DNA upstream and downstream from this region is common to pRSB107 and pAPEC-O1-ColBM. Thus, the tra region could have been acquired following homologous recombination between the two plasmids or by transposition. (iii) Of note, an RNA-directed DNA polymerase, present in two copies in pIP1206, had a high degree of similarity to a protein of Shewanella sp., a genus which is widespread in the environment and in wastewater. Furthermore, the hsdS gene of the restriction-modification system in pIP1206 was similar to that of S. frigidimarina and V. eiseniae. It therefore appears that pIP1206 was constructed by DNA exchange in the environment. (iv) All the antibiotic resistance genes were clustered in a fragment flanked by two IS26 copies that contained transposons Tn3, Tn10, and Tn21. However, Tn3 and Tn10 had truncated transposase genes and Tn21 had undergone several rearrangements, presumably due to sequential IS26 insertions. Association of several antibiotic resistance genes with those for two efflux pumps in a plasmid that originated from a human clinical isolate but is also likely to be present in environmental bacteria represents a major risk for dissemination of multidrug resistance. (v) The stabilization of the plasmid in the host, and thus vertical inheritance of genetic information, is ensured by various types of maintenance systems: one active partition (sop operon), two postsegregational killing (ccdA/ccdB and pemI/pemK genes), and one restriction-modification (hsd genes) system.
In summary, pIP1206 may have resulted from recombination between pRSB107 and a pAPEC-01-ColBM-like plasmid associated with structural rearrangements following acquisition of additional DNA by recombination and of mobile elements by transposition; its stabilization is due to the presence of four different systems.
Acknowledgments
We thank P. E. Reynolds for reading the manuscript and F. Kunst for encouraging us to initiate this project.
Footnotes
Published ahead of print on 5 May 2008.
REFERENCES
- 1.Abo, T., and E. Ohtsubo. 1995. Characterization of the functional sites in the oriT region involved in DNA transfer promoted by sex factor plasmid R100. J. Bacteriol. 177:4350-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, C., D. Krones, C. Ulmke, K. Schmid, and R. Benz. 1998. The porin RafY encoded by the raffinose plasmid pRSD2 of Escherichia coli forms a general diffusion pore and not a carbohydrate-specific porin. Eur. J. Biochem. 254:679-684. [DOI] [PubMed] [Google Scholar]
- 3.Argos, P. 1985. Evidence for a repeating domain in type I restriction enzymes. EMBO J. 4:1351-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aslanidis, C., K. Schmid, and R. Schmitt. 1989. Nucleotide sequences and operon structure of plasmid-borne genes mediating uptake and utilization of raffinose in Escherichia coli. J. Bacteriol. 171:6753-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aslanidis, C., and R. Schmitt. 1990. Regulatory elements of the raffinose operon: nucleotide sequences of operator and repressor genes. J. Bacteriol. 172:2178-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barcelona-Andres, B., A. Marina, and V. Rubio. 2002. Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J. Bacteriol. 184:6289-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birnboim, H. C., and J. Doly. 1979. Rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogaerts, P., M. Galimand, C. Bauraing, A. Deplano, R. Vanhoof, R. De Mendonca, H. Rodriguez-Villalobos, M. Struelens, and Y. Glupczynski. 2007. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J. Antimicrob. Chemother. 59:459-464. [DOI] [PubMed] [Google Scholar]
- 9.Bourdineaud, J. P., D. Heierli, M. Gamper, H. J. C. Verhoogt, A. J. M. Driessen, W. N. Konings, C. Lazdunski, and D. Haas. 1993. Characterization of the arcD arginine:ornithine exchanger of Pseudomonas aeruginosa. Localization in the cytoplasmic membrane and a topological model. J. Biol. Chem. 268:5417-5424. [PubMed] [Google Scholar]
- 10.Boyd, E. F., C. W. Hill, S. M. Rich, and D. L. Hartl. 1996. Mosaic structure of plasmids from natural populations of Escherichia coli. Genetics 143:1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chabbert, Y. A., M. R. Scavizzi, J. L. Witchitz, G. R. Gerbaud, and D. H. Bouanchaud. 1972. Incompatibility groups and classification of fi− resistance factors. J. Bacteriol. 112:666-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalmers, R., S. Sewitz, K. Lipkow, and P. Crellin. 2000. Complete nucleotide sequence of Tn10. J. Bacteriol. 182:2970-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, C. Y., L. L. Chang, Y. H. Chang, T. M. Lee, Y. H. Li, and S. F. Chang. 2000. Two new gene cassettes, drf17 (for trimethoprim resistance) and aadA4 (for spectinomycin/streptomycin resistance), inserted in an Escherichia coli class 1 integron. J. Antimicrob. Chemother. 46:87-89. [DOI] [PubMed] [Google Scholar]
