Abstract
The ribosomal protein (r-protein) S20 is a primary binding protein. As such it interacts directly and independently with the 5′ domain and the 3′ minor domain of 16S ribosomal RNA (rRNA) in minimal particles and the fully assembled 30S subunit. The interactions observed between r-protein S20 and the 5′ domain of 16S rRNA are quite extensive, while the interactions with the 3′ minor domain are significantly more limited. In this study directed hydroxyl radical probing mediated by Fe(II)-derivatized S20 proteins was used to monitor the folding of 16S rRNA during r-protein association and 30S subunit assembly. An analysis of the cleavage patterns in the minimal complexes [16S rRNA and Fe(II)-S20] and the fully assembled 30S subunit containing the same Fe(II)-derivatized proteins shows intriguing similarities and differences. These results suggest that the two domains, the 5′ and 3′ minor, are organized relative to S20 at different stages of assembly. The 5′ domain acquires, in a less complex ribonucleoprotein particle (RNP) than the 3′ minor domain, the same architecture as observed in mature subunits. These results are similar to what would be predicted of subunit assembly by the 5′ to 3′ direction assembly model.
Introduction
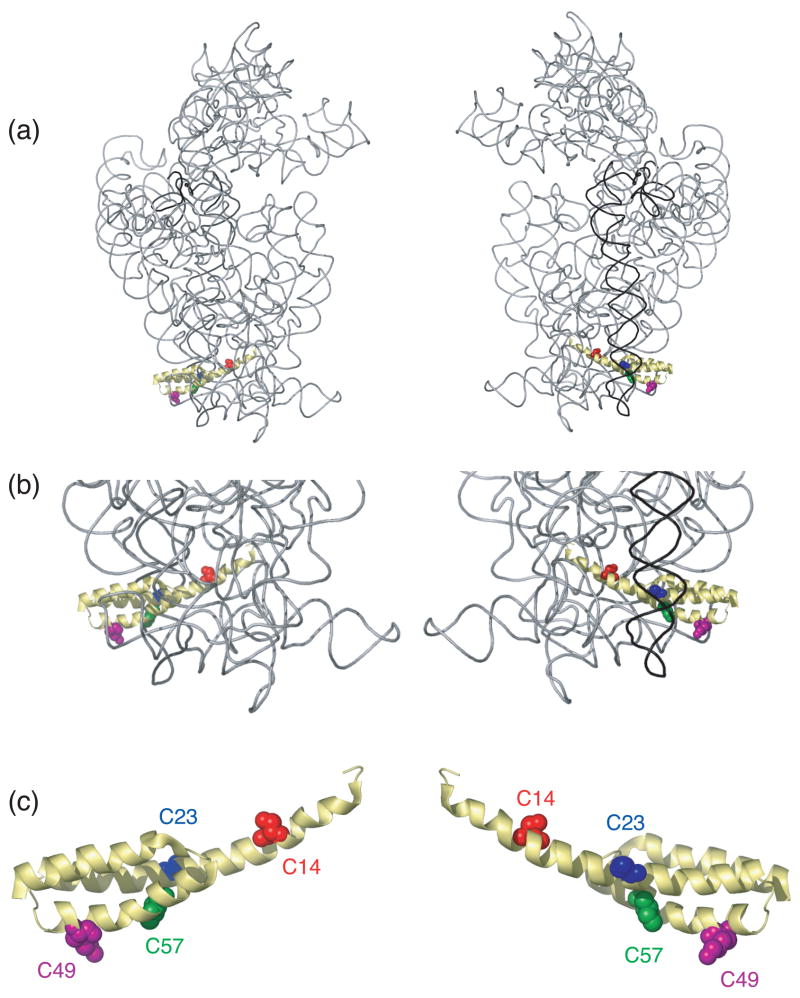
S20 is one of a few small subunit ribosomal proteins (r-proteins) that interacts with two different domains of 16S rRNA, the 5′ and the 3′ minor domains1– 4(Figure 1(a) and (b)). R-protein S20 is a primary binding protein and thus can interact directly and independently with 16S ribosomal RNA (rRNA)1; 5; 6. Footprinting experiments1 and the crystal structure of Escherichia coli (E. coli) 30S subunits 4 revealed that S20 binds several helices from the 5′ domain and the major helical element (helix 44: the penultimate stem) of the 3′ minor domain of 16S rRNA. The structure of r-protein S20 has not been determined in its free state, but in the assembled 30S subunit, S20 is a three-helix bundle located at the bottom of the body of the small subunit, where it is “sandwiched” between the 5′ domain and the penultimate stem, helix 44,4 (Figure 1). Additionally, it has been shown that the penultimate stem interacts extensively with the large subunit4 and thus appropriate positioning of helix 44 could be critical for accurate and rapid translation. Based on the model of 5′ to 3′ cotranscriptional assembly 7 and on the suggestion that the interaction between r-proteins and 16S rRNA occurs in discrete stages of 30S subunit assembly7; 8; 9, it is possible that S20 interacts differently with these two domains during the course of 30S subunit assembly.
Figure 1.
Three-dimensional structure of selected components of the E. coli 30S ribosomal subunit. (a) The entire16S rRNA is shown in light grey with the 3′ minor domain shown in black; S20 is shown in yellow with the cysteine substituted sites colored; position 14 is shown in orange, 23 in blue, 49 in magenta and 57 in green. Other small subunit r-proteins are omitted for clarity. Two views are presented for clarity. (b) Close-up of the position of S20 taken from (a). (c) Structure of S20 taken from panels (a) and (b). All the figures containing 3-D structures were prepared using Pymol31, and the pdb file 2AW74.
Recently, directed hydroxyl probing was used to explore changes in 16S rRNA elements surrounding the r-protein S15 during 30S subunit assembly 10; 11. Changes in 16S rRNA cleavage patterns generated by Fe(II)-S15 in ribonucleoprotein particles (RNPs) of different complexities were related to conformational changes in 16S rRNA as a result of r-protein binding 10; 11. Thus, roles of different r-proteins in the assembly process were elucidated. This is a major advantage of the directed hydroxyl radical probing strategy compared to traditional footprinting studies; much can be learned about the roles of many different r-proteins in one set of experiments and not just regarding the protein that is being footprinted or used as a probe. Another advantage of directed hydroxyl radical probing is the fact that the region of RNA that interacts with the area of the protein where the derivatized residue is present can be determined. Thus affording information about the orientation and directionality of the interactions. This approach made possible a better understanding of the assembly of the 30S subunit, by detailing the roles played by r-proteins and the conformational rearrangements that occur in nucleic acid-protein complexes during assembly and the high resolution structures of 30S subunits3 can be used to analyze the data in terms of achieving a known conformation or passing through an intermediary terrain. Directed hydroxyl radical probing offers a means for dissecting the potential differential interaction of S20 with 16S rRNA and a means of identifying specific r-proteins that facilitate these interactions.
Results from directed hydroxyl radical probing from four positions on r-protein S20 (see Figure 1(c)) in the 30S subunit have been published2. In fully formed 30S subunits, cleavage of the 5′ and the 3′ minor domains of 16S rRNA were observed. Consequently, the results of the directed hydroxyl radical probing experiments from r-protein S20 in more minimal complexes will make possible a better understanding of the interactions of the 5′ and 3′ minor domains with this protein. Toward this end, directed hydroxyl radical probing was effected in the minimal, binary Fe(II)–S20/16S rRNA complexes and the observed cleavage patterns were compared to the ones identified in the 30S subunits both as published 2 and reproduced herein. These studies also allow the cleavage data from Fe(II)-S20 to be examined in the context of the 30S subunit structure 2; 4. The results from this work can be grouped into three main categories. First, many of the cleavage sites especially those in the 5′ domain, are the same in the binary complex as in the fully assembled particle, although at positions where cleavage is observed in both complexes there can be differences in intensity. This suggests that much of the S20/16S rRNA environment from the 5′ domain is organized very early in assembly. Second, most of the cleavages in the penultimate stem are very weak or absent in the binary complexes, suggesting that helix 44 is not completely aligned until later in assembly. Third, there is a correlation between the location of the probing sites on S20 and the intensity and extent of the cleavage differences between the binary and complete complexes. This could indicate that different regions of S20 associate with 16S rRNA at different stages of assembly or that regions of S20 have stabilities or folding requirements that are influenced by the higher order structure of the assembling particle. Overall, these results are consistent with a polarity to the assembly of the 16S rRNA into the 30S subunit and with possible higher order organization of r-proteins as well.
