Abstract
Enterococcus faecalis N06-0364, exhibiting a vancomycin MIC of 8 μg/ml, was found to harbor a novel d-Ala-d-Ser gene cluster, designated vanL. The vanL gene cluster was similar in organization to the vanC operon, but the VanT serine racemase was encoded by two separate genes, vanTmL (membrane binding) and vanTrL (racemase).
Among enterococci, there are currently three characterized d-Ala-d-Ser operons that confer low-level vancomycin resistance, vanC, vanE, and vanG (1, 5, 6, 10, 14). The vanC operon is intrinsic to Enterococcus gallinarum (vanC) and Enterococcus casseliflavus/flavescens (vanC2/3), while vanE and vanG have been acquired by a small number of Enterococcus faecalis strains (21). Resistance is achieved by essentially the same mechanism in the three types: the ligases, encoded by vanC, vanE, or vanG, catalyze the formation of d-Ala-d-Ser, which is incorporated into peptidoglycan precursors, which subsequently have a low binding affinity for vancomycin. The vanC and vanE operons are organized similarly, vanC or vanE, vanXY (d,d-dipeptidase-d,d-carboxypeptidase), vanT (serine racemase), vanR (response regulator), and vanS (sensor histidine kinase) (1, 5, 14). The vanG operon begins with a three-component regulatory system, vanU (transcriptional regulator), vanR, vanS, vanY (d,d-carboxypeptidase) in the vanG operon which is absent in the vanG2 operon, vanW (unknown function), and then vanG, vanXY, and vanT (6, 10). In this report we describe a novel d-Ala-d-Ser gene cluster, designated vanL.
The primers used in this study are listed in Table 1. PCR and thermal asymmetric interlaced PCR (TAIL-PCR) were carried out with AmpliTaq Gold (Applied Biosystems, Foster City, CA) or the FailSafe PCR system (Epicenter Biotechnologies, Madison, WI) as previously described (6, 7, 12, 22). DNA sequencing was carried out by the National Microbiology Laboratory's Genomics Core Facility. Homology comparisons were made using the BLAST suite of programs at the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov/BLAST/). Vancomycin induction was studied as previously described (19). E. faecalis JH2-2 was used as a recipient in filter mating experiments with selection for transconjugants on 50 μg/ml rifampin, 100 μg/ml fusidic acid, and 2 μg/ml vancomycin. Antimicrobial susceptibilities were determined using broth microdilution according to CLSI guidelines (9) and by Etest (AB Biodisk, Solna, Sweden).
TABLE 1.
Primers used in this study
| Name | Sequence (5′ to 3′)a | Coordinates (bp)b | PCR no. (strand)c | Reference |
|---|---|---|---|---|
| vanA1 | GGGAAAACGACAATTGC | NAd | NA | 11 |
| vanA2 | GTACAATGCGGCCGTTA | NA | NA | 11 |
| vanB-F | AAGCTATGCAAGAAGCCATG | NA | NA | 12 |
| vanB-R | CCGACAATCAAATCATCCTC | NA | NA | 12 |
| ddlFm-1 | ATTACAAAGGCAGAAAACCG | NA | NA | This study |
| ddlFm-2 | TGTCAAAAAGAAATCGCACC | NA | NA | This study |
| ddlFs-1 | TTATTTTGTTGTATGGCGGC | NA | NA | This study |
| ddlFs-2 | AAAGTCAGTAAAACCAGGCA | NA | NA | This study |
| vanC1-93 | GAAAGACAACAGGAAGACCGC | NA | NA | 8 |
| vanC2-93 | ATCGCATCACAAGCACCAATC | NA | NA | 8 |
| vanD-U1 | TATTGGAATCACAAATCCGG | NA | NA | 7 |
| vanD-U2 | CGGCTGTGCTTCCTGATG | NA | NA | 7 |
| VANE1 | TGTGGTATCGGAGCTGCAG | NA | NA | 14 |
| VANE2 | GTCGATTCTCGCTAATCC | NA | NA | 14 |
| vanG5 | TTCGATTTCATCAACTCTGC | NA | NA | This study |
| vanG6 | CAGGAATACCTGTTGTTGG | NA | NA | This study |