- 14.Datta, N., and R. W. Hedges. 1971. Compatibility groups among Fi− R factors. Nature 234:222-223. [DOI] [PubMed] [Google Scholar]
- 15.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 16.Frangeul, L., P. Glaser, C. Rusniok, C. Buchrieser, E. Duchaud, P. Dehoux, and F. Kunst. 2004. CAAT-Box, Contigs-Assembly and Annotation Tool-Box for genome sequencing projects. Bioinformatics 20:790-797. [DOI] [PubMed] [Google Scholar]
- 17.Frost, L. S., K. Ippenihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuller-Pace, F. V., L. R. Bullas, H. Delius, and N. E. Murray. 1984. Genetic recombination can generate altered restriction specificity. Proc. Natl. Acad. Sci. USA 81:6095-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuller-Pace, F. V., and N. E. Murray. 1986. Two DNA recognition domains of the specificity polypeptides of a family of type I restriction enzymes. Proc. Natl. Acad. Sci. USA 83:9368-9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guardabassi, L., L. Dijkshoorn, J. M. Collard, J. E. Olsen, and A. Dalsgaard. 2000. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J. Med. Microbiol. 49:929-936. [DOI] [PubMed] [Google Scholar]
- 21.Jacoby, G. A., and L. Sutton. 1982. Restriction-modification systems determined by Pseudomonas plasmids. Plasmid 8:141-147. [DOI] [PubMed] [Google Scholar]
- 22.Jaffe, A., T. Ogura, and S. Hiraga. 1985. Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 163:841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakowec, M., P. Prentki, M. Chandler, and D. J. Galas. 1988. Mutational analysis of the open reading frames in the transposable element IS1. Genetics 120:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, T. J., S. J. Johnson, and L. K. Nolan. 2006. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J. Bacteriol. 188:5975-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi, I. 1998. Selfishness and death: raison d'etre of restriction, recombination and mitochondria. Trends Genet. 14:368-374. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi, I., A. Nobusato, N. Kobayashi-Takahashi, and I. Uchiyama. 1999. Shaping the genome—restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev. 9:649-656. [DOI] [PubMed] [Google Scholar]
- 27.Kulakauskas, S., A. Lubys, and S. D. Ehrlich. 1995. DNA restriction-modification systems mediate plasmid maintenance. J. Bacteriol. 177:3451-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehnherr, H., E. Maguin, S. Jafri, and M. B. Yarmolinsky. 1993. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233:414-428. [DOI] [PubMed] [Google Scholar]
- 29.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, C. D. 2006. Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains. Appl. Microbiol. Biotechnol. 70:261-272. [DOI] [PubMed] [Google Scholar]
- 31.Maas, W. K. 1994. The arginine repressor of Escherichia coli. Microbiol. Rev. 58:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machida, Y., C. Machida, H. Ohtsubo, and E. Ohtsubo. 1982. Factors determining frequency of plasmid cointegration mediated by insertion sequence IS1. Proc. Natl. Acad. Sci. USA 79:277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maghnouj, A., T. F. D. Cabral, V. Stalon, and C. Vander Wauven. 1998. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor ArgR. J. Bacteriol. 180:6468-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maki, S., S. Takiguchi, T. Miki, and T. Horiuchi. 1992. Modulation of DNA supercoiling activity of Escherichia coli DNA gyrase by F plasmid proteins. J. Biol. Chem. 267:12244-12251. [PubMed] [Google Scholar]
- 36.Maneewannakul, K., and K. Ippenihler. 1993. Construction and analysis of F plasmid traR, trbJ, and trbH mutants. J. Bacteriol. 175:1528-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manwaring, N. P., R. A. Skurray, and N. Firth. 1999. Nucleotide sequence of the F plasmid leading region. Plasmid 41:219-225. [DOI] [PubMed] [Google Scholar]