Results
Complexes and purification
Four positions for Fe(II)-derivatization of S20 via cysteine substitution (C14, C23, C49 and C57) have already been reported and as mentioned above these positions were used for probing the 16S rRNA environment surrounding r-protein S20 in the 30S subunits 2. Overall, the probing data correlate well with the positioning of rRNA elements as determined by the mapping of the data on the three dimensional structure of 16S rRNA from the 30S subunit, which is now available (see Figures 4 and 6). Thus, it appears that this system is poised to address folding of 16S rRNA during 30S subunit assembly. These same derivatization sites were used for probing the 16S rRNA environment of S20 in minimal, binary particles [Fe(II)-S20/16S rRNA)] and in the fully assembled small subunits containing Fe(II)-S20 to allow a more direct and accurate comparison of cleavage sites and intensity. For consistency, the 30S subunits were purified using the same method of size exclusion chromatography as for the binary complexes, which is different than in the previous 30S subunit probing experiments 2, but similar to previous studies using Fe(II)-directed probing of more minimal 16S rRNA containing RNPs10; 11. Sucrose gradient analysis and chemical footprinting demonstrated that the 30S subunits are intact after purification using this method (data not shown) and thus direct comparison of cleavage patterns between particles of different complexities is possible.
Figure 4.
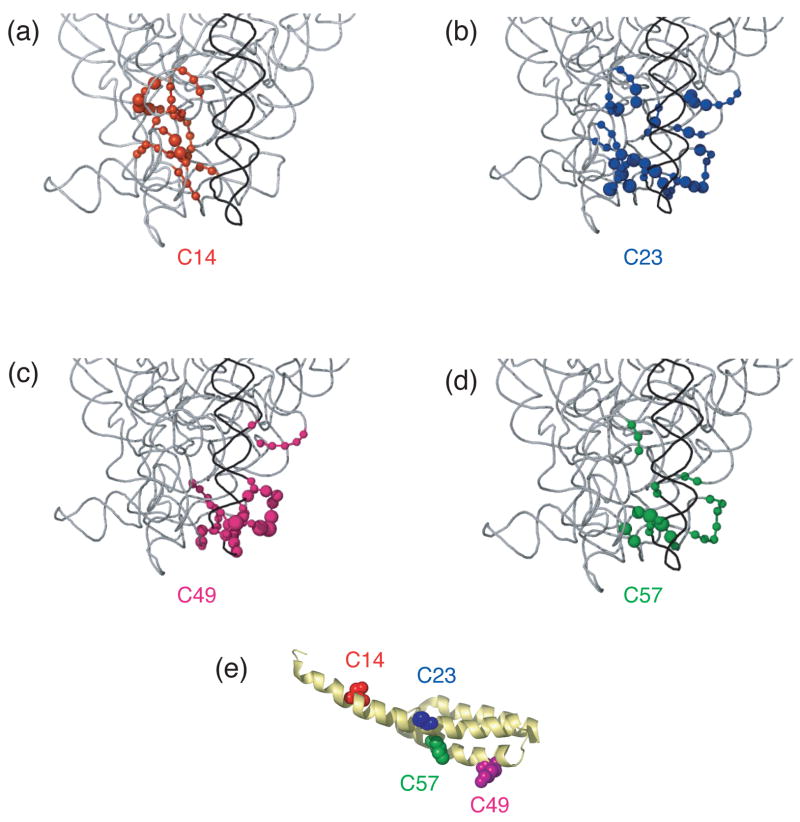
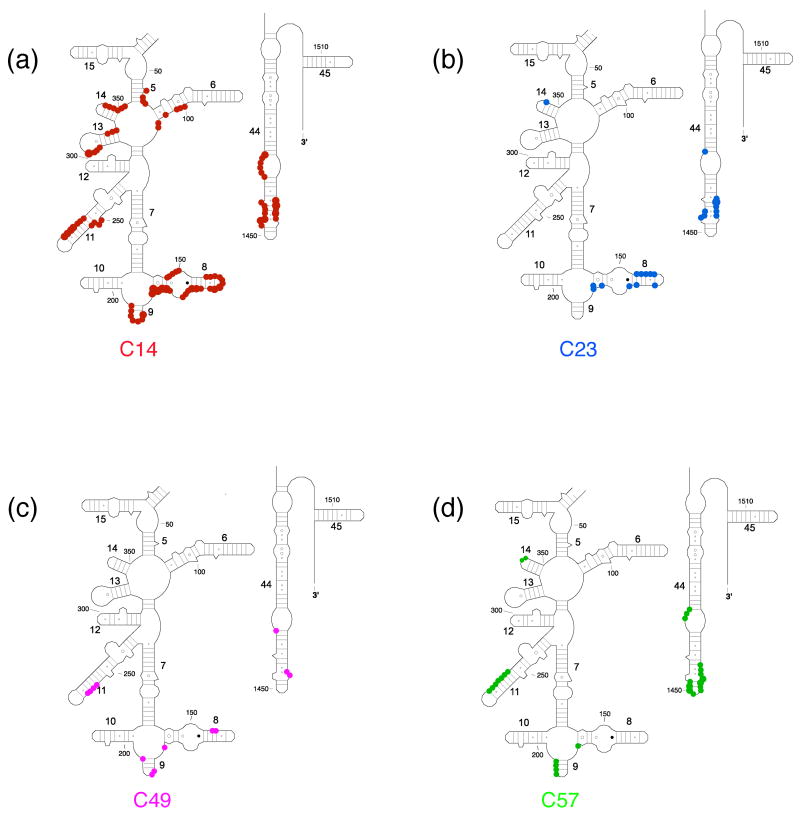
Directed hydroxyl radical cleavage sites from Fe(II)-S20 in minimal RNPs mapped on to the three-dimensional structure of E. coli 30S ribosomal subunit4. Only a limited portion of 16S rRNA (gray) is shown similar to Figure 1(b) and S20 is omitted for clarity. (a) Sites of cleavage from Fe(II)-C14-S20. (b) Sites of cleavage from Fe(II)-C23-S20. (c) Sites of cleavage from Fe(II)-C49-S20. (d) Sites of cleavage from Fe(II)-C57-S20. (e) S20 with tethered positions shown as in Figure 1(c). The position of the phosphate backbone that is cleaved is marked in the color corresponding to the Fe(II) attachment site (see Figure 1 and this figure panel (e)). Color coding is as in Figure 1.
Figure 6.
