Abstract
Variations in Hox protein sequences and functions have been proposed to contribute to evolutionary changes in appendage shape and number in crustaceans and insects. One model is that insect Hox proteins of the Ultrabithorax (UBX) ortholog class evolved increased abilities to repress Distal-less (Dll) transcription and appendage development in part through the loss of serine and threonine residues in casein kinase 2 (CK2) phosphorylation sites. To explore this possibility, we constructed and tested the appendage repression function of chimeric proteins with insertions of different CK2 consensus sites or phosphomimetics of CK2 sites in C-terminal regions of Drosophila melanogaster UBX. Our results indicate that CK2 sites C-terminal to the homeodomain can inhibit the appendage repression functions of UBX proteins, but only in the context of specific amino acid sequences. Our results, combined with previous findings on evolutionary changes in Hox protein, suggest how intra-protein regulatory changes can diversify Hox protein function, and thus animal morphology.
Keywords: Ultrabithorax, Evolution, Phosphorylation, Casein kinase 2, Homeotic, Transcription factor, Hox function, Hox modification
Introduction
Members of the Hox gene family contribute to the morphological diversification of structures that develop from the anterior–posterior axis on triploblastic animal embryos (McGinnis and Krumlauf 1992; Carroll et al. 2005). The different Hox genes encode homeodomain proteins that bind to cis-regulatory DNA sites, and they diversify morphology by the differential regulation of downstream target genes (Pearson et al. 2005). There is a growing body of correlative evidence that changes at many levels in Hox genetic pathways have contributed to the evolution of morphological diversity in triploblastic animals. These include changes in Hox expression patterns (Averof and Akam 1995; Carroll et al. 2005), changes in the regulation of Hox target genes (Jeong et al. 2006), changes in Hox protein function (Hsia and McGinnis 2003), and perhaps even changes in the number and variety of Hox genes in different animal lineages (Lemons and McGinnis 2006). However, there are few examples where experimental evidence supports strong associations between (a) specific evolutionary variations in Hox coding or regulatory DNA sequences, and (b) evolutionary changes in animal axial morphology that might plausibly have been influenced by the specific coding and/or regulatory sequence variations.
As regards Hox protein functional evolution, one of the best current examples is found in the evolutionary history of the Drosophila fushi tarazu (ftz) gene. Insect ftz appears to have evolved from a duplicated and diverged Antennapedia-like gene (a Hox gene of the Hox5/6 ortholog class) during an approximately 300-million-year period while crustacean and insect lineages were diversifying (Telford 2000; Papillon and Telford 2007). In Drosophila, the FTZ protein is expressed in multiple stripes and provides a pair-rule segmentation function in combination with a different sequence-specific DNA binding cofactor called FTZ-F1. In more “basal” insects and crustaceans, the ancestral version of FTZ is expressed in a Hox-like pattern (Hughes and Kaufman 2002; Papillon and Telford 2007).
During arthropod evolution, the loss of a Hox/homeotic function and a Hox-like expression pattern for the progenitor of Drosophila FTZ protein is correlated with the loss of a motif that contains variants of a tyrosine-proline-tryptophan-methionine (YPWM) amino acid sequence (Lohr et al. 2001; Lohr and Pick 2005). In many Hox proteins, the YPWM sequence is part of a key interaction surface with the EXD/PBX class of Hox cofactors (Mann and Carroll 2002). In contrast, the gain of segmentation function and/or a pair-rule-like segmentation expression pattern, in a Drosophila FTZ progenitor, is roughly correlated with the gain of a leucine-any-any-leucine-leucine (LXXLL) amino acid motif. Gain of an LXXLL motif is predicted to result in increased interactions with the FTZ segmentation cofactor FTZ-F1 (Schwartz et al. 2001; Yussa et al. 2001). These changes, along with the evolution of cis-regulatory sequences that deployed the FTZ protein in seven stripes in early embryos, contributed to the evolution of Drosophila FTZ from an apparent Hox precursor (Lohr and Pick 2005; Papillon and Telford 2007).
In Drosophila embryos, nascent limb primordia in the abdomen are repressed by the Hox proteins UBX and abdominal-A (ABD-A). In Drosophila and many hexapods, this is associated with complete transcriptional repression of the Distal-less (Dll) gene in embryonic abdominal cells that express UBX or ABD-A protein. There are exceptions to this generalization. Staining for an antigen common to the UBX and ABD-A proteins indicates that one or both proteins are expressed in the embryonic first abdominal (A1) segments of Folsomia candida (a springtail), Shistocerca americana (a grasshopper), and Tribolium castaneum (a beetle), and that these proteins do not completely repress Dll transcription in A1 (Palopoli and Patel, 1998; Bennett et al. 1999; Lewis et al. 2000). The Dll transcription observed in the A1 appendage primordia of these three hexapods is lower than in thoracic appendage primordia, and the resulting A1 appendages are smaller than the legs that develop in the thorax. In addition, Tribolium larvae that are Ubx mutants develop larger, leg-like appendages in A1. The simplest interpretation of all these data is that the Ubx gene partially represses Dll in the first abdominal segment of Folsomia, Schistocerca, and Tribolium. However, this does not necessarily imply that the UBX protein in these species is a poorer Dll or limb repressor, as subtle differences in the timing and amounts of UBX protein expression can result in dramatically different repressive effects on Dll transcription and limb development (Castelli-Gair and Akam, 1995; Tour et al. 2005).
UBX proteins of some non-hexapod arthropods are expressed in embryonic limb primordia but provide little or no repression of Dll and appendage development (Panganiban et al. 1995; Averof and Akam 1995; Grenier et al. 1997; Hughes and Kaufman 2002). In ectopic expression assays in Drosophila embryos, the reduced limb repression function of non-hexapod UBX proteins has been associated with two changes in protein sequence relative to hexapod UBX. One change is the presence of C-terminal serine and threonine residues, some of which map in consensus casein kinase 2 (CK2) phosphorylation sites (Jaffe et al. 1997; Ronshaugen et al. 2002; Shiga et al. 2002). However, it is not known whether CK2 sites in other amino acid contexts, or highly acidic regions that constitute phosphomimetics of CK2 sites, are sufficient to inhibit the ability of Hox proteins to repress Dll and appendage development in embryos. Another change associated with reduced UBX repression function is the absence of an alanine-rich region in C-terminal sequences (Galant and Carroll 2002). Deletions of the alanine-rich region have indicated that it provides part of the Drosophila UBX limb repression function, although mutants lacking the repeat develop to adulthood with no ectopic limbs (Hittinger et al. 2005). One reason for this weak phenotype, as shown by the rigorous genetic experiments of Hittinger et al. (2005), is that the abdominal limb repressive function encoded in the UBX glutamine-alanine repeat is redundantly supplied by the ABD-A Hox protein, which is expressed in many potential abdominal limb primordia with UBX protein.
To provide more insight into the evolution of UBX protein functions, we wished further assay the function of CK2 sites in the regulation of UBX repression of limb development. Therefore, we tested whether CK2 phosphorylation sites of different strengths, and in different sequence contexts, influenced the repressive function of Drosophila UBX on Dll transcription and embryonic appendage development.
