Abstract
Prostaglandin E2 blocks transforming growth factor TGF β1-induced CCN2/CTGF expression in lung and kidney fibroblasts. PGE2 levels are high in gingival tissues yet CCN2/CTGF expression is elevated in fibrotic gingival overgrowth. Gingival fibroblast expression of CCN2/CTGF in the presence of PGE2 led us to compare the regulation of CCN2/CTGF expression in fibroblasts cultured from different tissues. Data demonstrate that the TGFβ1-induced expression of CCN2/CTGF in human lung and renal mesangial cells is inhibited by 10 nm PGE2, whereas human gingival fibroblasts are resistant. Ten nm PGE2 increases cAMP accumulation in lung but not gingival fibroblasts, which require 1 μm PGE2 to elevate cAMP. Micromolar PGE2 only slightly reduces the TGFβ1-stimulated CCN2/CTGF levels in gingival cells. EP2 prostaglandin receptor activation with butaprost blocks the TGFβ1-stimulated expression of CCN2/CTGF expression in lung, but not gingival, fibroblasts. In lung fibroblasts, inhibition of the TGFβ1-stimulated CCN2/CTGF by PGE2, butaprost, or forskolin is due to p38, ERK, and JNK MAP kinase inhibition that is cAMP-dependent. Inhibition of any two MAPKs completely blocks CCN2/CTGF expression stimulated by TGFβ1. These data mimic the inhibitory effects of 10 nm PGE2 and forskolin that were dependent on PKA activity. In gingival fibroblasts, the sole MAPK mediating the TGFβ1-stimulated CCN2/CTGF expression is JNK. Whereas forskolin reduces TGFβ1-stimulated expression of CCN2/CTGF by 35% and JNK activation in gingival fibroblasts, micromolar PGE2-stimulated JNK in gingival fibroblasts and opposes the inhibitory effects of cAMP on CCN2/CTGF expression. Stimulation of the EP3 receptor with sulprostone results in a robust increase in JNK activation in these cells. Taken together, data identify two mechanisms by which TGFβ1-stimulated CCN2/CTGF levels in human gingival fibroblasts resist down-regulation by PGE2: (i) cAMP cross-talk with MAPK pathways is limited in gingival fibroblasts; (ii) PGE2 activation of the EP3 prostanoid receptor stimulates the activation of JNK.
Connective tissue growth factor (CTGF),2 or CCN2, is a 38-kDa secreted protein belonging to the CCN family of growth factors, and its expression is induced in cultured fibroblasts by TGFβ (1-4). CCN2/CTGF has been shown to promote the synthesis of various constituents of the extracellular matrix (5-9) and its overexpression is associated with the onset and progression of fibrosis in skin (scleroderma) (10, 11), kidney (12, 13), liver (14-18), brain (Alzheimer disease) (19), lung (20, 21), human gingiva (22-24), vasculature (atherosclerosis) (25-27), pancreas (chronic pancreatitis) (28), digestive system (inflammatory bowel disease) (29), and the eye (lens) (30). In scleroderma lesions of the skin CCN2/CTGF is overexpressed in the body of the lesion, whereas TGFβ is expressed in only the leading edges of the lesion and suggests that TGFβ initiates persistent CCN2/CTGF expression (10). A similar mechanism may be involved in promoting gingival overgrowth and fibrosis in response to the anti-convulsive drug phenytoin (Dilantin®). Immunohistologic analyses have revealed that CCN2/CTGF, and to a lesser degree TGFβ1, are overexpressed in gingival lesions in patients experiencing phenytoin- (Dilantin) induced gingival overgrowth and fibrosis (22-24).
More recent quantitative real time PCR analysis of RNA isolated from human gingival tissues indicate that CCN2/CTGF mRNA expression is increased 6-fold in phenytoin-induced gingival overgrowth tissues (24). We have found that TGFβ1 strongly up-regulates CCN2/CTGF expression in cultured primary human gingival fibroblasts (22-24). Among drug-induced gingival overgrowth lesions, those caused by phenytoin are more fibrotic and contain little to no leukocytic infiltration compared with lesions induced by cyclosporine-A or nifedipine (22). Taken together with the notion that CCN2/CTGF can itself enhance the production of extracellular matrix macro-molecules, these data suggest that phenytoin-induced gingival overgrowth results from the accumulation of fibrous constituents of the extracellular matrix that are likely stimulated by TGFβ1 and CCN2/CTGF.