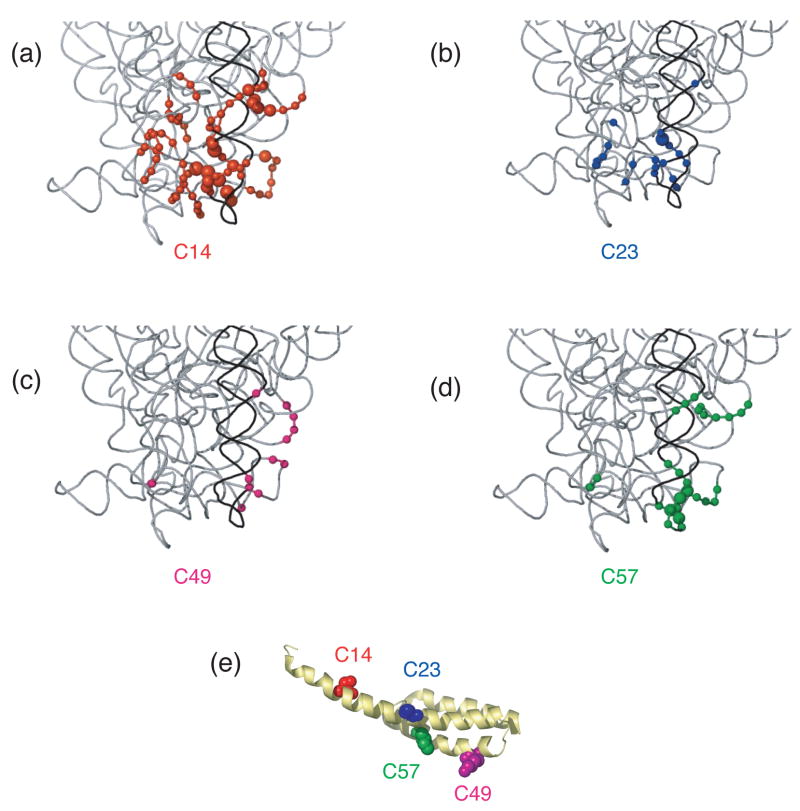
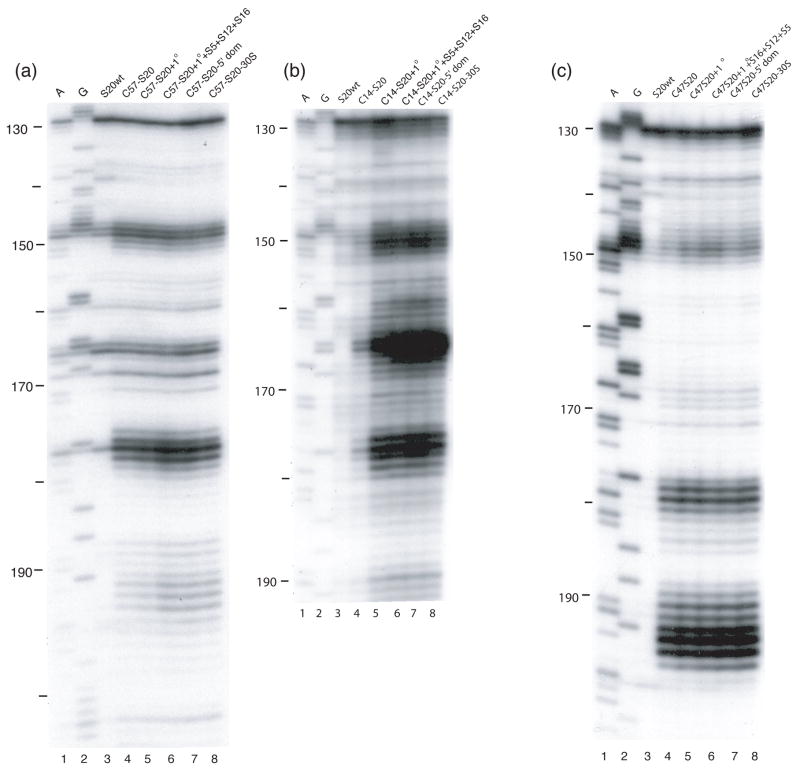
Differences in cleavage sites from Fe(II)-S20 in minimal RNPs as compared to 30S subunits mapped on the three-dimensional structure of E. coli 30S ribosomal subunit4. Nucleotides that are cleaved from Fe(II)-S20 with higher intensity in/or only in 30S subunits compared to the corresponding minimal particles are shown. The expanded region is the same as shown in Figure 4. (a) Sites cleaved from Fe(II)-C14-S20. (b) Sites cleaved from Fe(II)-C23-S20. (c) Sites cleaved from Fe(II)-C49-S20. (d) Sites cleaved from Fe(II)-C57-S20. The position of the phosphate backbone that is cleaved is marked in color corresponding to the Fe(II) attachment site (see Figure 1 and this figure panel (e)). Color coding is as in Figure 1.
Probing binary complexes of Fe(II)-S20/16S rRNA and comparison to probing of 30S subunits
The binary complexes containing Fe(II)-S20/16S rRNA were purified and probed with hydroxyl radicals generated from the tethered Fe(II). The cleavage sites were identified by primer extension analysis, and mapped on the secondary12 (Figure 3) and tertiary 4 (Figure 4) structures of 16S rRNA from E. coli 30S subunits, the latter in the context of the fully assembled small ribosomal subunit. The cleavage patterns observed for each of the minimal complexes were distinct from one another (Figures 2–4), and in the four complexes cleavage was exclusively localized to the 5′ domain and helix 44 in the 3′ minor domain of the 16S rRNA (Figures 2–4). There are distinct similarities and differences in the cleavage patterns when Fe(II) is tethered to different positions in S20 and there are also some notable differences between cleavage patterns observed in the minimal RNPs and the fully assembled 30S subunits. One highly plausible hypothesis is that differences in the dynamics of the minimal RNP relative to the fully assembled subunit account for the observed cleavage differences; this and other possible explanations of our data are further discussed below.
Figure 3.
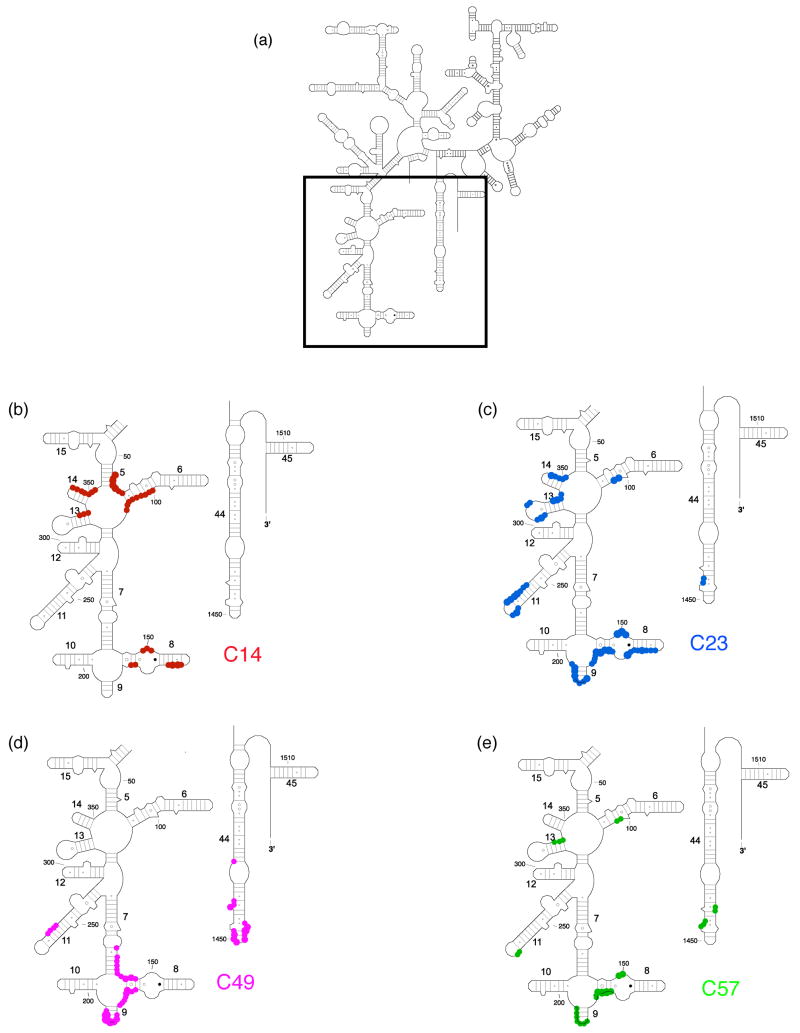
Directed hydroxyl radical cleavage sites from Fe(II)-S20 in minimal RNPs mapped on the secondary structure of 16S rRNA12. (a) Secondary structure of 16S rRNA with regions containing Fe(II)-S20 cleavage sites boxed. The highlighted domains are expanded in (b)-(e). (b) Sites cleaved from Fe(II)-C14-S20. (c) Sites cleaved from Fe(II)-C23-S20. (d) Sites cleaved from Fe(II)-C49-S20. (e) Sites cleaved from Fe(II)-C57-S20. The target nucleotides are marked by colored circles corresponding to the similarly colored cysteine-substituted sites on S20 shown in Figure 1. The size of the circle correlated with the strength of the observed cleavage. Some helix numbers are given12. Color coding is as in Figure 1.