| AD5 | NTCGASTWTSGWGTT | NA | 1 (+), 9 (−) | 22 |
| 364U4 | TTTCCGCCTAAATCAATTCC | 1081-1100 | 1 (−) | This study |
| 364U5 | GAGTTTCTAGTACACTTACTG | 1028-1048 | 1 (−) | This study |
| 364U6 | ATATTCCGAAGATTGACCTCC | 988-1008 | 1 (−) | This study |
| AD3 | AGWGNAGWANCAWAGG | NA | 2 (+) | 22 |
| 364U1 | AGCATCCTCTAGCTTATCTGG | 1537-1557 | 2 (−) | This study |
| 364U2 | TAGTGCGAATTATGAGCGTAG | 1409-1429 | 2 (−) | This study |
| 364U3 | CACTCTGAGCAAACTCATGC | 1371-1390 | 2 (−) | This study |
| V3 | GARGATGGITSCATMCARGGW | 1274-1293 | 3 (+) | 15 |
| V4 | MGTRAAICCIGGCAKRGTRTT | 1882-1902 | 3 (−) | 15 |
| 364D4 | AGCAAAGGTAAATATACCTGC | 1728-1748 | 4 (+) | This study |
| vanXY-U | CCNACRTANCKRAARTGCCA | 2468-2487 | 4 (−) | This study |
| vanXYL-D3 | GCACATGATATCATAGCACC | 2336-2355 | 5 (+) | This study |
| vanT-U | TDATNVDNGCNGGNARRTACCA | 2941-2963 | 5 (−) | This study |
| vanTL-D1 | TCTATCACTTTGGATATGTC | 2869-2888 | 6 (+) | This study |
| vanT-U2 | CCNARNCKRTGCAMNCCNGTRTC | 3939-3961 | 6 (−) | This study |
| vanTL-D2 | CTTATCCATTACGAACTTACC | 3843-3873 | 7 (+) | This study |
| RserU1 | AYRTCNGGNARCATNACRTC | 4887-4906 | 7 (−) | This study |
| 364RD1 | AAGGGTTTCAAGTAACAACC | 4804-4823 | 8 (+) | This study |
| SerU1 | GGNGTNYKNARRTCRTGNGC | 5863-5882 | 8 (−) | This study |
| 364SD4 | GAAGGGATATTGATTATCGGTGTG | 5605-5628 | 9 (+) | This study |
| 364SD5 | CCAGAACAGCGTGTTAAGCTATC | 5725-5747 | 9 (+) | This study |
| 364SD1 | TAATGCTGACAATCGCGCAG | 5799-5818 | 9 (+) | This study |
| ISvL-D1 | ATGGACTATTACGAGAGTACC | 7481-7502 | 10 (+) | This study |
| ISvL-D2 | TGATTCACAGGCTCCTTAGC | 7524-7543 | 10 (+) | This study |
| ISvL-D3 | TCAGAAGTTCAGTGAAGCTAC | 7568-7588 | 10 (+) | This study |
| AD1 | NGTCGASWGANAWGAA | NA | 10 (−) | 22 |
K is G or T, M is A or C, R is A or G, S is G or C, W is A or T, Y is C or T, D is A, G, or T, V is A, C, or G, I is inosine, and N is any base.
Coordinates are from accession no. EU250284.
PCR number as indicated in Fig. 1.
NA, not applicable.
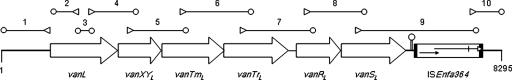
E. faecalis N06-0364 (vancomycin MIC, 8 μg/ml) was isolated from a single patient after a screen for vancomycin-resistant enterococci was carried out 2 days after hospitalization for a total hip resection. No vancomycin-resistant enterococci were isolated from this patient or any other in the hospital on follow-up screening 7 days later. E. faecalis N06-0364 was susceptible to teicoplanin, ampicillin, fluoroquinolones, erythromycin, gentamicin, streptomycin, linezolid, and tetracyclines. PCR analysis was positive for ddlE. faecalis but negative for ddlE. faecium, vanA, and vanB in a multiplex PCR (12). PCRs for vanD, vanC, vanE, and vanG-type ligases were negative. A ∼600-bp product amplified with the V3/V4 degenerate primers for d-Ala-d-Xxx ligases (15) was sequenced, and its putative translation product exhibited 57% identity to the VanC ligase. Using DNA sequence alignments of homologous genes from existing d-Ala-d-Ser operons, a number of degenerate primers from conserved regions were designed (Table 1). These primers and gene-specific primers were used in PCRs to obtain a total region of ∼4 kb downstream of the V3/V4 PCR product (Fig. 1). An additional ∼1.2-kb region upstream and ∼2.5-kb region downstream were obtained by TAIL-PCR (Fig. 1). All products were sequenced to generate 8,295 bp of contiguous DNA sequence from E. faecalis N06-0364 (Fig. 1).
