Abstract
Antifungal susceptibility testing of molds has been standardized in Europe and in the United States. Aspergillus fumigatus strains with resistance to azole drugs have recently been detected and the underlying molecular mechanisms of resistance characterized. Three hundred and ninety-three isolates, including 32 itraconazole-resistant strains, were used to define wild-type populations, epidemiological cutoffs, and cross-resistance between azole drugs. The epidemiological cutoff for itraconazole, voriconazole, and ravuconazole for the wild-type populations of A. fumigatus was ≤1 mg/liter. For posaconazole, the epidemiological cutoff was ≤0.25 mg/liter. Up till now, isolates susceptible to itraconazole have not yet displayed resistance to other azole drugs. Cross-resistance between azole drugs depends on specific mutations in cyp51A. Thus, a substitution of glycine in position 54 of Cyp51A confers cross-resistance between itraconazole and posaconazole. A substitution of methionine at position 220 or a duplication in tandem of a 34-bp fragment in the cyp51A promoter combined with a substitution of leucine at position 98 for histidine confers cross-resistance to all azole drugs tested. The results obtained in this study will help to develop clinical breakpoints for azole drugs and A. fumigatus.
Invasive mold infections have increased in recent decades, and those caused by Aspergillus fumigatus are the most common. The mortality rate of these infections is very high and is mainly determined by patient risk factors; however, other issues, such as the infection being caused by a resistant strain, can also affect the patient outcome. Although information on the clinical relevance of isolates exhibiting high MICs is limited, there have been several reports describing a poorer response by these kinds of strains to antifungal treatment (8, 18). The Clinical and Laboratory Standards Institute (CLSI) has standardized antifungal susceptibility testing for molds (14), and the Antifungal Susceptibility Testing Subcommittee of the European Committee on Antibiotic Susceptibility Testing (AFST-EUCAST) has also started to standardize antifungal susceptibility testing of molds, with the definitive standard now pending final approval (7). The discussion document is available at http://www.escmid.org/sites/index_f.aspx?par=2.4. Using these methodologies, A. fumigatus strains exhibiting high MICs to azole drugs and other antifungal agents have been described (2-6, 10, 11, 19). The molecular mechanisms underlying high MICs to azole drugs have been characterized and consist of point mutations in the cyp51A gene, involving substitutions for different amino acids, individually or in combination with a duplication in tandem of a 34-bp fragment in the promoter sequence of the gene (5, 6, 9-13). EUCAST has defined a procedure for harmonizing and defining breakpoints (http://www.srga.org/Eucastwt/bpsetting.htm). Although breakpoints for molds are not a current objective of AFST-EUCAST, the analysis of some of the parameters included in the procedure would help to define wild-type populations, epidemiological cutoffs, and cross-resistance between antifungal drugs.
The aim of this study is to define epidemiological cutoffs and cross-resistance for azole drugs using a large collection of clinical strains of A. fumigatus, including 32 isolates with well-known molecular mechanisms of resistance to azole drugs.
MATERIALS AND METHODS
Microorganisms.
Three hundred and ninety-three isolates of A. fumigatus were included in this study. All of them, except four, were isolated from human clinical samples. The cyp51A gene was sequenced in 75 strains out of 393 isolates. Forty-three had itraconazole MICs of ≤1 mg/liter, and 32 had itraconazole MICs of ≥8 mg/liter. Information about the sequenced strains, including the clinical sources, geographical origins, mutations of the cyp51A gene, other polymorphisms in the gene, and accession numbers to GenBank, when available, is provided in the supplemental material.
Antifungal susceptibility testing.
Microdilution testing was performed following the EUCAST standard methodology (7). This standard is similar to CLSI M38-A (14), with the following minor modifications: (i) RPMI 1640 medium was supplemented with glucose to reach a 2% concentration; (ii) the inoculum size was between 2 × 105 to 5 × 105 CFU/ml, and (iii) inoculum preparations were performed by means of counting spores in a hematocytometer (1, 15, 16). A. fumigatus ATCC 204305 and Aspergillus flavus ATCC 204304 were used as quality-control strains (14).