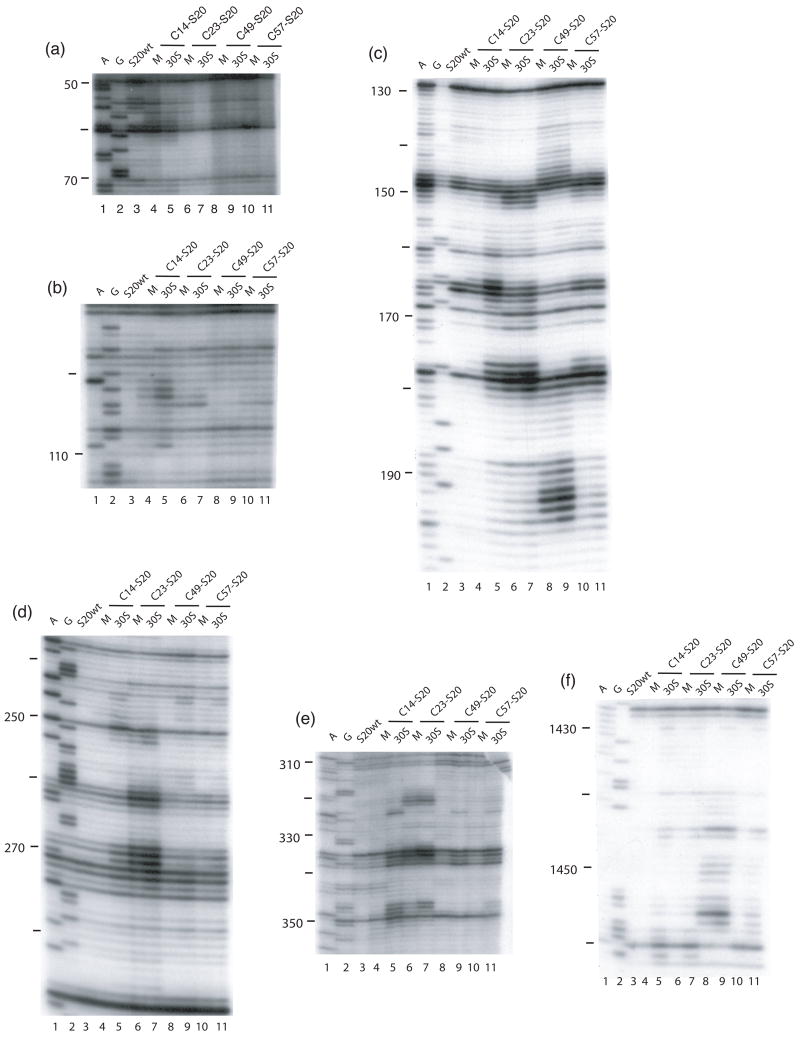
Figure 2.
Directed hydroxyl radical cleavage of 16S rRNA from Fe(II)-S20 in minimal RNPs detected by primer extension. All complexes contain natural 16S rRNA and Fe(II)-S20. For each gel there are two lanes for each Fe(II)-derivatized protein. M; minimal RNPs containing only Fe(II)-S20 and 16S rRNA; 30S; 30S subunits containing Fe(II)-S20. Lanes 1 and 2 are A and G, sequencing lanes respectively; lane 3, S20wt, wild-type S20 (cys-less). The different derivatized S20 proteins are Fe(II)-C14-S20, lanes 4 and 5; Fe(II)-C23-S20, lanes 6 and 7; Fe(II)-C49-S20, lanes 8 and 9; and Fe(II)-C57-S20, lanes 10 and 11. The primers used for extensions are 161 (a and b), 232 (c), 323 (d), 480 (e), and 1490 (f).
In the RNP containing Fe(II)-C14-S20, cleavage sites from the hydroxyl radicals are present in helices 5, 6, 8, 13, 14 all in the 5′ domain and no cleavage was observed in helix 44 (Figure 2, lane 4; Figure 3(b); Figure 4(a)). The most extensive cleavage by the radicals generated from Fe(II)-C14-S20, in the minimal complex, is observed at helices 5, 6 and 14, which are part of a multi-helical junction (Figure 2(a), (b) and (e), lane 4). In general, for the 5′ domain the cleavage patterns are quite similar in the minimal complex and the 30S subunits containing Fe(II)-C14-S20, but with much higher intensity observed in 30S subunits (Figure 2(a–c) and (e), compare lanes 4 and 5; Figure 5(a) and 6 (a)). Additionally, three cleavage sites are observed in helix 44: at nucleotide1430–1435, 1441–1447 and 1457–1461 in the context of 30S subunits, which were not detected in the binary complex (Figure 2(f), compare lanes 4 and 5; Figure 5(a) and 6(a)).
Figure 5.
Differences in cleavage sites from Fe(II)-S20 in minimal RNPs as compared to 30S subunits mapped on the secondary structure of 16S rRNA12. Nucleotides that are cleaved from Fe(II)-S20 with higher intensity in/or only in 30S subunits compared to the minimal particles are shown. The expanded region is the same as shown in Figure 3(a). (a) Sites cleaved from Fe(II)-C14-S20. (b) Sites cleaved from Fe(II)-C23-S20. (c) Sites cleaved from Fe(II)-C49-S20. (d) Sites cleaved from Fe(II)-C57-S20. The target nucleotides are marked by colored circles corresponding to the similarly colored cysteine-substituted sites on S20 shown in Figure 1. The size of the circle is correlated with the strength of the observed cleavage. Some helix numbers are given12.
The most pronounced cleavage for any of the Fe(II)-S20/16S rRNA complexes in the 5′ domain was observed from Fe(II)-C23-S20 (Figure 2, lane 6; Figure 3(c), Figure 4(b)). In the minimal complex containing Fe(II)-C23S20, helices 8 and 9 (Figure 2(c), lane 6) showed the most extensive cleavage with cleavage also observed in helices 6, 11, 13 and 14 (Figure 2(a), (b), (d), and (e), lane 6). Additionally, quite weak cleavage is observed in helix 44, at nucleotides 1444 and 1445 (Figure 2(f), lane 6). The cleavage patterns observed for the Fe(II)-C23-S20 containing 30S subunit are very similar to the ones observed when the minimal complex is probed in the case of the 5′ domain (Figure 2 (a–e), compare lanes 6 and 7; Figures 5(b) and 6(b)). Again, the intensity of the observed cleavage differs; for example cleavage observed in helix 8 of the 30S subunit is stronger than for the minimal complex (Figure 2(c), compare lanes 6 and 7). Meanwhile, two new patches of cleavage, 1430 and 1457–1461 and more intense cleavage at nucleotides 1444–1446 are detected in the 3′ minor domain in the fully assembled 30S subunit compared to the binary complex (Figure 2(f), compare lanes 6 and 7; Figures 5(b) and 6(b)).
When Fe(II)-C49-S20 is used as a probe in the minimal complex, the cleavage pattern was quite different from that observed with the other three derivatized proteins (Figures 2–4). In the minimal complex Fe(II)-C49-S20/16S rRNA, helices 7, 8, 9, and 11 are cleaved in the 5′ domain, while in helix 44 there are three discrete cleavage sites (Figure 2, lane 8; Figure 3(d) and Figure 4(c)). Some nucleotides around 1430 and 1440 are weakly targeted, while stronger cleavage takes place in the regions 1450 and 1460 (Figure 2(f), lane 8). The cleavage observed in helix 44 from the hydroxyl radicals generated by Fe(II) tethered C49-S20 is the strongest from all the minimal complexes (Figure 2 (f)). Thus, the cleavage pattern from Fe(II)-C49-S20 is very similar in the binary RNP and the 30S subunit for both the 5′ and 3′ minor domains (Figure 2, compare lanes 8 and 9; see Figures 5(c) and 6(c)).
The cleavage pattern observed in the complex containing Fe(II)-C57-S20 as a probe is similar to the one observed in the Fe(II)-C23-S20/16S rRNA RNP, but generally the cleavage is less intense from this probing position (Figure 2, compare lanes 6 and 10; Figures 3 and 4 compare (c) and (e)). These similar patterns are consistent with the relative locations of positions 23 and 57 (see Figure 1). Residues 23 and 57 are located on two different helices of S20, but they are very close to each other in the helical bundle structure of S20 (Figure 1). In helices 8 and 13 the different cleavage intensity from these two positions is quite obvious (Figure 2(c) and (e), compare lanes 6 and 10). Additionally the hydroxyl radicals generated from Fe(II)-C57-S20 cleave fewer overall residues (Figure 2, lane 10; Figure 3(e) and Figure 4(d)). Weak cleavage from Fe(II)-C57-S20 in 30S subunits was observed in helix 44, at nucleotides 1444–1446 and 1460–1461 (Figure 2(f), lane 11). Thus, results for probing in minimal particles from Fe(II)-C57-S20 are similar to those observed when positions C14 and C23 of S20 were used to tether Fe(II); small differences are observed in the cleavage patterns present for the minimal and the 30S subunit containing Fe(II)-C57-S20 for the 5′ domain, while major differences are detected for the 3′ minor domain (see Figures 5(d) and 6(d)).