Materials and methods
Recombinant constructs and transgenic flies
Drosophila melanogaster UBX (Dm-UBX) expression constructs were made by polymerase chain reaction (PCR) amplification using synthetic oligonucleotides, and were verified by DNA sequencing. The oligonucleotides used to generate chimeras Dm-UBX DE, SE, AE, and TE were as follows: Dm-UBX DE: CTG AAC GAA CAG GAG AAG GCC GCC GCC ACT GCT GCC GCG GAC AAG GCC GAC GAG GAG GAC GAT GAT GAA GAA GAG GAA CAA GGT GGA CAC TTA GAT, Dm-UBX SE: CTG AAC GAA CAG GAG AAG TCC GTT TCC ACA GCT GCT GAC AAG GCG GAC GAG GAG GAA GAG GAG GAA GAG GAG GAA GAA CAA GGT GGA CAC TTA GAT, Dm-UBX AE: CTG AAC GAA CAG GAG AAG GCC GTT GCC GCA GCT GCT GAC AAG GCG GAC GAG GAG GAA GAG GAG GAA GAG GAG GAA GAA CAA GGT GGA CAC TTA GAT, and Dm-UBX TE: CTG AAC GAA CAG GAG AAG ACC GCC GAC AGC CTG GGC GGA AAA GAG GAA AAG CGG GAA GAG ACA GAA GAG GAG AAG CAA GGT GGA CAC TTA GAT. A second PCR reaction was used to incorporate codons for the hemagglutinin antigen (HA) at the 3′ end of the Dm-UBX open reading frame sequences using the primer CAA GGT GGA CAC TTA GAT CAG TAC CCA TAC GAC GTC CCA GAC TAC GCT TAG. For the generation of transgenic Drosophila, chimeric gene constructs were sub-cloned into EcoRI/XhoI sites of the GAL4-inducible vector pUAST (Brand et al. 1994). Constructs were injected into white1118 embryos, and multiple transgenic lines were established and tested for ectopic protein expression levels and regulatory phenotypes. To induce ectopic expression in embryos, flies homozygous for UAS-chimera transgenes were crossed to flies homozygous for the arm-GAL4 driver, which provides ubiquitous GAL4 protein in embryos from stage 9 and thereafter. We report here only embryos from crosses that ectopically expressed the Dm-UBX chimeric proteins at near physiological levels to normal Dm-UBX in limb primordia, but other tested lines for each construct showed phenotypes consistent with those reported in this paper.
Cuticle preparations and counts of Keilin’s organs
To analyze the effect of chimeric UBX protein function on larval cuticular phenotypes, embryos that ectopically expressed various constructs were collected for 12 h and aged for an additional 30–35 h. After dechorionation in 50% bleach for 3 min and devitellinization with an equal volume mix of heptane and methanol, larval cuticles were mounted in Hoyer’s mounting media. For each line, 16 embryos were scored for the number of hairs of Keilin’s organs. Since the numbers of developing Keilin’s organ hairs per larvae were usually not a close fit to a normal distribution, we have reported the simple percentage of total Keilin’s organs developed by 15 larvae that ectopically expressed different variants of Dm-UBX.
In situ hybridization, immunostaining of embryos, and protein expression levels
In situ hybridizations to detect Ubx, Dll, Antp, en, and dpp transcripts were performed as described in Kosman et al. (2004). Ectopic levels of chimeric protein expression were detected as in Tour et al. (2005), first using either rat monoclonal antibodies (1:500 dilution) directed against the hemagglutinin antigen (anti-HA) from Roche, or mouse monoclonal antibodies FP3.38 (1:20 dilution) against a UBX protein epitope. The anti-HA and FP3.38 antibodies were then visualized with secondary antiserum, either anti-rat IgG or anti-mouse IgG (1:500 dilution) conjugated with Alexa Fluor 555 dye (Invitrogen), respectively. Unsaturated images of stage 11 embryos were taken using a Leica SP2 confocal microscope. Using the Leica Confocal software, we then compared protein staining intensities by measuring the average levels of pixel intensity in three areas in 16 embryos, encompassing the cells that accumulate Dll transcripts in the first, second, and third thoracic segments. The average and the standard error were calculated using Microsoft Excel software.
CK2 kinase assays
Protein coding sequences corresponding to different cDNA constructs were cloned into Sma1 restriction sites in pGEX-4T-1 to generate glutathione S-transferase (GST) fusion constructs, which were transformed into the E. coli BL21 strain. GST fusion proteins were induced and purified according to the manufacturer’s instructions (Pharmacia). In kinase assays, 1–3 μg of purified recombinant GST-Af-UBX, GST-Dm-UBX, GST-Dm-UBXΔQA, GST-Dm-UBX SE, GST-Dm-UBX AE, GST-Dm-UBX DE, and GST-Dm-UBX TE fusions proteins were used as substrates for in vitro CK2 phosphorylation. Kinase reactions were performed at 30°C for 30 min with 50 U of human CK2 (NEBiolabs) and 10 μCi of γ-32P-labeled ATP. SDS-polyacrylamide gels were used to separate reaction products, and gels were stained with Coomassie blue, washed in 1× destain buffer (40% methanol, 10% acetic acid), and dried. Dried gels were exposed to X-ray film for approximately 1 h. Relative protein amounts per Coomassie stained band and relative 32P incorporation per band were obtained by measuring the number and the intensity of pixels using the NIH Image program.
Results
Drosophila UBX chimeras
Although the homeodomain regions, YPWM motifs, and a few other blocks of sequence similarity are highly conserved among animal Hox orthologs, other amino acid sequences in Hox orthologs diverge between distant phyla. For example, the UBX1A ortholog of D. melanogaster and a UBX ortholog from a crustacean, Artemia franciscana, differ at more than half of their 300–400 amino acid residues and have many insertions/deletions relative to each other (Ronshaugen et al. 2002). A variety of experimental evidence supports the idea that casein kinase 2 site variation between distant Hox orthologs as well as Hox paralogs modifies their regulatory functions (Jaffe et al. 1997; Ronshaugen et al. 2002; Shiga et al. 2002; Hsia and McGinnis 2003). For example, a UBX protein of A. franciscana (Af-UBX) has an inhibitory region in the C terminus that includes a match to a consensus casein kinase 2 (CK2) phosphorylation site, the amino acid motif serine-aspartate-aspartate-glutamate (SDDE), with S the predicted phosphorylation site. When the Af-UBX C-terminal domain was substituted for the C-terminus of the Drosophila UBX protein, the resulting chimera had a reduced ability to repress Dll and embryonic limb development in Drosophila embryos, similar to the reduced repressive abilities of full-length Af-UBX when expressed in Drosophila embryos (Ronshaugen et al. 2002).
To test whether the consensus Af-UBX CK2 consensus site is indeed a substrate for CK2, we performed in vitro CK2 kinase assays with full-length wild-type Af-UBX protein, as well as a control Af-UBX protein in which the SDDE sequence was mutated to ADDE. As seen in Fig. 1b, increasing amounts of wild-type Artemia UBX are phosphorylated in vitro by CK2 in a substrate concentration-dependent fashion, while phosphorylation of the Artemia UBX ADDE mutant by CK2 was comparatively poor.
Fig. 1.