The antifungal agents used in the study were itraconazole (range, 0.015 to 8 mg/liter) (Janssen S.A., Madrid, Spain), voriconazole (range, 0.015 to 8 mg/liter) (Pfizer S.A., Madrid, Spain), ravuconazole (range, 0.015 to 8 mg/liter) (Bristol-Myers Squibb, Princeton, NJ), and posaconazole (range, 0.015 to 8 mg/liter) (Schering-Plough, Kenilworth, NJ).
The endpoints were recorded at 48 h and defined as the antifungal concentrations that produced a complete inhibition of visual growth (17). The MICs of strains with high MICs to itraconazole were determined at least twice on different days (range, 2 to 11 times). The MICs of strains with itraconazole MICs of ≤1 mg/liter were determined once, except for 18 strains that were tested at least twice.
Sequencing of the A. fumigatus cyp51A gene.
The full coding sequence of cyp51A was amplified by using specific primer sets: P450-A1 (5′-ATGGTGCCGATGCTATGG-3′) and P450-A2 (5′-CTGTC-TCACTTGGATGTG-3′). The amplifications were performed in a 50-μl volume of PCR mixture as previously described (9). The PCR products were analyzed by electrophoresis on agarose gels and stained with ethidium bromide. Sequence analysis of the cyp51A genes showed some point mutations and polymorphisms. To verify that these point changes were not due to errors in the PCR amplification of the genes, the cyp51A genes from all isolates were newly amplified and sequenced a second time. Exactly the same point mutations were found again, indicating that they were not artificially introduced during the PCR.
Definitions. (i) Wild-type microorganism.
A microorganism was defined as a wild type when it did not have any acquired or mutational resistance mechanisms to the drug in question. A microorganism was categorized as the wild type for a species by applying the appropriate cutoff value in a defined phenotypic test system.
(ii) Epidemiological cutoff.
The epidemiological cutoff is the value obtained from an analysis of a MIC distribution after taking into consideration the mode of distribution and the inherent reproducibility of the MICs. The mode ±1 twofold dilution was calculated for each antifungal. The epidemiological cutoffs were based on the MICs of susceptible and resistant strains.
Statistical calculations.
Statistical analysis was done with the Statistical Package for the Social Sciences (version 15.0) (SPSS S.L., Madrid, Spain). Both on-scale and off-scale results were included in the analysis. The off-scale MICs were converted upwards or downwards to the next concentration. In order to approach a normal distribution, the MICs were transformed into log2 values.
The log2 values of the MICs for susceptible and resistant strains were compared by one-way analysis of variance. In this case, all MICs obtained for resistant strains were included in the analysis. The test for homogeneity of variance was run to analyze equal variances, in order to choose the most adequate test for estimating differences. As the results showed unequal variances, the Tamhane T2 test was used to test the null hypotheses. A test was considered significant when the P value was less than 0.01.
GenBank accession numbers.
GenBank accession numbers for each type of mutation and polymorphism (in parentheses) of the cyp51A gene in various strains were as follows: CM-1244 (G54E), EU626227; CM-0796 (G54V), EU626228; CM-2097 (G54R), EU626229; CM-3500 (G54W), EU626230; CM-1245 (M220V), EU626231; CM-2159 (M220K), EU626232; CM-2164 (M220T), EU626233; CM-4593 (M220I), EU626234; CM-2627 (tandem repeat [TR]), EU626235; CM-2733 (F46Y, M172V, and E426K), EU626236; and CM-3248 (N248K), EU626237.