The cleavage patterns observed in the 30S subunits when S20 is used as a probe are very similar to the ones previously published (2; see Figure 2), with a few exceptions: the extent and intensity of cleavage are higher in the experiments performed in this study, which may arise from a better quality or more accurate concentration of the starting materials, such as commercially available BABE (1-(p-bromoacetamidobenzyl) ethylenediaminetetraacetate) (Dojindo Labs) or fresh hydrogen peroxide; and from the different and more streamlined purification regime used in the current study (see Material and Methods and above). In the current study, overall more cleavage is observed from Fe(II)-C49-S20, which could be due to the quality of BABE or the purification protocol. Also, it appears that in the previous work2 two of the samples were misloaded or mislabeled (Fe(II)-C14-S20 and Fe(II)-C23-S20 in 30S subunits in the primer extension using primer 232; see Figure 2(c) lanes 5 and 7 and compare these with data in reference 2). We established at which stage of the experiments the error was introduced by recloning the genes encoding these cysteine substituted proteins (C14-S20 and C23-S20), purifying the proteins expressed from these clones, and repeating the probing experiments multiple times (see Materials and Methods). This appears to be a single aberration in the previous work as all other data are consistent between this work and the previously published results2. Thus, it appears that the 16S rRNA elements surrounding S20 are altered between the more minimal complex and 30S subunits, but not to the extent seen for S15 and its rRNA environment changes during 30S subunit 10; 11.
R-proteins involved in organizing the 16S rRNA environment of S20
To determine if the addition of specific r-proteins could account for differences in the cleavage patterns observed between the binary and fully assembled RNPs containing Fe(II)-S20, r-protein addition experiments were performed. Specific individual or groups of r-proteins were reconstituted with Fe(II)-S20 and 16S rRNA to generate more elaborate RNPs. Directed probing and primer extension analysis of the RNPs followed in an attempt to understand the folding of the 5′ and 3′ minor domains of 16S rRNA relative to r-protein S20.
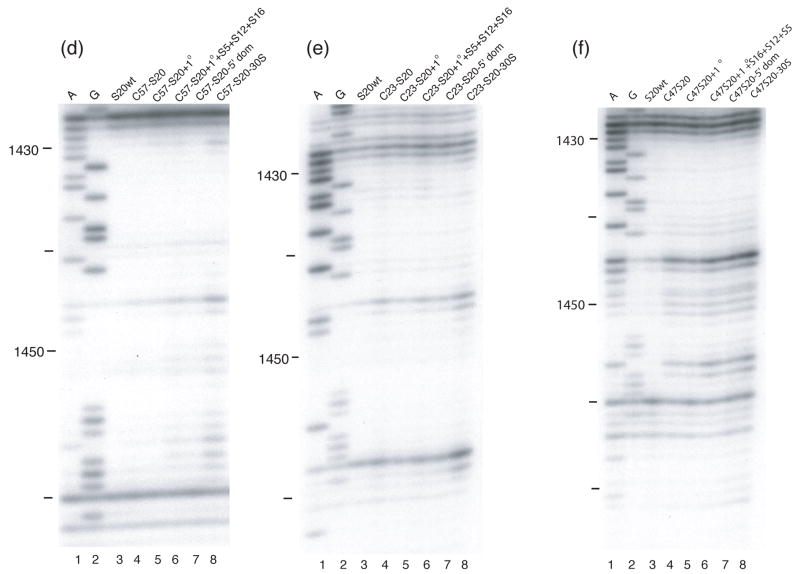
For three of the four probing positions on S20 (C23, 49 and 57) only slight differences were observed between the minimal RNPs and fully assembled 30S subunits (see Figures 5 and 6 (b–d)), in particular for the 5′ domain. The subtle nature of these differences is reflected in the various r-protein combinations that can be added to result in the Fe(II)-S20-30S subunit probing results (Figure 7). Addition of the other r-proteins that bind the 5′ domain (S4, S5, S8, S12, S16, and S17) or all of the other primary binding r-proteins (S4, S7, S8, S15 and S17 ) result in the 30S-like probing patterns from C23, C49 and C57 of S20. The presence of r-proteins S15 and S7 does not seem to be important for the organization of the 5′ domain. Only the presence of the primary and secondary r-proteins that bind or facilitate binding of proteins specific for the 5′ domain appears to give rise to the same cleavage patterns as for the fully assembled 30S subunit However, for the significant differences observed in helix 44 between minimal and complete particles (in the case of C14, C23 and C57), no limited additions appeared to be sufficient (Figure 7 (d and e)). The full complement of r-proteins appears to be required to position the penultimate stem. This is in agreement with the data observed when probing from Fe(II)-S15 and suggests that the ultimate orientation of helix 44 occurs very late in assembly.
Figure 7. Directed hydroxyl radical cleavage of 16S rRNA from Fe(II)-S20 in different RNPs detected by primer extension.
All complexes contain natural 16S rRNA and Fe(II)-S20. Lanes 1 and 2 are A and G, sequencing lanes respectively; lane 3, S20wt, wild-type S20 (cys-less); (a) lane 4, Fe(II)-C57-S20. All other lanes contain Fe(II)-C57-S20 and lane 5, 1°; the five additional primary binding proteins (S4, S7, S8, S15 and S17); lane 6, 1° + S5+ S12+S16, same as lane 5 with S5, S12 and S16. Lane 7, 5′ domain, S4, S5, S8, S12, S16, and S17. Lane 8, 30S, the completely assembled 30S subunit. Primer 232 was used for detection. (b) Same as panel (a) except Fe(II)-C14-S20 is used in place of Fe(II)-C57-S20. (c) Same as panel (a) except Fe(II)-C47-S20 is used in place of Fe(II)-C57-S20. (d) Same as panel (a) except primer 1490 is used for detection. (e) Same as panel (d) except Fe(II)-C23-S20 is used in place of Fe(II)-C57-S20. (f) Same as panel (d) except Fe(II)-C47-S20 is used in place of Fe(II)-C57-S20.
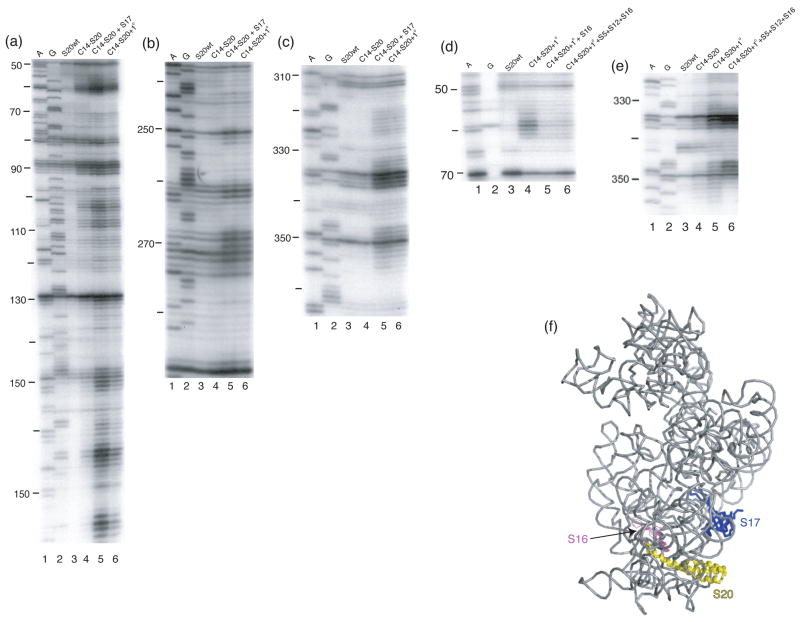
Dissection of the more substantial differences observed between minimal and complete subunits containing Fe(II)-C14-S20 yields more specific results. The majority of the cleavages observed in Fe(II)-C14-S20/30S subunits which are absent in the minimal RNP can be observed by the simple addition of r-protein S17. S17 is another primary binding protein that binds in the 5′ domain1; 4 and actually has some overlapping footprints with S201. Thus, a role for S17 in organizing elements of the 5′ domain in the vicinity of S20 is consistent with much other data. One very interesting observation was made during the r-protein addition experiments using Fe(II)-C14-S20. For the first time in our experience, the appearance of cleavage sites in an intermediate RNP was observed. Cleavage from Fe(II)-C14-S20 was not observed in helix 5 and at the base of helix 6 in the minimal particle nor in the 30S subunit (Figure 2(a), lanes 4 and 5). However, upon addition of the remaining primary binding proteins (S4, S7, S8, S15 and S17) to the minimal particle cleavage at nucleotides 56–63 and 354–357 was observed (Figure 8(d) and (e), lane 4). Furthermore, it was shown that addition of r-protein S16 to this intermediate RNP was sufficient to block cleavage and return the pattern to that observed in 30S subunits (Figure 8(d) and (e))), lane 5 compare to lane 4 and compare with Figure 2(a), lane 5). Thus, it appears that as the primary binding proteins assemble there are conformational rearrangements that render helix 5 and the base of helix 6 susceptible to cleavage and that the addition of S16 can compensate for this change. It is plausible that r-protein S16 is responsible for the change that protects the aforementioned nucleotides from cleavage, since in the three-dimensional structure of the 30S subunit it seems that S16 is located close to S20 and the cleaved nucleotides, 4 however the role of S16 in modulating the cleavage of these sites is likely indirect (see below and Figure 8). These observations suggest that although cleavage patterns are similar in the minimal RNP and the fully assembled subunit interesting r-protein-directed conformational changes may occur that await detection.