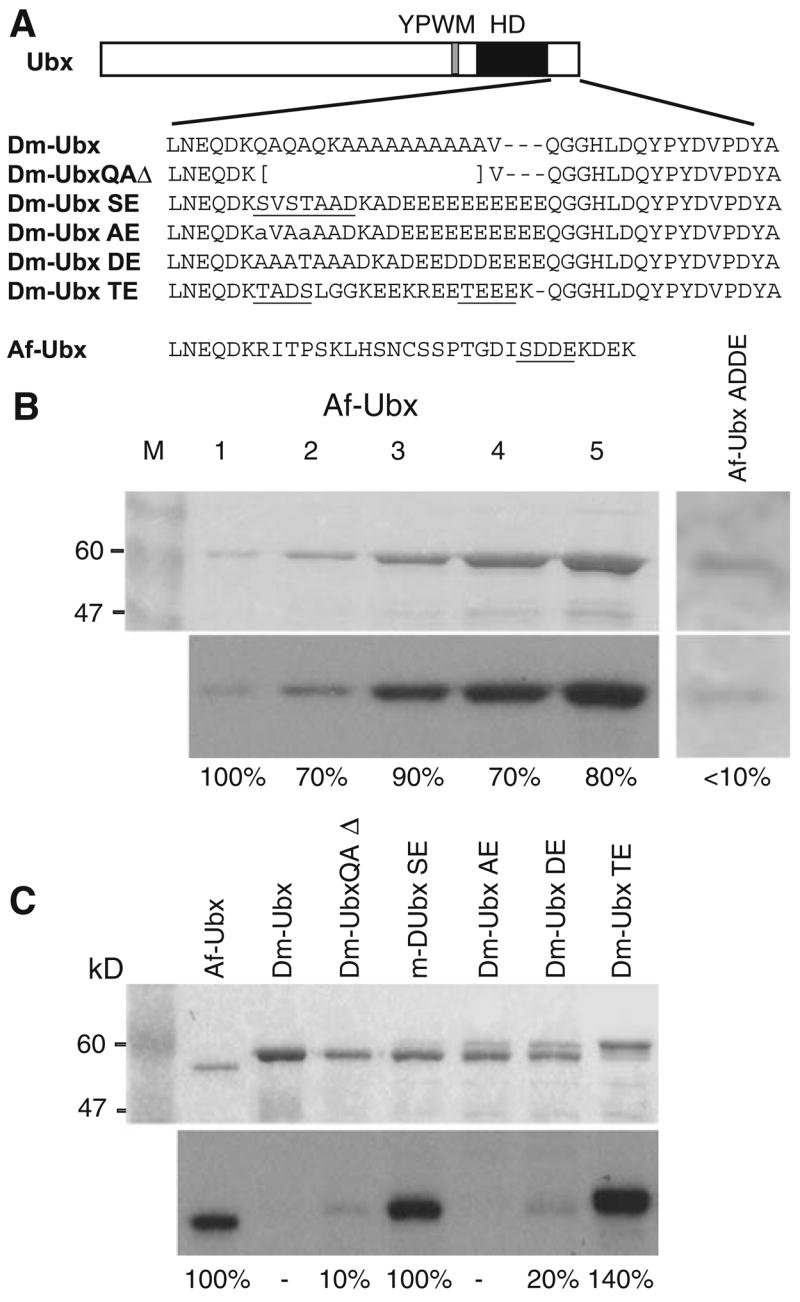
Drosophila melanogaster UBX (Dm-UBX) chimeric proteins. a At top is a diagram of the Dm-UBX protein, with the positions of the YPWM motif and homeodomain (HD) indicated. The C-terminal sequence of Dm-UBX and the substituted amino acid sequences in the chimeras are shown. The predicted CK2 sites are underlined. Af-UBX: Artemia franciscana UBX C-terminal sequence. b Af-UBX is a substrate for CK2 in vitro. Kinase reactions were performed as described in “Materials and methods”, using 0.5 U of CK2 enzyme per reaction. Reactions were then subjected to SDS-polyacrylamide gel electrophoresis. The top panel shows Coomassie staining of the gels, with the lanes containing increasing amounts of Af-UBX protein. Lanes 1–5 have 0.24, 0.54, 0.96, 1.5 and 3.6 μg, respectively. Lane M: protein size markers. The bottom panel in (b) shows an autoradiograph of the gel reflecting the amount of phosphorylation per protein band. The percentage number is the signal intensity per protein band normalized to the Af-UBX standard in lane 1, averaged to the nearest 10%, which was the approximate standard error over three measurements. The dose response indicates that the amount of CK2 enzyme used was not limiting in the protein concentration range of the phosphorylation reactions shown in this figure. c In vitro phosphorylation of the Dm-UBX chimeras. Kinase reactions were performed as described in “Materials and methods”. The top panel shows a Coomassie stained gel for the different Dm-UBX hybrid proteins; the bottom panel, an autoradiograph of the radiolabeled, phosphorylated proteins. Relative protein amounts and phosphorylation levels were estimated as described in (b)
The protein isoforms of wild-type Drosophila UBX have only one poor match to a consensus CK2 site (TTQD), near the N-terminus, and this is consistent with it being a poor substrate for CK2 in vitro (Fig. 1c). We constructed a set of chimeric proteins designed to test whether heterologous CK2 phosphorylation sites or a string of acidic amino acid residues without a CK2 site (potential phosphomimetic CK2 sites) were sufficient to inhibit the ability of Drosophila UBX to repress Dll and appendage development in the thorax of Drosophila embryos. Since CK2 optimal sites are serine or threonine residues embedded in strings of aspartate or glutamate residues, acidic amino acid repeats can often functionally mimic chains of phosphorylated CK2 sites (Ghose et al. 2004). The association of high affinity CK2 phosphorylation sites with adjacent acidic amino acid residues also means that the effects of acidic amino acid residues cannot be completely separated from CK2 phosphorylation itself.
The chimeric UBX proteins we constructed had the entire N-terminal region, homeodomain, UbdA region, and the extreme C-terminus of Drosophila UBX1A (hereafter called Dm-UBX). However, the chimeras had the middle of the Dm-UBX C-terminal region replaced with naturally evolved C-terminal Hox protein sequences from other animals (Fig. 1a). Naturally evolved C-terminal Hox sequences were used instead of synthetic sequences to reduce the possibility of amino acid sequences that would promote misfolding or degradation (Fig. 1a). The first chimera, Dm-UBX SE, contained a fragment of wild-type mouse HoxA7 protein sequence that encoded two consensus CK2 sites, which are predicted to be sequentially phosphorylated (Meggio and Pinna 2003). The second chimera, Dm-UBX AE, had the same extent of mouse HoxA7 sequence, but the serine and threonine codons of the consensus CK2 sites were mutated to alanine codons. The third chimera, Dm-UBX DE, had substituted C-terminal sequences from the human HOXA7 protein with no predicted CK2 sites, but had three aspartic acid residues embedded in a run of glutamic acid residues, providing at least one phosphomimetic of the SDDE CK2 site in the C-terminus of Af-UBX (Fig. 1a). The fourth chimeric protein, Dm-UBX TE, had substituted C-terminal sequences from the human HOXC6 protein, which included two consensus CK2 sites, one of high predicted affinity (TEEE, Fig. 1a).
In vitro CK2 phosphorylation of UBX chimeric proteins
We used in vitro kinase assays to determine the relative extent of phosphorylation of predicted CK2 sites in the chimeric proteins, setting A. franciscana UBX (Af-UBX) protein as a standard. In the dose response controls shown in Fig. 1b, CK2 was incubated with increasing amounts of Af-UBX protein to eliminate the possibility that the amount of enzyme was limiting in the concentration range of protein we used in vitro. Similar reaction conditions were then used for the different Dm-UBX hybrid proteins (Fig. 1c). The Dm-UBX protein with the C-terminal glutamine-alanine repeat deleted (Dm-UBXQAΔ) is phosphorylated at ~10% relative to Af-UBX, but at much higher levels that wt Dm-UBX, which is not detectably phosphorylated. The higher level of phosphorylation in the deleted version of Dm-UBX suggests that the glutamine-alanine region inhibits CK2 phosphorylation of Dm-UBX in some manner, perhaps by sequestering low affinity CK2 phosphorylation sites elsewhere in the Dm-UBX protein. The Dm-UBX SE protein is phosphorylated at a level comparable to Af-UBX. By comparison, the Dm-UBX AE protein, derived from Dm-UBX SE but with alanine substitutions in the predicted CK2 phosphorylation sites, was not detectably phosphorylated in vitro (Fig. 1c). Although it has no good matches to consensus CK2 sites (Meggio and Pinna 2003), Dm-UBX DE protein is phosphorylated by CK2 to a level of ~20% compared to Af-UBX. Finally, under these in vitro reaction conditions, the Dm-UBX TE protein is phosphorylated to approximately 140% the levels of the Af-UBX standard, making it the highest affinity protein target for CK2 that we tested (Fig. 1c).
Expression levels of chimeric proteins and assays for thoracic appendage development
Drosophila evolved from insects that developed walking appendages on thoracic segments during the transition from embryos to first larval (nymphal) stage. However, Drosophila larvae have highly reduced external thoracic appendages, called Keilin’s organs. The strong reduction in the external appendages of fly larvae presumably evolved as an adaptation that increased the efficiency of their burrowing feeding habits. In cuticular preparations, the Keilin’s organs are represented by a bump studded with three hairs (Fig. 2a,d). The embryonic cells that give rise to the Keilin’s organs are only part of the thoracic limb primordium, which also contains cells destined for the leg and wing imaginal disks (Bolinger and Boekhoff-Falk 2005). Ectopic expression of wild-type Dm-UBX transforms thoracic cuticular structures toward abdominal cuticle types, in the process repressing the development of Keilin’s organs, and the limb promoting gene, Distal-less (Gonzalez-Reyes and Morata 1990).