RESULTS
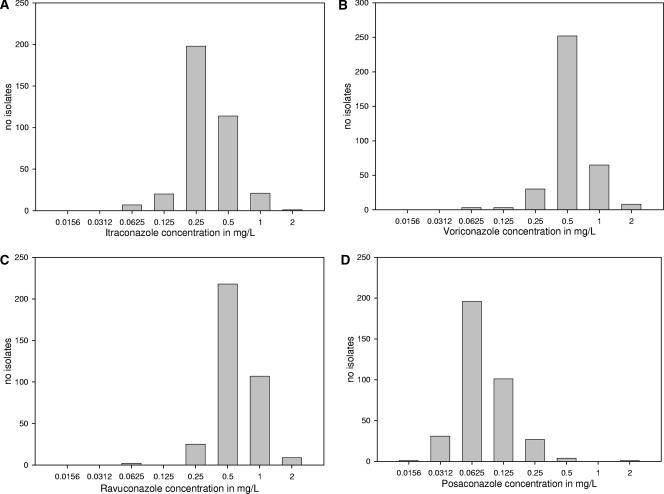
Figure 1 shows the MIC distributions for itraconazole, voriconazole, ravuconazole, and posaconazole for all strains except those with an itraconazole MIC of ≥8 mg/liter. The modal MICs (percentages of MICs) for itraconazole, voriconazole, ravuconazole, and posaconazole were, respectively, 0.25 mg/liter (54.8%), 0.5 mg/liter (69.8%), 0.5 mg/liter (60.4%), and 0.06 mg/liter (54.3%). The modal MICs ±1 twofold dilution encompassed 91.9% of the strains for itraconazole, 96.1% for voriconazole, 96.9% for ravuconazole, and 90.9% for posaconazole. Quality-control strain A. fumigatus ATCC 204305 was tested 127 times. The modal MICs ±1 twofold dilution for itraconazole, voriconazole, ravuconazole, and posaconazole ranged from, respectively, 0.12 to 0.5 mg/liter, 0.25 to 1 mg/liter, 0.25 to 1 mg/liter, and 0.03 to 0.12 mg/liter. Quality-control strain MICs were in range 96.1% of the time. The epidemiological cutoffs (itraconazole, ≤1 mg/liter; voriconazole, ≤1 mg/liter; ravuconazole, ≤1 mg/liter; posaconazole, ≤0.25 mg/liter) were chosen taking into consideration the MIC distributions and the inherent variability of the antifungal susceptibility testing. These cutoffs encompassed 99.7% of the itraconazole MICs, 97.8% of the voriconazole MICs, 97.5% of the ravuconazole MICs, and 98.6% of the posaconazole MICs. In this group of isolates, strains with a MIC of 2 mg/liter for itraconazole, voriconazole, and ravuconazole could be considered outliers. A total of 18 strains showed such MICs of 2 mg/liter. Five of those showing such MICs for voriconazole or ravuconazole were available for further testing and were tested two or three more times for a total of 26 MIC values. Only once was a value of 2 mg/liter obtained on repeat testing; the other MICs were below the epidemiological cutoffs. A set of 12 strains with MICs below the epidemiological cutoffs were repeated at least one more time. The highest MIC obtained was below or equal to the epidemiological cutoff. For posaconazole, strains with MICs of 0.5 mg/liter could also be considered outliers. There were four isolates with such MICs (1.1%), but none were available for further testing. However, 55 MICs obtained after repeated testing of a set of 17 strains with MICs below or equal to the epidemiological cutoffs were always ≤0.25 mg/liter.
FIG. 1.
MIC distribution for itraconazole, voriconazole, ravuconazole, and posaconazole for all strains except those with an itraconazole MIC of ≥8 mg/liter.
The cyp51A gene was sequenced in 75 strains. Forty-three had azole drug MICs below or equal to the epidemiological cutoffs, and 32 had itraconazole MICs of ≥8 mg/liter. Gene cyp51A was identical in 35 (81.4%) strains with azole drug MICs below or equal to the epidemiological cutoffs, but 8 (18.6%) had polymorphisms in the cyp51A gene that produced amino acid changes. One of them was the quality-control strain A. fumigatus ATCC 204305. It had a polymorphism in the cyp51A gene that produced a change from isoleucine to valine at position 242 (I242V). In addition, six strains had three amino acid changes: phenylalanine to tyrosine (F46Y), methionine to valine (M172V), and glutamic acid to lysine (E426K). The last one had only one change: asparagine to lysine (N248K). Those strains were tested at least twice, and all azole drug MICs were below or equal to the epidemiological cutoffs. Therefore, some strains with itraconazole MICs of ≤1 mg/liter may have polymorphisms in the cyp51A gene capable of causing a change in amino acid but with a wild-type azole drug phenotypic pattern.
The mechanisms of resistance underlying an itraconazole MIC of ≥8 mg/liter have been previously characterized (5, 6, 9-13). Three mechanisms are involved, and all affect the cyp51A gene: (i) G54 mutation (at position 54, nine strains had substituted glycine for valine [three], glutamic acid [three], arginine [one], or tryptophan [two]), (ii) M220 mutation (at position 220, six strains had substituted methionine for valine [three], lysine [one], threonine [one], or isoleucine [one]), and (iii) TR-L98H (at position 98, 17 strains had substituted leucine for histidine and two copies of a 34-bp sequence in tandem in the promoter of the cyp51A gene were present) (5, 11, 12).