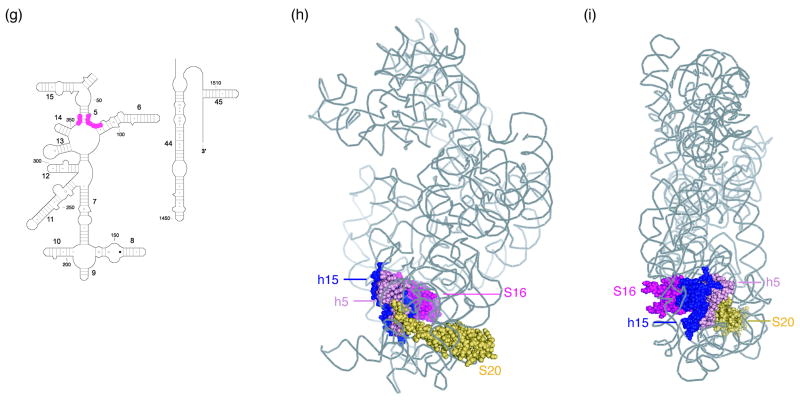
Figure 8. Directed hydroxyl radical cleavage of 16S rRNA from Fe(II)-C14-S20 in different RNPs detected by primer extension.
All complexes contain natural 16S rRNA and Fe(II)-S20. Lanes 1 and 2 are A and G, sequencing lanes respectively; lane 3, S20wt, S20 (cys-less); (a-c) lane 4, Fe(II)-C13-S20. All other lanes contain Fe(II)-C14-S20 and lane 5, C14-S20 + S17, r-protein S17; lane 6, C14-S20 + 1°, the five additional primary binding proteins (S4, S7, S8, S15 and S17). (d-e) Lane 4, C14-S20 + 1°; Fe(II)-C14-S20 and the five additional primary binding proteins (S4, S7, S8, S15 and S17); lane 5, C14-S20 + 1° + S16, lane 4 plus r-protein S16; lane 6, C14-S20 + 1° +S5+S12+S16, lane 5 with r-proteins S5 and S12. The primers used for extensions are 161 (a and d), 323 (c), and 480 (c and e). (f) Three-dimensional structure of selected components of the E. coli 30S ribosomal subunit4. 16S rRNA is shown in light grey, S20 is shown in yellow, S17 is shown in blue and S16 is shown in pink. The remaining proteins are omitted for clarity. (g) Sites that are protected by S16 as shown in panels (d) and (e) are shown on a portion of the secondary structure12 of 16S rRNA in pink balls. (h) Three-dimensional structure of selected components of the E. coli 30S ribosomal subunit4. 16S rRNA is shown in light grey with helix 15 (h15) shown in blue and sites of cleavage in helix 5 (h5) are shown in light pink, S20 is shown in yellow, and S16 is shown in magneta. The remaining proteins are omitted for clarity. (i) same as panel (h) but turned 90°.
Discussion
Directed hydroxyl radical probing using complexes containing Fe(II)-S20 and 16S rRNA reveal aspects of rRNA domain organization and 30S subunit assembly. The results of the directed hydroxyl radical probing in the minimal Fe(II)-S20/16S rRNA complexes and fully assembled subunits correlate well with the relative positions of the derivatized residues in S20 (Figures 4 and 6) and the cleavage sites and their intensity correlate well with the placement of the respective probe in the structure of r-protein S20 and of the 30S subunit (Figures 1, 4 and 6). Our data are consistent with a substantial folding and architectural organization of the 5′ domain early in assembly (see 9; 13; 14; 15; 16) and with the final alignment of helix 44 occurring later in the assembly cascade9; 11. These results, taken together with our earlier work suggest that there are different means by which domains or elements of 16S rRNA are organized.
Overall, our data from Fe(II)-S20 probing in 30S subunits is in good agreement with the structures of 30S subunits3,4. A large majority of the cleavage sites are within the accepted 40 Å range for tethered cleavage from r-proteins32. However, there are nuances in the data, such as cleavage of helical faces that do not absolutely agree with the relative positioning to the S20 probing sites, which need to be addressed. A plausible explanation for data such as these is that the minimal RNPs and 30S subunits are dynamic and can adopt multiple conformations, as has been observed crystallographically4, 32. Thus, it seems likely that in our probing experiments we are examining an ensemble of 30S subunit or RNP structures and thus our experiments may allow us to access more conformations than could be allowed in a crystal lattice. This idea is not unprecedented as differences in solvent accessibility observed in crystal structures and reactivity of rRNA to hydroxyl radicals has been observed34. In this work, it was hypothesized, after several other possible hypotheses were examined and refused, that these differences were due to the existence of different conformations in solution and that the crystal structure may represent only one of these possible conformations. It should also be mentioned that the addition of our BABE-linker could change some local conformations or dynamics and thereby result in some of the subtle discrepancies between our data and what would be predicted from the crystal structures. Nonetheless, the level at which our data correlates with structural information is striking and sufficient to suggest that S20 is binding at a similar site in the minimal RNPs, the 30S subunits studied herein and in the subunits studies by crystallography, yet differences in overall architecture, dynamics and details of the interactions are possible.
Three of the four derivatized Fe(II)-S20 proteins have essentially the same cleavage patterns in the 5′ domain of 16S rRNA, in the binary particle and fully assembled 30S subunit (see Figure 6). For Fe(II)-C23, -C49 and -C57, the 5′ domain cleavage patterns only differ slightly in these rather distinct complexes. In contrast, when Fe(II)-C14-S20 is used as a probe the cleavage patterns in the minimal complex and the 30S subunit are quite distinct although there is an underlying similarity (Figure 4(a) and 6(a)). Many of the cleavage sites from Fe(II)-C14-S20 observed in the small subunit are present in the binary complex however the intensity of cleavage is significantly weaker. Verification of these data (the weak cleavage sites) required much additional repetition of probing experiments and primer extension from multiple, overlapping primers. Base-specific footprinting demonstrated that this derivatized protein does bind 16S rRNA under the isolation and purification conditions used herein and thus the “lack” of cleavage is not due simply to the absence of this protein in the minimal complexes. Some of the differences observed when probing is initiated from position 14 of the r-protein can be explained by its location in S20 and in the 30S subunit, and by the structural changes that likely take place in S20 upon further assembly (i.e. binding of r-protein S17). Amino acid 14 of r-protein S20 is located on its N-terminal helix, the longest helix of the three that compose S20 (Figure 1(c)). This portion of the N-terminal helix is not part of the helical bundle, but is somewhat extended and isolated from other protein elements. Unlike the other primary binding r-proteins (S4, S7, S8, S15 and S17) whose structures in the unliganded state have been determined by NMR, crystallography or both17–25, the structure of S20 has not been determined in the free state. Moreover, it has been shown that free S20 has only about 36% helical content26, compared to its structure in the 30S subunit where it is almost completely helical4. It is possible that changes in S20 conformation particularly in the N-terminal helix, upon binding of additional r-proteins such as S17, could explain changes in the cleavage patterns. The N-terminal helix of r-proteins S20 in the structure of the 30S subunit is enclosed in a channel formed by the tertiary interaction between helix 8 and helix 14. The cleavage sites in helix 11, 13 and 14 appear for Fe(II)-C14-S20 only after binding of S17. We can envision that binding of S17 helps the interaction between helix 14 and helix 8, and both the r-protein S20 and the rRNA surrounding it adopt the appropriate conformation. Thus, a combination of the structure and dynamics of the N-terminal helix of S20 and the 16S rRNA surrounding it, may account for the lower intensity of cleavage in the minimal complex from position C14 and further experiments will be required to fully understand these observations.