Fig. 2.
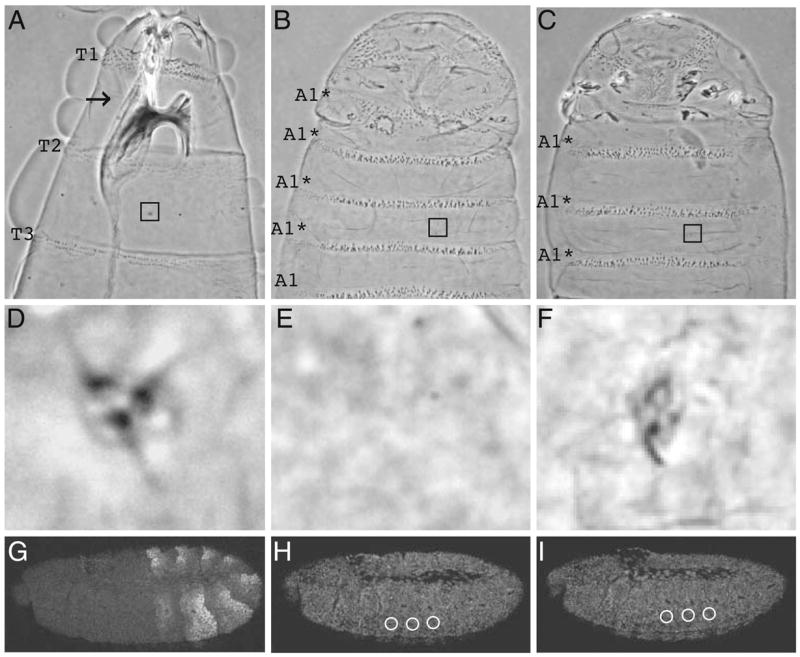
Dm-UBX chimeric protein expression levels and their effects on larval thoracic appendage (Keilin’s organ) development. Phase contrast micrographs of the anterior and ventral surfaces of wild-type first instar larval cuticles (wt). a Cuticle from a wild-type larva. b Cuticle from a larva in which Dm-UBX was ectopically expressed. c Cuticle from a larva in which Dm-UBX TE was ectopically expressed. T1, T2, and T3 denote the first, second, and the third thoracic segments, respectively. A1* denotes the ectopic first abdominal denticle identities induced by ectopic UBX protein expression. Embryos expressing the Dm-UBX positive control and Dm-UBX TE (c), as well as other Dm-UBX proteins with the exception of Dm-UBX DE, promote variable transformation of thoracic denticle belts towards abdominal identities, as well as suppression of T1 beard formation and disruption of head involution. The squares in (a), (b), and (c) indicate positions of the thoracic Keilin’s organs, shown in higher magnification in inserts in (d), (e), and (f), respectively. Note that the thoracic Keilin’s organs are affected in embryos expressing Dm-UBX TE under arm-GAL4; this Keilin’s organ would be scored as possessing one hair. g Staining pattern of DM-UBX protein in stage 11 embryos, detected with anti-UBX FP3.38 in the posterior thorax and anterior abdomen. h, i The respective panels show stage 11 embryos ectopically expressing Dm-UBX and Dm-UBX TE proteins, detected with anti-hemagglutinin antibodies. The staining levels in the ventral–lateral thorax (white circles) for 16 embryos were measured for each line (see “Materials and methods”) to determine the average percentage of ectopic protein compared to the Dm-UBX standard. In embryos, anterior is to the left and dorsal up
We generated transgenic Drosophila lines with GAL4-inducible expression constructs for the different chimeric UBX proteins, which were all labeled with a hemagglutinin (HA) epitope at the extreme C-terminus. The HA addition has no detectable influence on the function of UBX protein in ectopic expression assays (Ronshaugen et al. 2002). Previous studies have shown that the amount of Keilin’s organ repression by wild-type UBX is highly sensitive to the levels of ectopic protein, and that the number of hairs of Keilin’s organs showed the best correlation with the repression strength of UBX protein on Dll transcript levels (Tour et al. 2005). Therefore, we chose transgenic lines that, when induced by arm-GAL4 drivers, produced ectopic chimeric proteins in limb primordia (examples in Fig. 2h,i) at levels that were within 10–20% of wild-type UBX levels—defined as the fluorescent anti-UBX antiserum signal in the anterior ventral–lateral region of the first abdominal segment (see Fig. 2g and Tour et al. 2005). The expression pattern of Dll in the thorax is a more accurate measure of appendage primordia than Keilin’s organ development, since the Dll cells include almost all of the larval and imaginal thoracic limb primordia, while the Keilin’s organs are derived from only a few neural and support cells in the central part of the Dll domain (Bolinger and Boekhoff-Falk 2005).
Ectopic ubiquitous expression of the wild-type Dm-UBX control protein at normal levels completely suppressed Keilin’s organ development (Fig. 2b,e). The chimeric protein that behaved most like wild-type UBX was Dm-UBX TE. Dm-UBX TE usually removed Keilin’s organs completely, but occasionally allowed the development of an organ with one or two hairs (Fig. 2f). The quantitative levels of Keilin’s organ hair development allowed by Dm-UBX TE and the other UBX chimeras are reported in graphs that follow.
Dm-UBX chimeric protein regulation of embryonic limb primordia
The first pair of chimeric UBX protein we tested for function in embryos were Dm-UBX SE and Dm-UBX AE. These chimeras differed by only two amino acids, both alanine residues that replaced serine or threonine residues in C-terminal CK2 sites (Fig. 1). The effects of ectopic embryonic expression of both chimeras on the number of Keilin’s organ hairs, and on Dll transcription, were compared with wild-type embryos and embryos ectopically expressing the normal UBX protein. In wild-type stage 11 embryos, Dll transcripts accumulate in cells that correspond to the head and thoracic appendage primordia (Fig. 3b). In the thorax, Dll transcripts accumulate in about 20 cells per hemisegment in stage 11 embryos. The ectopic expression of normal UBX in thoracic cells at levels normally found in the first abdominal segment repressed thoracic Dll transcripts below levels of detection (Fig. 3c). This is correlated with the removal of all Keilin’s organ hairs from larvae that develop from ectopic UBX embryos (Figs. 2b and 3a, graph).
Fig. 3.