Strains with itraconazole MICs of ≥8 mg/liter were tested at least twice (range, 2 to 11 times). A total of 270 MICs were obtained (90 for each antifungal). Itraconazole MICs were ≥8 mg/liter every time. For voriconazole and ravuconazole, the repetition of MIC testing showed a difference of ≤2 twofold dilutions. The repetition of MIC testing for posaconazole showed that 30 strains had MICs with ≤1 twofold dilution difference, but for 2 strains, the difference was 4 twofold dilutions. MIC testing for one of the two strains was repeated three times, and the resulting values were 2, 4, and 16 mg/liter, respectively. Testing for the other strain was repeated twice, with MICs of 1 and 8 mg/liter. In summary, in 91.6% of the cases, the difference in the MICs was ≤1 twofold dilution; in 93.7% of the cases, the difference was ≤2 twofold dilutions.
The geometric means (GM) and ranges for the azole drugs with the wild-type strains and those strains with a mechanism of resistance are shown in Table 1. The GMs for voriconazole, ravuconazole, and posaconazole were different, depending on the underlying mechanism of resistance. The log2 MICs for each antifungal were compared for the wild-type isolates and those with a mechanism of resistance. All differences between the wild-type strains and those showing itraconazole MICs of ≥8 mg/liter were significant (P < 0.001). Strains with G54 mutations had voriconazole and ravuconazole GMs lower than the GMs for the wild-type populations (Table 1). Analysis of variance of the log2 MICs showed no differences between the voriconazole and ravuconazole MICs for the wild-type populations and those for strains with G54 mutations. This was not the case for posaconazole, for which the differences were statistically significant (P < 0.001), suggesting cross-resistance to itraconazole and posaconazole for the G54-mutated strains (Table 1).
TABLE 1.
Geometric means and ranges of MICs obtained at 24 and 48 h for A. fumigatus isolates susceptible and resistant to azole drugs
| Isolate type and mechanism of resistance | Itraconazole
|
Voriconazole
|
Ravuconazole
|
Posaconazole
|
||||
|---|---|---|---|---|---|---|---|---|
| GM (range) | MIC90 | GM (range) | MIC90 | GM (range) | MIC90 | GM (range) | MIC90 | |
| Susceptible (n = 361) | 0.32 (0.06-2) | 0.5 | 0.53 (0.06-2) | 1 | 0.60 (0.06-2) | 1 | 0.08 (0.015-2) | 0.125 |
| Resistant (n = 32) | ||||||||
| G54 mutationa (n = 9) | 16 (16-16) | 0.42 (0.12-1) | 0.32 (0.12-1) | 1.6 (0.25-16) | ||||
| M220 mutationb (n = 6) | 16 (16-16) | 1.0 (0.5-2) | 1.85 (1-4) | 0.65 (0.25-2) | ||||
| TRc (n = 17) | 16 (16-16) | 3.7 (2-8) | 6.8 (4-16) | 0.68 (0.5-8) | ||||
At position 54, nine strains had a substitution of glycine for valine (three), glutamic acid (three), arginine (one), or tryptophan (two).
At position 220, six strains had a substitution of methionine for valine (three), lysine (one), threonine (one), or isoleucine (one).
At position 98, 17 strains had a substitution of leucine for histidine and two copies of a 34-bp sequence in tandem in the promoter of the cyp51A gene were present.
Although the number of strains is limited, the posaconazole MICs depended on the amino acid change. Thus, strains with tryptophan instead of glycine (G54W) had posaconazole GMs of 16 mg/liter, whereas those with valine (G54V) or glutamic acid (G54E) had GMs of 0.59 and 0.60 mg/liter, respectively. One strain with arginine instead of glycine (G54R) had a posaconazole MIC GM of 2 mg/liter.
The GMs for voriconazole and ravuconazole with M220 amino acid substitutions or TR changes were higher than those obtained with the G54-mutated strains. Although the posaconazole GM was lower than that obtained for the G54-mutated strains, it was statistically different from the GMs of the susceptible strains (P < 0.001) (Table 1). Therefore, M220 substitutions or TR mutations generate strains with statistically higher MICs to azole drugs than those obtained for susceptible strains, indicating cross-resistance.