- 38.Masuda, Y. J., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuhierl, B., M. Thanbichler, F. Lottspeich, and A. Bock. 1999. A family of S-methylmethionine-dependent thiol/selenol methyltransferases. Role in selenium tolerance and evolutionary relation. J. Biol. Chem. 274:5407-5414. [DOI] [PubMed] [Google Scholar]
- 40.Ogura, T., and S. Hiraga. 1983. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 80:4784-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogura, T., and S. Hiraga. 1983. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell 32:351-360. [DOI] [PubMed] [Google Scholar]
- 42.Perez-Casal, J. F., A. E. Gammie, and J. H. Crosa. 1989. Nucleotide sequence analysis and expression of the minimum REPI replication region and incompatibility determinants of pCOLV-K30. J. Bacteriol. 171:2195-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Périchon, B., P. Courvalin, and M. Galimand. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob. Agents Chemother. 51:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price, C., J. Lingner, T. A. Bickle, K. Firman, and S. W. Glover. 1989. Basis for changes in DNA recognition by the EcoR124 and EcoR124/3 type-I DNA restriction and modification enzymes. J. Mol. Biol. 205:115-125. [DOI] [PubMed] [Google Scholar]
- 45.Pullinger, G. D., and A. J. Lax. 1992. A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol. Microbiol. 6:1631-1643. [DOI] [PubMed] [Google Scholar]
- 46.Sandvang, D. 1999. Novel streptomycin and spectinomycin resistance gene as a gene cassette within a class 1 integron isolated from Escherichia coli. Antimicrob. Agents Chemother. 43:3036-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szczepanowski, R., S. Braun, V. Riedel, S. Schneiker, I. Krahn, A. Puhler, and A. Schluter. 2005. The 120,592 bp lncF plasmid pRSB107 isolated from a sewage-treatment plant encodes nine different antibiotic-resistance determinants, two iron-acquisition systems and other putative virulence-associated functions. Microbiology 151:1095-1111. [DOI] [PubMed] [Google Scholar]
- 48.Thanbichler, M., B. Neuhierl, and A. Bock. 1999. S-methylmethionine metabolism in Escherichia coli. J. Bacteriol. 181:662-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsutsui, H., A. Fujiyama, T. Murotsu, and K. Matsubara. 1983. Role of nine repeating sequences of the mini-F genome for expression of F-specific incompatibility phenotype and copy number control. J. Bacteriol. 155:337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tyndall, C., H. Lehnherr, U. Sandmeier, E. Kulik, and T. A. Bickle. 1997. The type IC hsd loci of the enterobacteria are flanked by DNA with high homology to the phage P1 genome: implications for the evolution and spread of DNA restriction systems. Mol. Microbiol. 23:729-736. [DOI] [PubMed] [Google Scholar]
- 51.Uga, H., F. Matsunaga, and C. Wada. 1999. Regulation of DNA replication by iterons: an interaction between the ori2 and incC regions mediated by RepE-bound iterons inhibits DNA replication of mini-F plasmid in Escherichia coli. EMBO J. 18:3856-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulmke, C., J. W. Lengeler, and K. Schmid. 1997. Identification of a new porin, RafY, encoded by raffinose plasmid pRSD2 of Escherichia coli. J. Bacteriol. 179:5783-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verhoogt, H. J. C., H. Smit, T. Abee, M. Gamper, A. J. M. Driessen, D. Haas, and W. N. Konings. 1992. arcD, the first gene of the arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa, encodes an arginine-ornithine exchanger. J. Bacteriol. 174:1568-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamane, K., J. Wachino, S. Suzuki, K. Kimura, N. Shibata, H. Kato, K. Shibayama, T. Konda, and Y. Arakawa. 2007. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51:3354-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan, R. 1981. Structure and mechanism of multifunctional restriction endonucleases. Annu. Rev. Biochem. 50:285-315. [DOI] [PubMed] [Google Scholar]
- 56.Zienkiewicz, M., I. Kern-Zdanowicz, M. Golebiewski, J. Zylinska, P. Mieczkowski, M. Gniadkowski, J. Bardowski, and P. Ceglowski. 2007. Mosaic structure of p1658/97, a 125-kilobase plasmid harboring an active amplicon with the extended-spectrum beta-lactamase gene blaSHV-5. Antimicrob. Agents Chemother. 51:1164-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zzaman, S., M. M. Abhyankar, and D. Bastia. 2004. Reconstitution of F factor DNA replication in vitro with purified proteins. J. Biol. Chem. 279:17404-17410. [DOI] [PubMed] [Google Scholar]