FIG. 1.
Schematic diagram of the VanL operon and flanking regions. Regions amplified and sequenced are indicated at the top and are numbered (see Table 1), with a triangle indicating a sequence-specific primer and an oval indicating a degenerate primer. The putative stem-loop region between vanSL and ISEnfa364 is shown. The inverted repeats of ISEnfa364 are indicated by black bars, the two overlapping reading frames coding for tnpA are shown as arrows, and the in-frame stop codon is indicated by a vertical line. Coordinates are from GenBank accession number EU250284.
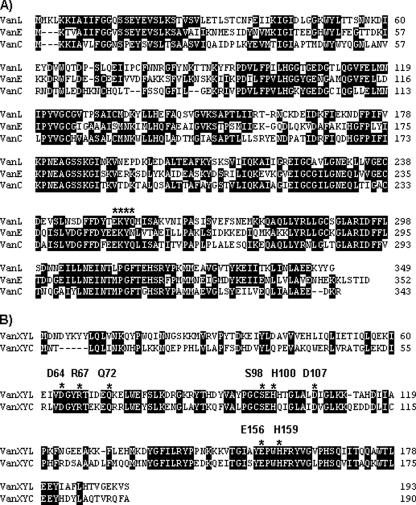
Analysis of the genes and their putative proteins indicated the presence of a novel d-Ala-d-Ser gene cluster, designated vanL. The percent identities of the vanL gene cluster proteins with the corresponding proteins from other d-Ala-d-Ser operons are shown in Table 2. The vanL gene cluster was organized essentially the same as the vanC and vanE operons; however, serine racemase activity appears to be encoded by two genes, vanTmL and vanTrL (discussed below). The putative VanL exhibited 51 and 49% identity to the VanE and VanC ligases, respectively, and the EKYQ motif involved in substrate recognition was conserved at residues 252 to 255 of VanL (Fig. 2A) (13).
TABLE 2.
Extents of identity of proteins from the VanL operon with the corresponding proteins from other D-Ala-D-Ser operons
| Protein | % Identity with protein from other indicated operon
|
|||||
|---|---|---|---|---|---|---|
| VanL | VanXYL | VanTmLa | VanTrLa | VanRL | VanSL | |
| VanC | 49 | 46 | 45 | 51 | 72 | 52 |
| VanE | 51 | 41 | 44 | 53 | 59 | 39 |
| VanG1 | 42 | 38 | 35 | 41 | 60 | 40 |
| VanG2 | 42 | 38 | 34 | 40 | 62 | 40 |
As aligned with the corresponding domain of VanT proteins.
FIG. 2.
Alignment of the VanL, VanE, and VanC proteins (A) and the VanXYL and VanXYC proteins (B). Identical residues are boxed in black. The EKYQ motif putatively involved in substrate binding is overlined by asterisks in panel A. The residues conserved in VanX, VanY, and VanXY proteins are indicated in panel B.
The putative VanXYL exhibited 46% identity to VanXYC and contained all the conserved active site residues of other VanXY proteins and also those found in VanX and VanY proteins (Fig. 2B) (20).