DISCUSSION
AFST-EUCAST has approved a discussion standard methodology to detect the resistance of molds to antifungals. This standard is pending final approval but can be downloaded at http://www.escmid.org/sites/index_f.aspx?par=2.4. In addition, AFST-EUCAST set up a system to obtain breakpoints for antifungal agents. The process consists of 10 steps, 1 step being the establishment of MIC distributions and epidemiological cutoffs. Breakpoints for molds are not a current priority for AFST-EUCAST; however, for the most frequent mold involved in invasive infections, A. fumigatus, the definition of the wild-type populations and the epidemiological cutoffs is important in order to have a clear concept about which MIC can be considered microbiologically susceptible. This exercise has been facilitated by the fact that strains with secondary resistance to azole drugs have been detected and their molecular mechanisms of resistance characterized. In this study, the MICs of a large set of susceptible and resistant strains have been analyzed in order to obtain epidemiological cutoffs to define the wild-type populations and cross-resistance between azole drugs.
The epidemiological cutoffs chosen encompassed ≥90% of the strains. For obvious reasons, cyp51A was not sequenced in all strains phenotypically susceptible to azole. Forty-three susceptible strains (12%) were sequenced, and 81.4% had identical cyp51A genes; however, in eight (18.6%), some polymorphisms were detected that produced amino acid changes. On the other hand, all resistant strains had itraconazole MICs of ≥8 mg/liter. The resistance mechanisms of those strains had been previously characterized. The transformation of a control susceptible strain with a mutated copy of the cyp51A gene always reproduced the resistance phenotype (5, 9-11). In addition, the targeted disruption of the cyp51A gene of resistant strains decreased the azole drug MICs below or equal to the epidemiological cutoffs (12).
In summary, itraconazole, as in the case of fluconazole and Candida spp., seems to be the guard for azole resistance detection in A. fumigatus. Therefore, this antifungal must be included each time an isolate of A. fumigatus is tested.
Cross-resistance in this group of antifungals is another matter of concern. Consequently, a comparison was run between the MICs of the wild-type populations and isolates with itraconazole MICs of ≥8 mg/liter. As Table 1 shows, cross-resistance depends on the mechanism of resistance. Thus, those strains with a G54 mutation show cross-resistance to itraconazole and posaconazole but not to voriconazole and ravuconazole. The rest of the mutations, M220 and TR, produced a cross-resistance pattern to all azole drugs tested. The resistant strains were tested a sufficient number of times to conclude that the EUCAST antifungal susceptibility testing for molds is reproducible enough to identify resistant strains and their patterns of cross-resistance without the need for sequencing cyp51A to detect the underlying mutation. Therefore, the MIC phenotype obtained by means of a standardized methodology is the most reliable technique for identifying resistant strains.
In summary, the wild-type populations of A. fumigatus and the epidemiological cutoffs for the azole drugs have now been defined. Isolates susceptible to itraconazole have not so far displayed resistance to other azole agents. Cross-resistance between azole drugs depends on the specific mutation of cyp51A. The results obtained in this study will help to develop breakpoints for azole drugs and A. fumigatus.
Supplementary Material
Acknowledgments
This study was funded in part by grants: MPY1175/06 and PI05/32 from the Instituto de Salud Carlos III, SAF2005-06541 from the Ministerio de Educacion y Ciencia, and REIPI RD06/0008 from the Spanish Network for Research into Infectious Diseases.
L.A.-F. holds a postdoctoral contract with the EU-STREP project (LSHM-CT-2005-518199). A.A.-I. holds a predoctoral fellowship with the Fondo de Investigaciones Sanitarias (grant FI05/00856).