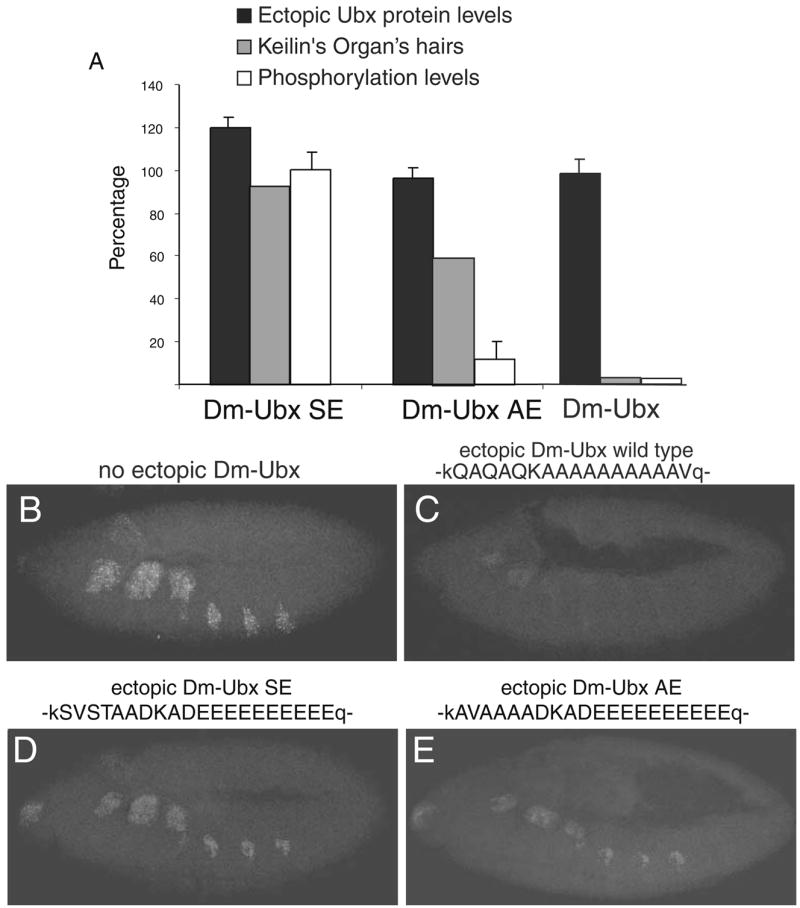
A C-terminal CK2 site in a heterologous context can inhibit the appendage repression function of Dm-UBX. a A plot of Keilin’s organ suppression versus average ectopic protein expression levels and average phosphorylation levels in vitro by CK2. The Dm-UBX control is not phosphorylated and suppresses development of all Keilin’s organ hairs. Dm-UBX SE is efficiently phosphorylated and develops a normal number of Keilin’s organ hairs. Dm-UBX AE is weakly phosphorylated by CK2 in vitro and develops an intermediate number (~60%) of Keilin’s organ hairs. The percentage of Keilin’s organ hairs is relative to the number that develops in wild-type first instar larvae. b The pattern of Dll transcripts that are detected in typical stage 11, wild-type embryos. c Dll transcripts in embryos that ectopically express Dm-UBX, d Dll transcripts in embryos that ectopically express Dm-UBX SE, and e Dll transcripts in embryos that ectopically express Dm-UBX AE. Embryonic orientation is anterior to the left, dorsal up. Single letter codes above (b)–(e) show the amino acid sequences substituted into the UBX C-terminal region. Capital letters denote insert sequences; small case letters denote flanking Dm-UBX amino acids. For more details, see Fig. 1
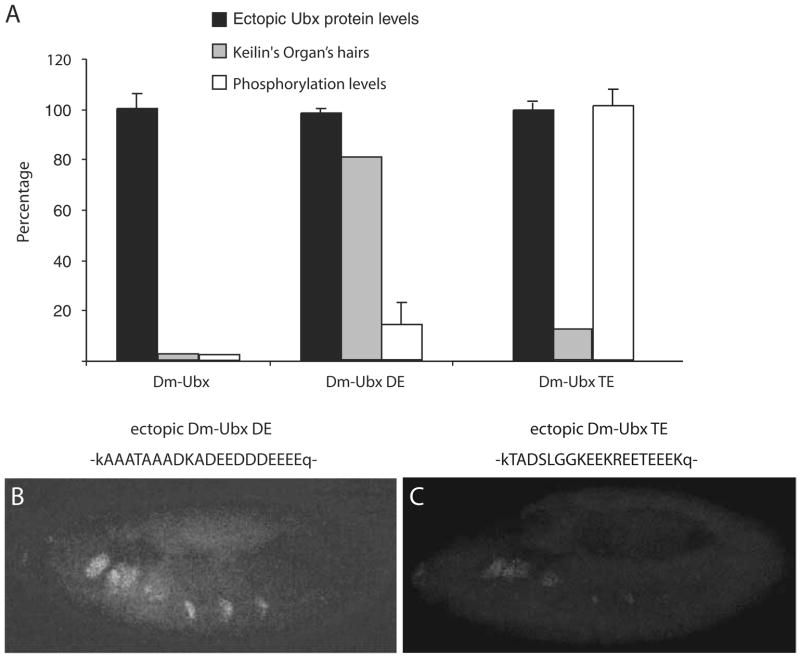
Larvae that developed from embryos that expressed the Dm-UBX SE protein developed normal numbers of hairs of Keilin’s organs, although in stage 11 embryos there were fewer cells that accumulated Dll transcripts (Fig. 3d). In contrast, embryos that ectopically expressed the Dm-UBX AE protein only developed about half the normal numbers of Keilin’s organ hairs (Fig. 3a), and this was associated with a much stronger Dll repression function, as only a few embryonic thoracic cells accumulated Dll transcripts (Fig. 3e). Ectopic expression of Dm-UBX DE in embryos had only a modest repressive effect on the number of Keilin’s organ hairs and Dll transcript levels. For Dm-UBX DE, the number of Keilin’s organ hairs was reduced to about 80% of normal levels (Fig. 4a), and Dll transcript levels were reduced to levels that were intermediate between the levels observed in the SE and AE embryos (Fig. 4b). Finally, the Dm-UBX TE protein was both a strong repressor of Keilin’s organ hairs and Dll transcript levels (Fig. 4a,c), almost as strong a repressor as wild-type UBX protein.
Fig. 4.
C-terminal CK2 phosphorylation sites are not sufficient to inhibit the appendage repression function of Dm-UBX. a A plot of Keilin’s organ suppression compared to average ectopic protein expression levels and average in vitro CK2 phosphorylation levels. The Dm-UBX control is not phosphorylated and is a strong suppressor of Keilin’s organ hair development. Dm-UBX DE is weakly phosphorylated, but has little effect on Keilin’s organ hair development. Dm-UBX TE is strongly phosphorylated, but is also a strong suppressor of Keilin’s organ hairs. b The pattern of Dll transcripts that are detected in stage 11 embryos that ectopically express Dm-UBX DE (compare with the wild-type pattern in Fig. 3b). c Dll transcripts in embryos that ectopically express Dm-UBX TE. Embryonic orientation is anterior to the left, dorsal up. Single letter codes above (b) and (c) show the amino acid sequences substituted into the UBX C-terminal region. Capital letters denote insert sequences; small case letters denote flanking Dm-UBX amino acids. For more details, see Fig. 1
Dm-UBX chimeric protein regulation of Antp transcript abundance
Antennapedia (Antp) is a Drosophila Hox gene that contributes to thoracic morphological identity, and its pattern of expression is limited (largely) to thoracic primordia by transcriptional repression of Antp exerted by UBX and ABDA. To further assay the regulatory effects of the Dm-UBX chimeras, we tested their ability to repress nascent Antp transcript levels after ectopic expression in embryos. We used a probe that detects nascent Antp P1 transcripts (the chromosomal sites of Antp transcription—the green “nuclear dots” in Fig. 5a) in stage 11 embryos. Antp P1 is normally activated in epidermal nuclei of embryonic parasegments 4 and 5 (from the posterior compartment of the first thoracic segment (T1) to the anterior compartment of the third thoracic segment (T3; Fig. 5a). Ectopic expression of normal Dm-UBX strongly repressed Antp P1 nascent transcription (Fig. 5b). Embryos ectopically expressing the Dm-UBX TE protein showed the strongest repressive effect on Antp P1 transcription, whereas embryos with ectopic Dm-UBX DE protein showed very little repression of Antp P1. The Dm-UBX SE and AE proteins repressed Antp P1 in a manner similar to their effects on Dll, with SE behaving as a moderate repressor, and AE repressing Antp P1 in more thoracic cells.
Fig. 5.