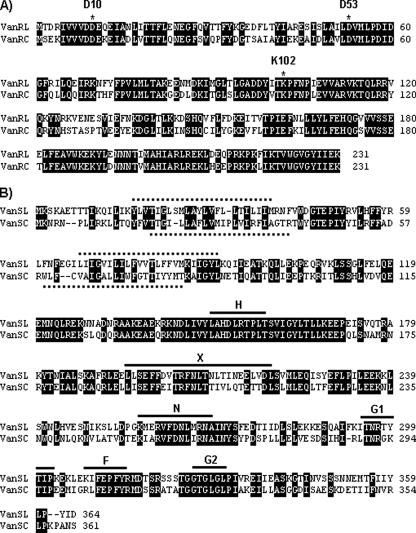
The third and fourth open reading frames, vanTmL and vanTrL, respectively, appear to encode serine racemase activity (Fig. 1). vanTmL and vanTrL are in different reading frames, and a single-base insertion between bases 3497 and 3564 (coordinates are from accession no. EU250284) would lead to a single 2,085-bp putative vanTL gene. Whether extant vanT genes arose in this way or whether vanTmL and vanTrL evolved by a deletion in a vanTC-like gene is purely speculative. An alignment of VanTmL and VanTrL with the VanTC protein is shown in Fig. 3. VanTmL exhibits 45% identity with the VanTC N-terminal region, which is postulated to function in the transport of l-serine (2, 3). The VanTrL protein exhibits 51% identity to the VanTC C-terminal region and shares all the residues important in structure and function of pyridoxal 5′-phosphate-dependent alanine racemases (2) (Fig. 3). Since l-serine transport and racemase activities are not codependent for function (3), having these two domains separately encoded would not necessarily compromise VanT activity in E. faecalis N06-0364. The vanRL and vanSL genes follow vanTrL (Fig. 1). Alignments of VanRL and VanRC and of VanSL and VanSC are shown in Fig. 4. The deduced VanRL exhibited 72% identity to VanRC and contained the conserved residues (D10, D53, and K102) found in response regulators from gram-positive two-component systems (Fig. 4A) (18). The deduced VanSL exhibited 52% identity to VanSC and contained the six conserved amino acid motifs (H, X, N, G1, F, and G2 boxes) of the C-terminal transmitter module of VanS proteins (16) (Fig. 4B). In addition, the VanSL N-terminal region contained two transmembrane regions characteristic of the sensor proteins in two-component systems (Fig. 4B) (4). VanSL has none of the substitutions (R200, D312, and G320) shown in VanSC to be associated with constitutive vancomycin resistance (19). This is consistent with growth experiments which revealed that vancomycin resistance was inducible in E. faecalis N06-0364 (data not shown). Blast searches of the GenBank database with the ∼950 bp upstream of vanL failed to return any matches. Beginning 110 bp downstream of vanSL is a 61-bp region with the potential to form a large stem-loop structure with a ΔG of −15.64 kcal/mol which may act as a transcriptional terminator (Fig. 1). Inserted 5 bp downstream of the putative stem-loop region is a 1,055-bp insertion sequence, designated ISEnfa364, with ends that are defined by 25-bp inverted repeats (IR) and that is most closely related to elements in the IS30 family (17). The ISEnfa364 transposase appears to be coded for by two overlapping reading frames whose products would presumably become linked through translational frameshifting (17). However, an in-frame stop codon would presumably lead to an inactive truncated protein. Typically, IS30 family members are flanked by short 2- to 4-bp direct repeats created upon insertion (17). ISEnfa364 is flanked by a single adenine residue. However, it was noted that the 4 bp flanking IR-L, ATGA, are repeated 2 bp downstream of IR-R.
FIG. 3.
Alignment of VanTC with the VanTmL and VanTrL proteins. Identical residues are boxed in black, hydropobic transmembrane domains are indicated by solid lines, and the pyridoxal phosphate attachment site (PPAS) is labeled.
FIG. 4.
Alignments of the VanRLand VanRC proteins (A) and the VanSL and VanSC proteins (B). Identical residues are boxed in black. The conserved aspartate and lysine residues of gram-positive response regulators are labeled in panel A. The transmembrane regions are indicated by hatched lines, and the conserved amino acid motifs H, X, N, G1, F, and G2 of VanS proteins are labeled and indicated by solid lines in panel B.
The vanL region has a G+C content of 32%, while the E. faecalis genome has a G+C content of 38%, indicating acquisition from an organism with a more-AT-rich genome.
Despite several attempts, transfer of the vanL gene cluster could not be demonstrated in mating experiments with E. faecalis JH2-2 as a recipient. Plasmids were not visualized on gels from multiple plasmid preparations from E. faecalis N06-0364 (data not shown). It appears likely that the vanL gene cluster is located in the chromosome. All acquired d-Ala-d-Ser gene clusters identified to date, vanE, vanG, and vanG2, are chromosomally located (5, 6, 10, 14). Definitive proof of vanL functionality awaits cloning and transfer experiments and biochemical analysis of cell wall precursors.
vanL expands the so-called van alphabet and highlights the importance of characterization of E. faecalis and Enterococcus faecium isolates exhibiting low-level vancomycin resistance. As both the origin of the vanL gene cluster by this strain and the way this strain was acquired by this patient are unknown, the clinical significance of this finding remains to be established.
Nucleotide sequence accession number.
The vanL gene cluster characterized in this study has been assigned accession number EU250284 in the GenBank Database.
Acknowledgments
We gratefully acknowledge the expert technical assistance of Romeo Hizon.
Footnotes
Published ahead of print on 5 May 2008.