Footnotes
Published ahead of print on 12 May 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Aberkane, A., M. Cuenca-Estrella, A. Gomez-Lopez, E. Petrikkou, E. Mellado, A. Monzon, and J. L. Rodriguez-Tudela. 2002. Comparative evaluation of two different methods of inoculum preparation for antifungal susceptibility testing of filamentous fungi. J. Antimicrob. Chemother. 50:719-722. [DOI] [PubMed] [Google Scholar]
- 2.Chen, J., H. Li, R. Li, D. Bu, and Z. Wan. 2005. Mutations in the cyp51A gene and susceptibility to itraconazole in Aspergillus fumigatus serially isolated from a patient with lung aspergilloma. J. Antimicrob. Chemother. 55:31-37. [DOI] [PubMed] [Google Scholar]
- 3.Dannaoui, E., E. Borel, M. F. Monier, M. A. Piens, S. Picot, and F. Persat. 2001. Acquired itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 47:333-340. [DOI] [PubMed] [Google Scholar]
- 4.Dannaoui, E., D. Garcia-Hermoso, J. M. Naccache, I. Meneau, D. Sanglard, C. Bouges-Michel, D. Valeyre, and O. Lortholary. 2006. Use of voriconazole in a patient with aspergilloma caused by an itraconazole-resistant strain of Aspergillus fumigatus. J. Med. Microbiol. 55:1457-1459. [DOI] [PubMed] [Google Scholar]
- 5.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2003. A point mutation in the 14α-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard, S. J., I. Webster, C. B. Moore, R. E. Gardiner, S. Park, D. S. Perlin, and D. W. Denning. 2006. Multi-azole resistance in Aspergillus fumigatus. Int. J. Antimicrob. Agents 28:450-453. [DOI] [PubMed] [Google Scholar]
- 7.Lass-Flörl, C., M. Cuenca-Estrella, D. W. Denning, and J. L. Rodriguez-Tudela. 2006. Antifungal susceptibility testing in Aspergillus spp. according to EUCAST methodology. Med. Mycol. 44:319-325. [DOI] [PubMed] [Google Scholar]
- 8.Lass-Florl, C., K. Griff, A. Mayr, A. Petzer, G. Gastl, H. Bonatti, M. Freund, G. Kropshofer, M. P. Dierich, and D. Nachbaur. 2005. Epidemiology and outcome of infections due to Aspergillus terreus: 10-year single centre experience. Br. J. Haematol. 131:201-207. [DOI] [PubMed] [Google Scholar]
- 9.Mellado, E., T. M. Diaz-Guerra, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2001. Identification of two different 14-α sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 39:2431-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellado, E., G. Garcia-Effron, L. Alcazar-Fuoli, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2004. Substitutions at methionine 220 in the 14α-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48:2747-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellado, E., G. Garcia-Effron, L. Alcázar-Fuoli, W. J. G. Melchers, P. E. Verweij, M. Cuenca-Estrella, and J. L. Rodríguez-Tudela. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51:1897-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellado, E., G. Garcia-Effron, M. J. Buitrago, L. Alcazar-Fuoli, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2005. Targeted gene disruption of the 14-α sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob. Agents Chemother. 49:2536-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nascimento, A. M., G. H. Goldman, S. Park, S. A. E. Marras, G. Delmas, U. Oza, K. Lolans, M. N. Dudley, P. A. Mann, and D. S. Perlin. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents Chemother. 47:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi. Approved standard M38-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 15.Petrikkou, E., J. L. Rodriguez-Tudela, M. Cuenca-Estrella, A. Gomez, A. Molleja, and E. Mellado. 2001. Inoculum standardization for antifungal susceptibility testing of filamentous fungi pathogenic for humans. J. Clin. Microbiol. 39:1345-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Tudela, J. L., E. Chryssanthou, E. Petrikkou, J. Mosquera, D. W. Denning, and M. Cuenca-Estrella. 2003. Interlaboratory evaluation of hematocytometer method of inoculum preparation for testing antifungal susceptibilities of filamentous fungi. J. Clin. Microbiol. 41:5236-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Tudela, J. L., L. Alcazar-Fuoli, A. Alastruey-Izquierdo, A. Monzon, E. Mellado, and M. Cuenca-Estrella. 2007. Time of incubation for antifungal susceptibility testing of Aspergillus fumigatus: can MIC values be obtained at 24 hours? Antimicrob. Agents Chemother. 51:4502-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinbach, W. J., J. R. Perfect, W. A. Schell, T. J. Walsh, and D. K. Benjamin, Jr. 2004. In vitro analyses, animal models, and 60 clinical cases of invasive Aspergillus terreus infection. Antimicrob. Agents Chemother. 48:3217-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trama, J. P., E. Mordechai, and M. E. Adelson. 2005. Detection of Aspergillus fumigatus and a mutation that confers reduced susceptibility to itraconazole and posaconazole by real-time PCR and pyrosequencing. J. Clin. Microbiol. 43:906-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.