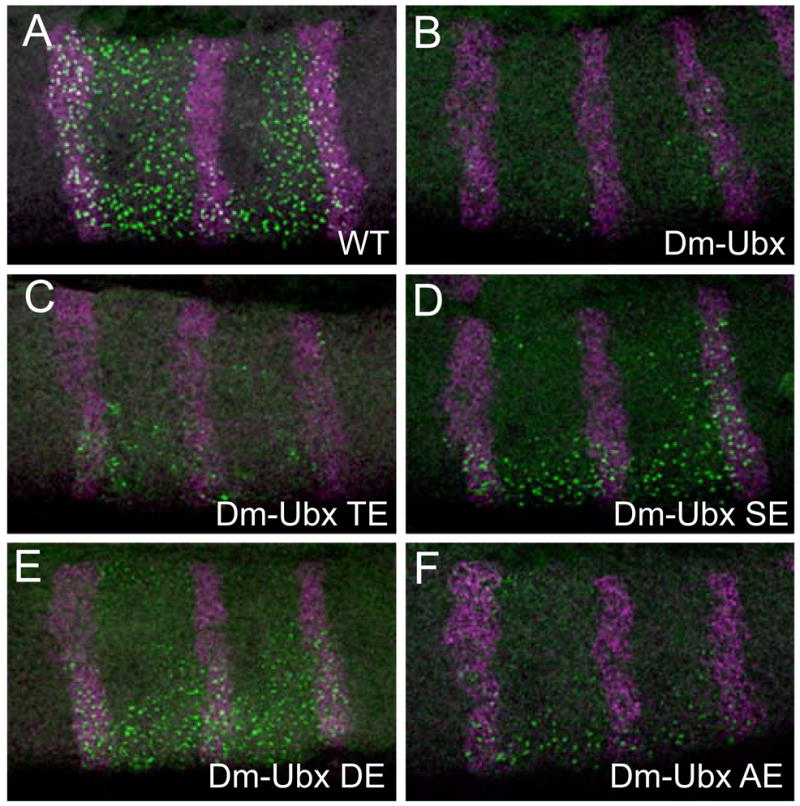
UBX chimera repression of Antennapedia (Antp) transcripts. a–f Micrographs of the thorax of stage 11 embryos hybridized with a probe to Antp P1 transcripts (green signal) and engrailed transcripts (magenta signal). Note that at this stage, most of the Antp P1 signal is observed at the nuclear sites of transcription (nascent transcripts; Kosman et al. 2004). a Wild-type embryo; the expression pattern of Antp P1 transcripts includes a region from the posterior compartment of T1 to the posterior compartment of T3. Embryos ectopically expressing Dm-UBX (b) or Dm-UBX TE (c) exhibit strong repression of Antp P1 transcripts. d Embryos that ectopically express Dm-UBX DE show only a slight repression of Antp P1 transcripts, whereas embryos that ectopically express Dm-UBX SE (e) and Dm-UBX AE (f) chimeras exhibit stronger repression of Antp P1 nascent transcript levels. Note that Antp P1 is repressed more efficiently in the dorsal part of the T2–T3 segments than in the ventral region
Dm-UBX chimeric protein regulation of dpp transcription in the visceral mesoderm
In parasegment 7 of the embryonic visceral mesoderm, UBX is required to activate the transcription of the decapentaplegic (dpp) gene, and ectopic expression of UBX protein can activate dpp transcription in a more extensive domain of the visceral mesoderm (Tremml and Bienz 1989; Sun et al. 1995; Capovilla and Botas 1998; Stultz et al. 2006). For example, Fig. 6a shows dpp transcript expression in a stage 14 wild-type embryo, and Fig. 6b shows dpp expression in an embryo where Dm-UBX was ectopically expressed. To test whether the changes in function of the chimeric UBX proteins affected both repression and activation functions, we tested their transcription activation functions on dpp. When the hybrid proteins Dm-UBX SE, AE, and TE were ectopically expressed, dpp transcripts accumulated in visceral mesoderm cells anterior to parasegment 7 (Fig. 6). However, only Dm-UBX TE was capable of activating dpp transcripts posterior to parasegment 7, in a manner similar to Dm-UBX, and the TE protein only weakly activated dpp in posterior visceral mesoderm cells (Fig. 6f). In embryos that ectopically expressed the Dm-UBX DE protein, there was no detectable ectopic activation of dpp transcripts (Fig. 6c).
Fig. 6.
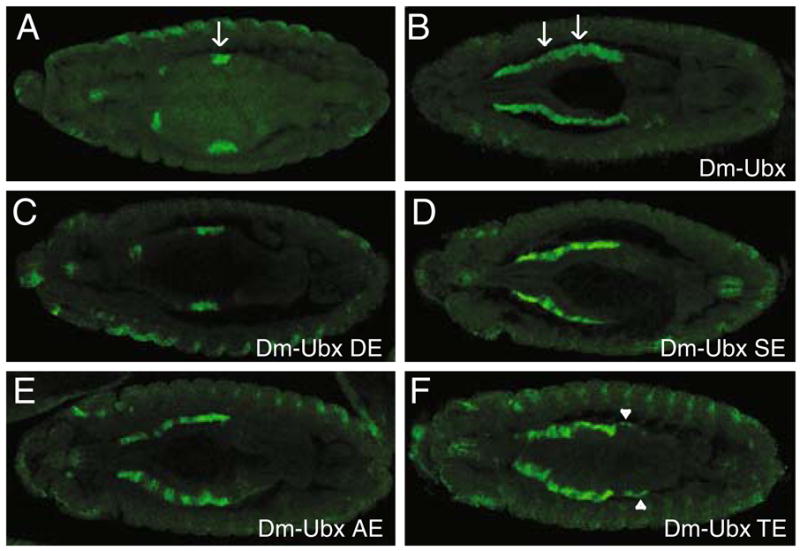
Effects of ectopic expression of Dm-UBX chimeras on decapentaplegic (dpp) transcription in the visceral mesoderm. a–f A dorsal view of stage 13 embryos, hybridized with a probe that detects dpp transcripts. a The wild-type pattern of dpp transcripts in stage 13 embryos includes the visceral mesoderm of parasegment 7 (arrow). b Embryos that ectopically express Dm-UBX activate dpp expression in anterior regions of the visceral mesoderm, as well as in some visceral mesoderm posterior to parasegment 7. c Embryos that ectopically express Dm-UBX DE do not influence the dpp pattern of transcription. d Embryos that ectopically express Dm-UBX SE robustly activate dpp expression in anterior regions of the visceral mesoderm, but not in visceral mesoderm posterior to parasegment 7. e Embryos that ectopically express Dm-UBX AE activate dpp expression in anterior regions of the visceral mesoderm, but more weakly on average than Dm-UBX SE, and not in the visceral mesoderm posterior to parasegment. f Embryos that ectopically express Dm-UBX TE activate dpp expression in anterior regions of the visceral mesoderm, and weakly in the visceral mesoderm posterior to parasegment 7 (arrowhead) when compared with wild-type Dm-UBX
Discussion
Previous studies have suggested that CK2 sites are associated with some of the different functional outputs that distinguish the ANTP and UBX Hox protein paralogs in Drosophila (Jaffe et al. 1997), as well as the variation in function that has evolved between evolutionarily diverged Hox protein orthologs (Ronshaugen et al. 2002; Shiga et al. 2002). In this study, we directly tested the in vitro phosphorylation levels of wild-type and chimeric UBX proteins with different CK2 consensus sites and tested the context dependence of those CK2 consensus sites on UBX protein regulatory functions in embryos.
Our results indicate that in some amino acid contexts, CK2 consensus sites can inhibit a UBX function required for repression. For example, the Dm-UBX SE chimera was efficiently phosphorylated by CK2 in vitro, and this was associated with this chimera having only a weak repressive effect on embryonic thoracic appendage development and Dll or Antp P1 transcript levels. In Dm-UBX AE, with the two Dm-UBX SE CK2 consensus sites mutated, in vitro phosphorylation was abolished, and the AE protein acquired an increased repressive strength on appendage development and Dll transcripts in embryos. Dm-UBX AE was not as strong a repressor as the parental protein Dm-UBX, so other sequences in the C-terminal tail of SE and AE may be supplying an inhibitory effect on UBX function. For Dm-UBX SE and AE, tests of three downstream target genes suggest that both activation and repression functions can be inhibited by CK2 consensus sites and adjacent residues in the C-terminal region.
Our studies of Drosophila UBX protein deleted for the C-terminal QA repeat showed that it had an increased ability to be phosphorylated by CK2 in vitro, when compared to wild-type UBX protein. This is correlated with diminished repressive effect on Dll transcript levels of the QA deleted protein when tested in Drosophila embryos (Hittinger et al. 2005), as well as an ability of the QA region to increase the repression function of an onychophoran version of UBX in Drosophila embryos. Thus, it is possible that the mechanism through which the QA repeat operates is to negatively regulate covert CK2 phosphorylation sites in Drosophila UBX, or in onychophoran UBX, thereby enhancing the abilities of these proteins to repress Dll and Keilin’s organ development in fly embryos.