R-proteins S16 and S17 appear to play interesting roles in altering the cleavage patterns from Fe(II)-C14-S20. The cleavage that is observed in helix 5 of 16S rRNA in a complex containing Fe(II)-C14-S20 and other r-proteins including S17 is almost completely diminished upon addition of S16 and this cleavage is absent in the 30S subunit. While S16 is proximal to helix 5 it seems unlikely that the reduced cleavage is direct since S16 does not contact helix 5 in the 30S subunit3,4. Helix 15 is sandwiched between S16 and helix 5 and S16 is in direct contact with helix 15 (Figure 8 (h) and (i)). Thus, it is possible that upon S16 association, helix 15 is organized and packs against helix 5 thus resulting in the changes observed in this work. The effect of S16 on helix 5 cleavage would be indirect, and this is also likely for the majority of changes associated with S17 binding given the large distance from S17 to many of the sites. These observations suggest that there are concerted 16S rRNA folding events that are mediated by r-proteins resulting in a hierarchical assembly cascade.
The results of cleavage when Fe(II) is tethered to position C49 of S20 are almost the opposite of what was just mentioned for Fe(II)-C14-S20. Very similar patterns of cleavage are observed when probing is initiated from Fe(II)-C49-S20 in both the binary and fully assembled complexes. Amino acid 49 is very near the junction of the helical bundle of S20 where the three helical elements come together and is the furthest probing position from C14. It is plausible that the N-terminal helix where C14 is localized is unstructured, as explained above. Nevertheless it is possible that the helical region near amino acid 49 of S20 is well formed upon interaction with 16S rRNA and that its environment is not changed significantly after formation of the binary complex. Additionally, many of the sites that are cleaved from Fe(II)-C49-S20 are unique to this tethering position and/or they are more on the periphery of 16S rRNA as it is folded in the 30S subunit4.
Additionally, given the strength of the cleavages in helix 44 from Fe(II)-C49-S20 it is possible that they are less sensitive to changes in helix 44 alignment and positioning. It is interesting that cleavage of helix 44 is so strong only from C49 in the minimal complex, although in the 30S subunit structure amino acids 23 and 57 are as close to these elements. Interestingly in the 30S subunit there are not significant differences in the distance between the three different probing sites and the most remote or proximal cleavage sites in helix 44: for C23 the range is 8–43 Å, for C49 it is 14–39 Å and for C57 it is 11–42 Å. Thus proximity as measurable in the available 30S subunit structure cannot explain the differences in probing results. Again, these differences could be due to strength of cleavage, S20 structure and folding, quenching from other 16S rRNA elements and/or dynamics. This portion of r-protein S20 and the elements of the rRNA cleaved from it may fold earlier in assembly or simply be more accessible and therefore fewer differences in cleavage patterns are observed between minimal and fully assembled RNPs.
Significant differences are observed in the cleavage patterns in helix 44 for binary complexes and the 30S subunit for three of the derivatized proteins Fe(II)-C14-S20, Fe(II)-C23-S20 and Fe(II)-C57-S20, while for Fe(II)-C49-S20 there are small variations. Most intriguing are the results obtained when Fe(II)-C23-S20 and Fe(II)-C57-S20 are used as probes, since significant differences in the cleavage patterns between the binary complexes and 30S subunits were observed. As mentioned above for these two probing positions only small differences in intensity were detected in the 5′ domain (see Figure 5 and 6) suggesting that derivatization of these positions does not interfere with S20 binding, which is consistent with footprinting data (data not shown). Residues 23 and 57 of r-protein S20 are located close to helix 44 in the fully assembled 30S subunit (Figure 1), and four hydrogen bonds between S20 and 16S rRNA are present spatially close to these derivatized residues (Arg28-A1437, Lys32-G1439, Ser25-G1459, Thr39-G1458)4; 27. In the binary complexes containing Fe(II)-C23-S20 or Fe(II)-C57-S20, little cleavage was observed in helix 44, while three and four cleavage sites, respectively were observed for the 30S subunit (Figures 4 and 6 (b) and (d)). Therefore, we cannot unequivocally rule out the possibility that the derivatization of these S20 residues interferes with the orientation of the penultimate stem, and that in the fully assembled subunit this interference is overcome. However, the cleavage patterns from Fe(II)-C49-S20 similar for both RNPs studied here (see Figure 6) and the S20-specific footprints observed in helix 44 in the S20/16S rRNA complexes do not support this hypothesis. Also, similar late docking of helix 44 was observed in tethered probing experiments using Fe(II)-S1510, which does not directly contact helix 444. Thus, it seems likely that the penultimate stem does not interact fully with S20 until late in assembly. Distance measurements within the 30S subunit support this idea. Position C23 of S20 is only 8 Å from some of its cleavage sites in helix 44 but these sites are not cleaved in any of the minimal RNPs tested. Similarly, the distance from C57 and some helix 44 cleavage sites is on the order of 12 Å. These data suggest that either S20 or helix 44, or both, are in an alternative environment or conformation in the minimal RNPs as compared with the fully assembled subunit. The results presented in this manuscript and the previous ones using Fe(II)-S15 as a probe 10 suggest that the positioning of this functionally important rRNA element can occur late in 30S subunit assembly.
Based on our data and that available in the literature 10; 15, we suggest that in the binary complex of S20 and 16S rRNA interaction is mainly between the 5′ domain of 16S rRNA and S20, with minimal interaction with the penultimate stem. The footprints observed in the minimal complex of S20/16S rRNA by chemical probing1 could probably arise from base pairing or RNA-RNA interactions since the protected nucleotides are not directly involved in 16S rRNA/S20 interactions in the fully assembled 30S subunit4; 27. Somewhat later in assembly, in the presence of other r-proteins, the interaction between helix 44 and S20 becomes complete and thus their interaction aids in positioning of this important rRNA element. The 5′ domain seems to be fairly well organized even in the absence of r-proteins15 and binding of S20, may aid this rRNA domain in adopting an architecture very similar to the one in the fully assembled 30S subunit. In contrast, the other three domains of 16S rRNA appear to require the presence of r-proteins (in various degrees) to achieve their functional structure. In the minimal, binary Fe(II)-S15/16S rRNA complex the number of cleavage sites was small and localized to a small region in the central domain of 16S rRNA10; 11. Probing from Fe(II)-S15 in the fully assembled 30S subunit yielded significantly more cleavage sites throughout the central domain and extending to others including the 3′ minor domain. Many of these cleavage sites were present in RNPs of higher complexity. The 3′ minor domain, which interacts mainly with r-protein S20, seems to adopt its final structure late in the assembly. The results presented in this work highlight the complexity of RNP assembly, particularly that of the small ribosomal subunit.
Materials and Methods
Mutagenesis, expression and purification of S20
The gene-encoding r-protein S20 was cloned from Escherichia coli (E. coli) MRE600 genomic DNA into pET24b vector (Novagen)28. Site-directed mutagenesis was used to introduce cysteine residues at three non-conserved positions (Ser 14, Ser 23, and Lys 49). The mutation was confirmed by sequence analysis. The mutated S20 proteins were expressed individually in E. coli pRBL21 and purified as described for wild-type protein28. The fourth cysteine-substituted S20 protein (Ile 57) was used as previously purified2.
Derivatization of S20 proteins
The fluorescent reagent 7-diethylamino-3-((4′-(iodoacetyl)amino)phenyl)-4-methylcoumarin (DCIA; Molecular Probes) was used to test the accessibility of each introduced cysteine residue for derivatization as previously described (methods chapter). Non-specific derivatization at positions other than the cysteine residues was assessed by using the wild-type S20, in parallel with the cysteine-containing S20 mutant proteins. Derivatization of cysteine-containing S20 proteins by DCIA and Fe(II)–BABE [1-(p-bromoacetamidobenzyl) ethylenediaminetetraacetate] was done essentially as described29 (data not shown).
Formation of Fe(II)–S20 containing RNPs
Natural 16S rRNA, isolated from natural 30S subunits as described previously30, was pre-incubated at 42°C for 15 minutes in buffer A (20 mM K+-Hepes (pH 7.6), 20 mM MgCl2). The Fe(II)–S20 containing RNPs were formed by mixing the 16S rRNA (40 pmoles) with the Fe(II)-S20 (either 60, 120 or 200 pmoles), and the other necessary r-proteins (320 pmoles each). In all cases with the varying amounts of Fe(II)-S20 the cleavage sites remained the same although there was slight variation in intensity. Most experiments were performed using a 5-fold molar excess of Fe(II)-S20 to 16S rRNA. The KCl concentration was adjusted to 330 mM in each of the reactions in a final volume of 100 μl and they were incubated at 42°C for 60 minutes. The reaction mixtures were kept on ice for 10 minutes before purification on spin columns and directed probing (see below).