REFERENCES
- 1.Arias, C. A., P. Courvalin, and P. E. Reynolds. 2000. vanC cluster of vancomycin-resistant Enterococcus gallinarum BM4174. Antimicrob. Agents Chemother. 46:95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias, C. A., M. Martin-Martinez, T. L. Blundell, M. Arthur, P. Courvalin, and P. E. Reynolds. 1999. Characterization and modelling of VanT: a novel, membrane-bound, serine racemase from vancomycin-resistant Enterococcus gallinarum BM4174. Mol. Microbiol. 31:1653-1664. [DOI] [PubMed] [Google Scholar]
- 3.Arias, C. A., J. Peña, D. Panesso, and P. Reynolds. 2003. Role of the transmembrane domain of the VanT serine racemase in resistance to vancomycin in Enterococcus gallinarum BM4174. J. Antimicrob. Chemother. 51:557-564. [DOI] [PubMed] [Google Scholar]
- 4.Arthur, M., C. Molinas, and P. Courvalin. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 173:2582-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D. A., T. Cabral, P. Van Caseele, J. Wylie, and M. R. Mulvey. 2002. Molecular characterization of the vanE gene cluster in vancomycin-resistant Enterococcus faecalis N00-410 isolated in Canada. Antimicrob. Agents Chemother. 46:1977-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, D. A., T. Du, R. Hizon, B. Kaplen, T. Murphy, S. Tyler, S. Brown, F. Jamieson, K. Weiss, M. R. Mulvey, et al. 2006. VanG-type vancomycin-resistant Enterococcus faecalis strains isolated in Canada. Antimicrob. Agents Chemother. 50:2217-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, D. A., P. Kibsey, D. Roscoe, and M. R. Mulvey. 2004. Enterococcus faecium N03-0072 carries a new VanD-type vancomycin resistance determinant: characterization of the VanD5 operon. J. Antimicrob. Chemother. 54:680-683. [DOI] [PubMed] [Google Scholar]
- 8.Clark, N. C., R. C. Cooksey, B. C. Hill, J. M. Swenson, and F. Tenover. 1993. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob. Agents Chemother. 37:2311-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Depardieu, F., M. G. Bonora, P. E. Reynolds, and P. Courvalin. 2003. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 50:931-948. [DOI] [PubMed] [Google Scholar]
- 11.Dukta-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptides resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elsayed, S., N. Hamilton, D. Boyd, and M. Mulvey. 2001. Improved primer design for multiplex PCR analysis of vancomycin-resistant Enterococcus spp. J. Clin. Microbiol. 39:2367-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evers, S., B. Casadewall, M. Charles, S. Dutka-Malen, M. Galimand, and P. Courvalin. 1996. Evolution of structure and substrate specificity in D-alanine:D-alanine ligases and related enzymes. J. Mol. Evol. 42:706-712. [DOI] [PubMed] [Google Scholar]
- 14.Fines, M., B. Périchon, P. Reynolds, D. F. Sahm, and P. Courvalin. 1999. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob. Agents Chemother. 43:2161-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gold, H. S., S. Unal, E. Cercenado, C. Thauvin-Eliopoulos, G. M. Eliopoulos, C. B. Wennersten, and R. C. Moellering, Jr. 1993. A gene conferring resistance to vancomycin but not teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes of enterococci. Antimicrob. Agents Chemother. 37:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsing, W., F. D. Russo, K. K. Bernd, and T. J. Silhavy. 1998. Mutations that alter kinase and phosphatase activities of the two-component sensor EnvZ. J. Bacteriol. 180:4538-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Msadek, T., F. Knust, and G. Rapopport. 1993. Two-component regulatory systems, p. 729-745. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC.
- 19.Panesso, D., L. Abadía-Patiño, N. Vanegas, P. E. Reynolds, P. Courvalin, and C. A. Arias. 2005. Transcriptional analysis of the vanC cluster from Enterococcus gallinarum strains with constitutive and inducible vancomycin resistance. Antimicrob. Agents Chemother. 49:1060-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds, P. E., C. A. Arias, and P. Courvalin. 1999. Gene vanXYC encodes D,D-dipeptidase (VanX) and D,D-carboxypeptidase (VanY) activities in vancomycin-resistant Enterococcus gallinarum BM4174. Mol. Microbiol. 34:341-349. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds, P. E., and P. Courvalin. 2005. Vancomycin resistance in enterococci due to synthesis of precursors terminating in d-alanyl-d-serine. Antimicrob. Agents Chemother. 49:21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singer, T., and E. Burke. 2003. High-throughput TAIL-PCR as a tool to identify DNA flanking insertions. Methods Mol. Biol. 236:241-271. [DOI] [PubMed] [Google Scholar]