Dramatic evidence that amino acid context is required for CK2 site regulation of Hox protein function is seen in the behavior of Dm-UBX TE. Although this chimera was very efficiently phosphorylated by CK2 in vitro, it was a potent repressor of appendage development as well as Dll and Antp transcription, almost as potent as wild-type Dm-UBX. Thus, high affinity C-terminal CK2 phosphorylation sites are not sufficient to inhibit Hox protein function, but require a specific amino acid context for their inhibitory function. The potency of repression of Antp P1 transcription was more pronounced in the dorsal regions than in the ventral regions; this correlates with the normally lower levels of Antp P1 transcripts in dorsal epidermal cells at stage 11.
The least informative chimera was Dm-UBX DE. The acidic C-terminal sequences inserted into this protein appeared to abolish nearly all regulatory functions. The expression of this chimera had no detectable activation effect on dpp transcript levels and had only weak repressive effects on Dll and Antp transcript levels. Since Dm-UBX DE was also the only chimera that did not transform thoracic denticle belts toward abdominal denticle morphologies, it is possible that the novel C-terminal sequences in Dm-UBX DE interfered with proper folding, attenuated almost all DNA binding functions, or otherwise disabled most functions of Dm-UBX DE.
These results are consistent with the idea that phosphorylation of Hox and other homeodomain proteins modulates their regulatory functions in development and evolution, an idea that is supported by much previous evidence (Gay et al. 1988; Gavis and Hogness 1991; Bourbon et al. 1995; Jaffe et al. 1997; Berry and Gehring 2000; Ronshaugen et al. 2002), but emphasizes the importance of neighboring amino acid sequences. There are a few different mechanisms, not mutually exclusive, that might explain how the evolution and loss of phosphorylation sites alter Hox regulatory functions.
One potential mechanism is that the phosphate groups, in themselves, that are added to CK2 site serine or threonine residues are sufficient to alter the conformation of HOX proteins, and/or their binding interactions with DNA or other regulatory proteins. Consistent with this, it is known that in vitro phosphorylation (or substitutions of acidic residues that mimic the phosphorylated state) of proteins in the homeodomain family can influence their in vitro interactions with either DNA and/or protein cofactors. For example, CK2 phosphorylation of a large fragment of the Drosophila EN homeodomain protein resulted in an enhancement of EN in vitro DNA binding function (Bourbon et al. 1995). In contrast, phosphomimetic (glutamate) residues at CK2 phosphorylation sites in a large fragment of the Drosophila ANTP homeodomain protein resulted in an inhibition of ANTP-EXD protein–protein interactions on heterodimer DNA binding sites in vitro (Jaffe et al. 1997).
Another potential mechanism is based on the recent finding that some kinases can act at DNA cis-regulatory regions as transcriptional cofactors (Pokholok et al. 2006). In this view, CK2 or other kinases may function as directly bound nuclear cofactors for HOX proteins in the nucleus and thereby regulate the balance of HOX activation and repression activities. This would be consistent with the isolation of CK2 using ANTP protein as “bait” in a two-hybrid assay (Jaffe et al. 1997), and also with evidence that CK2 is a stably associated subunit of several chromatin remodeling complexes (Poole et al. 2005). If true, this would make the evolution of CK2 “phosphorylation-binding” sites on Hox proteins more akin to the cofactor interaction motifs that evolved in FTZ protein during its transition from a Hox to a segmentation function (Lohr and Pick 2005), or the MCM1 protein interaction motif that evolved in the S. cerevisiae (and other closely related yeast species) α-2 proteins that allowed α-2 to regulate yeast mating type (Tsong et al. 2006).
The current evidence indicates that the Hox gene cluster evolved in the lineage leading to cnidarians after the split between the sponge and cnidarian lineages (Larroux et al. 2007: Lemons and McGinnis 2006). In ancient cnidarian or triploblastic animals, it appears that Hox proteins evolved functions allowing them to diversify morphology on the anterior–posterior axis of embryos, in the primordia of the posterior head and trunk (McGinnis and Krumlauf 1992; Carroll et al. 2005). At some unknown point in animal evolution, Hox proteins evolved the ability to diversify appendage morphology, a function that is most obvious in extant arthropods (Hughes and Kaufman 2002). In the appendage primordia of proto-arthropods, it seems likely that most Hox proteins, including UBX were modifiers of an anterior antennal-like appendage identity, and later, some Hox proteins evolved the ability to repress Dll and appendage development in entire trunk segments or in subregions of trunk segments (Stuart et al. 1991; Panganiban et al. 1995; Grenier et al. 1997; Grenier and Carroll 2000; Shiga et al. 2002).
In the case of UBX protein, we propose that it first evolved a partial ability to repress Dll and limbs by loss of high affinity CK2 sites; this is consistent with the known UBX protein sequences from crustaceans and insects (Ronshaugen et al. 2002, unpublished results). CK2 site loss associated with a gain of Dll transcriptional repression function may also have occurred to the ANTP Hox protein in the lineage leading to the crustacean Daphnia (Shiga et al. 2002). Next, we propose that the alanine repeat in the C-terminus expanded, a repeat that is found in all known hexapod UBX proteins, with the exception of the basal hexapod Folsomia (Ronshaugen et al. 2002; Galant and Carroll 2002). This expanded alanine repeat sequence has some autonomous ability to act as a repression domain when appended to other transcription factor DNA binding domains (Galant and Carroll 2002), but as we find in this study, inhibits the phosphorylation of even weak CK2 sites in Drosophila UBX. It is possible that the alanine repeat sequence can also inhibit the CK2 phosphorylation of other insect UBX proteins, some of which have weak consensus sites for CK2 phosphorylation (unpublished results). In this view, even within the UBX protein sequence, regulatory functions have been evolved atop each other, the alanine repeat increasing UBX repression strength and simultaneously inhibiting the inhibition of repression function exerted by CK2 sites. We propose that the result of these successive functional changes led to insect UBX proteins that can partially or completely repress Dll and appendage development, if produced at sufficient levels at the appropriate developmental stages in appendage primordia (Castelli-Gair and Akam 1995; Warren et al. 1994; Galant and Carroll 2002; Tour et al. 2005).
Acknowledgments
We are grateful to Rob White for providing antiserum directed against UBX protein, to Dave Kosman for the help with confocal microscopy and staining, and to the members of the McGinnis lab for intellectual and practical assistance in many areas.