Purification of RNPs from Fe(II)–S20
The complexes containing Fe(II)–S20 proteins were purified to remove any unbound protein with spin columns, prior to directed hydroxyl radical probing. This was done as described by Culver and Noller29, the only difference being the centrifugation speed, which in our case was 6. The purified RNPs were cooled on ice for 10 minutes before the hydroxyl radical probing.
Directed hydroxyl radical probing and primer extension
Directed hydroxyl radical probing of 16S rRNA from Fe(II)–S20, in the different RNPs and the subsequent primer extension analysis were done as described by Culver and Noller29. Each primer extension was performed multiple times with primers designed to redundantly cover appropriate regions of 16S rRNA. The intensity of cleavage was evaluated by visual inspection.
Acknowledgments
This work was supported by the National Institute of Health grant GM62432 to GMC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stern S, Changchien LM, Craven GR, Noller HF. Interaction of proteins S16, S17 and S20 with 16 S ribosomal RNA. J Mol Biol. 1988;200:291–299. doi: 10.1016/0022-2836(88)90241-0. [DOI] [PubMed] [Google Scholar]
- 2.Culver GM, Noller HF. Directed hydroxyl radical probing of 16S ribosomal RNA in ribosomes containing Fe(II) tethered to ribosomal protein S20. RNA. 1998;4:1471–1480. doi: 10.1017/s1355838298981201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 4.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from Escherichia coli. Nature. 1970;226:1214–1218. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- 6.Traub P, Nomura M. Studies on the assembly of ribosomes in vitro. Cold Spring Harb Symp Quant Biol. 1969;34:63–67. doi: 10.1101/sqb.1969.034.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Culver GM. Assembly of the 30S ribosomal subunit. Biopolymers. 2003;68:234–249. doi: 10.1002/bip.10221. [DOI] [PubMed] [Google Scholar]
- 8.Powers T, Noller HF. A temperature-dependent conformational rearrangement in the ribosomal protein S4.16 S rRNA complex. J Biol Chem. 1995;270:1238–1242. doi: 10.1074/jbc.270.3.1238. [DOI] [PubMed] [Google Scholar]
- 9.Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J Mol Biol. 1993;232:362–374. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 10.Jagannathan I, Culver GM. Ribosomal protein-dependent orientation of the 16 S rRNA environment of S15. J Mol Biol. 2004;335:1173–1185. doi: 10.1016/j.jmb.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 11.Jagannathan I, Culver GM. Assembly of the central domain of the 30 S ribosomal subunit: roles for the primary binding ribosomal proteins S15 and S8. J Mol Biol. 2003;330:373–383. doi: 10.1016/s0022-2836(03)00586-2. [DOI] [PubMed] [Google Scholar]
- 12.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinf. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes KL, Culver GM. Analysis of conformational changes in 16S rRNA during the course of 30S subunit assembly. J Mol Biol. 2005;354:340–357. doi: 10.1016/j.jmb.2005.09.056. [DOI] [PubMed] [Google Scholar]
- 14.Holmes KL, Culver GM. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat Struct Mol Biol. 2004;11:179–186. doi: 10.1038/nsmb719. [DOI] [PubMed] [Google Scholar]
- 15.Adilakshmi T, Ramaswamy P, Woodson SA. Protein-independent folding pathway of the 16S rRNA 5′ domain. J Mol Biol. 2005;351:508–519. doi: 10.1016/j.jmb.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Dutca LM, Jagannathan I, Grondek JF, Culver GM. Temperature-dependent RNP conformational rearrangements: analysis of binary complexes of primary binding proteins with 16 S rRNA. J Mol Biol. 2007;368:853–869. doi: 10.1016/j.jmb.2007.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayers EW, Gerstner RB, Draper DE, Torchia DA. Structural preordering in the N-terminal region of ribosomal protein S4 revealed by heteronuclear NMR spectroscopy. Biochemistry. 2000;39:13602–13613. doi: 10.1021/bi0013391. [DOI] [PubMed] [Google Scholar]
- 18.Ramakrishnan V, White SW. Ribosomal protein structures: insights into the architecture, machinery and evolution of the ribosome. Trends Biochem Sci. 1998;23:208–212. doi: 10.1016/s0968-0004(98)01214-6. [DOI] [PubMed] [Google Scholar]
- 19.Clemons WM, Jr, Davies C, White SW, Ramakrishnan V. Conformational variability of the N-terminal helix in the structure of ribosomal protein S15. Structure. 1998;6:429–438. doi: 10.1016/s0969-2126(98)00045-8. [DOI] [PubMed] [Google Scholar]
- 20.Wimberly BT, White SW, Ramakrishnan V. The structure of ribosomal protein S7 at 1.9 A resolution reveals a beta-hairpin motif that binds double-stranded nucleic acids. Structure. 1997;5:1187–1198. doi: 10.1016/s0969-2126(97)00269-4. [DOI] [PubMed] [Google Scholar]
- 21.Hosaka H, Nakagawa A, Tanaka I, Harada N, Sano K, Kimura M, Yao M, Wakatsuki S. Ribosomal protein S7: a new RNA-binding motif with structural similarities to a DNA architectural factor. Structure. 1997;5:1199–1208. doi: 10.1016/s0969-2126(97)00270-0. [DOI] [PubMed] [Google Scholar]
- 22.Jaishree TN, Ramakrishnan V, White SW. Solution structure of prokaryotic ribosomal protein S17 by high-resolution NMR spectroscopy. Biochemistry. 1996;35:2845–2853. doi: 10.1021/bi951062i. [DOI] [PubMed] [Google Scholar]
- 23.Golden BL, Hoffman DW, Ramakrishnan V, White SW. Ribosomal protein S17: Characterization of the three-dimensional structure by proton and nitrogen-15 NMR. Biochemistry. 1993;32:12812–12820. doi: 10.1021/bi00210a033. [DOI] [PubMed] [Google Scholar]
- 24.Agalarov SC, Sridhar Prasad G, Funke PM, Stout CD, Williamson JR. Structure of the S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain. Science. 2000;288:107–113. doi: 10.1126/science.288.5463.107. [DOI] [PubMed] [Google Scholar]
- 25.Davies C, Gerstner RB, Draper DE, Ramakrishnan V, White SW. The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: one domain shows structural homology to the ETS DNA-binding motif. EMBO J. 1998;17:4545–4558. doi: 10.1093/emboj/17.16.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paterakis K, Littlechild J, Woolley P. Structural and functional studies on protein S20 from the 30-S subunit of the Escherichia coli ribosome. Eur J Biochem. 1983;129:543–548. doi: 10.1111/j.1432-1033.1983.tb07083.x. [DOI] [PubMed] [Google Scholar]
- 27.Allers J, Shamoo Y. Structure-based analysis of protein–RNA interactions using the program ENTANGLE. J Mol Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- 28.Culver GM, Cate JH, Yusupova GZ, Yusupov MM, Noller HF. Identification of an RNA-protein bridge spanning the ribosomal subunit interface. Science. 1999;285:2133–2136. doi: 10.1126/science.285.5436.2133. [DOI] [PubMed] [Google Scholar]
- 29.Culver GM, Noller HF. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol. 2000;318:461–475. doi: 10.1016/s0076-6879(00)18070-x. [DOI] [PubMed] [Google Scholar]
- 30.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 31.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- 32.Lancaster L, Culver GM, Yusupova GZh, Cate JH, Yusupov MM, Noller HF. The location of protein S8 and surrounding elements of 16S rRNA in the 70S ribosome from combined use of directed hydroxyl radical probing and x-ray crystallography. RNA. 2000;6:717–729. doi: 10.1017/s1355838200000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noller HF, Baucom A. Structure of the 70S ribosome: implications for movement. Biochem Soc Trans. 2002;30:1159–1161. doi: 10.1042/bst0301159. [DOI] [PubMed] [Google Scholar]
- 34.Nguyenle T, Laurberg M, Brenowitz M, Noller HF. Following the dynamics of changes in solvent accessibility of 16S and 23S rRNA during ribosomal subunit association using synchrontron-generated hydroxyl radicals. J Mol Biol. 359:1235–1248. doi: 10.1016/j.jmb.2006.04.030. [DOI] [PubMed] [Google Scholar]