References
- Averof M, Akam M. Hox genes and the diversification of insect and crustacean body plans. Nature. 1995;376:420–423. doi: 10.1038/376420a0. [DOI] [PubMed] [Google Scholar]
- Bennett RL, Brown JS, Denell RE. Molecular and genetic analysis of the Tribolium Ultrabithorax ortholog, Ultrathorax. Dev Genes Evol. 1999;209:608–619. doi: 10.1007/s004270050295. [DOI] [PubMed] [Google Scholar]
- Berry M, Gehring W. Phosphorylation status of the SCR homeodomain determines its functional activity: essential role for protein phosphatase 2A,B¢. EMBO J. 2000;19(12):2946–2957. doi: 10.1093/emboj/19.12.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger RA, Boekhoff-Falk G. Distal-less functions in subdividing the Drosophila thoracic limb primordium. Dev Dyn. 2005;232(3):801–816. doi: 10.1002/dvdy.20329. [DOI] [PubMed] [Google Scholar]
- Bourbon HM, Martin-Blanco E, Rosen D, Kornberg TB. Phosphorylation of the Drosophila engrailed protein at a site outside its homeodomain enhances DNA binding. J Biol Chem. 1995;270(19):11130–11139. doi: 10.1074/jbc.270.19.11130. [DOI] [PubMed] [Google Scholar]
- Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. In: Goldstein LSB, Fyrberg E, editors. Methods in cell biology. Academic; New York: 1994. [DOI] [PubMed] [Google Scholar]
- Capovilla M, Botas J. Functional dominance among Hox genes: repression dominates activation in the regulation of Dpp. Development. 1998;125(24):4949–4957. doi: 10.1242/dev.125.24.4949. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity. 2. Blackwell Science; London: 2005. [Google Scholar]
- Castelli-Gair J, Akam M. How the Hox gene Ultrabithorax specifies two different segments: the significance of spatial and temporal regulation within metameres. Development. 1995;121:2973–2982. doi: 10.1242/dev.121.9.2973. [DOI] [PubMed] [Google Scholar]
- Galant R, Carroll SB. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 2002;415:910–913. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Hogness DS. Phosphorylation, expression and function of the Ultrabithorax protein family in Drosophila melanogaster. Development. 1991;112(4):1077–1093. doi: 10.1242/dev.112.4.1077. [DOI] [PubMed] [Google Scholar]
- Gay NJ, Poole SJ, Kornberg TB. The Drosophila engrailed protein is phosphorylated by a serine-specific protein kinase. Nucleic Acids Res. 1988;16(14A):6637–6647. doi: 10.1093/nar/16.14.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose R, Malik M, Huber PW. Restricted specificity of Xenopus TFIIIA for transcription of somatic 5S rRNA genes. Mol Cell Biol. 2004;24(6):2467–2477. doi: 10.1128/MCB.24.6.2467-2477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Morata G. The developmental effect of overexpressing a Ubx product in Drosophila embryos is dependent on its interactions with other homeotic products. Cell. 1990;61:515–522. doi: 10.1016/0092-8674(90)90533-k. [DOI] [PubMed] [Google Scholar]
- Grenier JK, Carroll SB. Functional evolution of the Ultra-bithorax protein. Proc Natl Acad Sci USA. 2000;97(2):704–709. doi: 10.1073/pnas.97.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier JK, Garber TL, Warren R, Whitington PM, Carroll S. Evolution of the entire arthropod Hox gene set predated the origin and radiation of the onychophoran/arthropod clade. Curr Biol. 1997;7:547–553. doi: 10.1016/s0960-9822(06)00253-3. [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Stern DL, Carroll SB. Pleiotropic functions of a conserved insect-specific Hox peptide motif. Development. 2005;132(23):5261–5270. doi: 10.1242/dev.02146. [DOI] [PubMed] [Google Scholar]
- Hsia CC, McGinnis W. Evolution of transcription factor function. Curr Opin Genet Dev. 2003;13(2):199–206. doi: 10.1016/s0959-437x(03)00017-0. [DOI] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC. Hox genes and the evolution of the arthropod body plan. Evol Dev. 2002;4(6):459–499. doi: 10.1046/j.1525-142x.2002.02034.x. [DOI] [PubMed] [Google Scholar]
- Jaffe L, Ryoo H, Mann RS. A role for phosphorylation by casein kinase II in modulating Antennapedia activity in Drosophila. Genes Dev. 1997;11:1327–1340. doi: 10.1101/gad.11.10.1327. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rokas A, Carroll SB. Regulation of body pigmentation by the abdominal-B hox protein and its gain and loss in Drosophila evolution. Cell. 2006;125(7):1387–99. doi: 10.1016/j.cell.2006.04.043. [DOI] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305(5685):846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Larroux C, Fahey B, Degnan SM, Adamski M, Rokhsar DS, Degnan BM. The NK homeobox gene cluster predates the origin of Hox genes. Curr Biol. 2007;17(8):706–710. doi: 10.1016/j.cub.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Lemons D, McGinnis W. Genomic evolution of Hox gene clusters. Science. 2006;313(5795):1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- Lewis DL, DeCamillis M, Bennett RL. Distinct roles of the homeotic genes Ubx and abd-A in beetle embryonic abdominal appendage development. Proc Natl Acad Sci U S A. 2000;97(9):4504–4509. doi: 10.1073/pnas.97.9.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr U, Pick L. Cofactor-interaction motifs and the cooption of a homeotic Hox protein into the segmentation pathway of Drosophila melanogaster. Curr Biol. 2005;15(7):643–649. doi: 10.1016/j.cub.2005.02.048. [DOI] [PubMed] [Google Scholar]
- Lohr U, Yussa M, Pick L. Drosophila fushi tarazu: a gene on the border of homeotic function. Curr Biol. 2001;11(18):1403–1412. doi: 10.1016/s0960-9822(01)00443-2. [DOI] [PubMed] [Google Scholar]
- Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12(5):592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2. FASEB J. 2003;17(3):349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- Palopoli MF, Patel NH. Evolution of the interaction between Hox genes and a downstream target. Curr Biol. 1998;8(10):587–590. doi: 10.1016/s0960-9822(98)70228-3. [DOI] [PubMed] [Google Scholar]
- Panganiban G, Sebring A, Nagy L, Carroll S. The development of crustacean limbs and the evolution of arthropods. Science (Wash D C) 1995;270(5240):1363–1366. doi: 10.1126/science.270.5240.1363. [DOI] [PubMed] [Google Scholar]
- Papillon D, Telford MJ. Evolution of Hox3 and ftz in arthropods: insights from the crustacean Daphnia pulex. Dev Genes Evol. 2007;217(4):315–22. doi: 10.1007/s00427-007-0141-8. [DOI] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6(12):893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313(5786):533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- Poole A, Poore T, Bandhakavi S, McCann RO, Hanna DE, Glover CV. A global view of CK2 function and regulation. Mol Cell Biochem. 2005;274(1–2):163–170. doi: 10.1007/s11010-005-2945-z. [DOI] [PubMed] [Google Scholar]
- Ronshaugen M, McGinnis N, McGinnis W. Hox protein mutation and macroevolution of the insect body plan. Nature. 2002;415:914–917. doi: 10.1038/nature716. [DOI] [PubMed] [Google Scholar]
- Schwartz CJE, Sampson HM, Hlousek D, Percival-Smith A, Copeland JWR, Simmonds AJ, Krause HM. FTZ-factor1 and Fushi tarazu interact via conserved nuclear receptor and coactivator motifs. EMBO. 2001;20:510–519. doi: 10.1093/emboj/20.3.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiga Y, Yasumoto R, Yamagata H, Hayashi S. Evolving role of Antennapedia protein in arthropod limb patterning. Development. 2002;129(15):3555–3561. doi: 10.1242/dev.129.15.3555. [DOI] [PubMed] [Google Scholar]
- Stuart JJ, Brown SJ, Beeman RW, Denell RE. A deficiency of the homeotic complex of the beetle Tribolium. Nature. 1991;350(6313):72–74. doi: 10.1038/350072a0. [DOI] [PubMed] [Google Scholar]
- Stultz BG, Jackson DG, Mortin MA, Yang X, Beachy PA, Hursh DA. Transcriptional activation by extradenticle in the Drosophila visceral mesoderm. Dev Biol. 2006;290(2):482–494. doi: 10.1016/j.ydbio.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Sun B, Hursh DA, Jackson D, Beachy PA. Ultrabithorax protein is necessary but not sufficient for full activation of decapentaplegic expression in the visceral mesoderm. EMBO. 1995;14:520–535. doi: 10.1002/j.1460-2075.1995.tb07028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford MJ. Evidence for the derivation of the Drosophila fushi tarazu gene from a Hox gene orthologous to lophotrochozoan Lox5. Curr Biol. 2000;10(6):349–352. doi: 10.1016/s0960-9822(00)00387-0. [DOI] [PubMed] [Google Scholar]
- Tour E, Hittinger CT, McGinnis W. Evolutionarily conserved domains required for activation and repression functions of the Drosophila Hox protein Ultrabithorax. Development. 2005;132(23):5271–5281. doi: 10.1242/dev.02138. [DOI] [PubMed] [Google Scholar]
- Tremml G, Bienz M. Homeotic gene expression in the visceral mesoderm of Drosophila embryos. EMBO J. 1989;8:2677–2685. doi: 10.1002/j.1460-2075.1989.tb08408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443(7110):415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- Warren RW, Nagy L, Selegue J, Gates J, Carroll S. Evolution of homeotic gene regulation and function in flies and butterflies. Nature. 1994;372:458–461. doi: 10.1038/372458a0. [DOI] [PubMed] [Google Scholar]
- Yussa M, Lohr U, Su K, Pick L. The nuclear receptor Ftz-F1 and homeodomain protein Ftz interact through evolutionarily conserved protein domains. Mech Dev. 2001;107(1–2):39–53. doi: 10.1016/s0925-4773(01)00448-8. [DOI] [PubMed] [Google Scholar]