Published studies investigating the TGFβ1-induced expression of CCN2/CTGF in human fetal lung fibroblasts (IMR90) and normal rat kidney fibroblasts indicate that CTGF expression is highly sensitive to PGE2-mediated inhibition via a cAMP-dependent mechanism (8, 31, 32). CCN2/CTGF overexpression occurs in gingival overgrowth tissues, in response to phenytoin, where PGE2 levels are persistently high due to normal wounding and inflammation in the oral environment that results from normal masticatory function (33, 34). We therefore conducted comparative analyses of human fibroblasts from different tissues to investigate the effects of PGE2 on the TGFβ1-stimulated expression of CCN2/CTGF. We suspected that gingival fibroblasts utilize unique signal transduction pathways to promote CCN2/CTGF expression in response to TGFβ1 that make these cells resistant to the inhibitory effects of PGE2.
Mitogen-activated protein kinases (MAPK) pathways stimulated by TGFβ1 are involved in the tissue-specific expression of CCN2/CTGF in human fibroblasts (35). Our findings demonstrate that the tissue-specific effect of PGE2 on the TGFβ1-stimulated expression of CCN2/CTGF in human fibroblasts depends primarily on the differences in cAMP-mediated inhibition of MAPK signaling. These data support the need for developing therapeutic approaches to regulate the TGFβ1-induced expression of CCN2/CTGF and fibrosis on the basis of the tissue-specific signal transduction pathways.
EXPERIMENTAL PROCEDURES
Materials
SB203580, SP600125, PD98059, forskolin, KT5720, and cAMP ELISA kits were obtained from Sigma; RNeasy mini-RNA purification kits purchased from Qiagen; TGFβ1 from Peprotech; prostaglandin E2 and mouse anti-β-actin monoclonal antibody from Calbiochem; butaprost and sulprostone from Cayman; rabbit anti-human CCN2/CTGF polyclonal antibody was kindly provided by FibroGen Corporation, South San Francisco, CA; U0126, LumiGLO chemiluminescent detection system, rabbit anti-MAPK antibodies against phospho-SAPK/JNK (Thr-183, Tyr-184; catalog number 9251), total SAPK/JNK (catalog number 9252), phospho-ERK1/2 (p44/42) (Thr-202, Tyr-204; catalog number 9101), total ERK1/2 (catalog number 9102), phospho-p38 (Thr-180, Tyr-182; catalog number 9211), total p38 (catalog number 9212) antibodies, and secondary anti-rabbit horseradish peroxidase-conjugated antibodies were obtained from Cell Signaling. Dulbecco’s modified Eagle’s medium, phosphate-buffered saline, trypsin, non-essential amino acids, and antibiotics (penicillin/streptomycin) from Invitrogen; all solutions, TaqMan probes, and instruments for real time PCR were obtained from Applied Biosystems; 100-mm and 6-well cell culture plates from Fisher; Restore Western blot Stripping Solution from Pierce. Nuclear extraction kits were obtained from Active Motif.
Cell Culture
IMR90 (human fetal lung) fibroblasts and primary human renal mesangial cells were purchased from ATCC and Cambrex, respectively. Primary human gingival fibroblasts were obtained from donors without gingival overgrowth from the Clinical Research Center at Boston University Goldman School of Dental Medicine as described previously (23). Gingival cells from two different adult donors were used and data obtained were fully reproducible. Data shown are from one donor (HCT-11). Fibroblasts from all tissues were grown in Dulbecco’s modified Eagle’s medium supplemented with 0.1 mm non-essential amino acids, 10% fetal bovine serum, and 100 units/ml penicillin, and 0.1 mg/ml streptomycin at 37 °C and 5% CO2 in a fully humidified incubator in 100-mm cell culture dishes. Cells were grown to 80% confluence and then placed in serum-free medium containing 0.1% bovine serum albumin for a minimum of 12 h prior to treatment. Specified inhibitors were next added in serum-free bovine serum albumin medium for 30 or 60 min prior to the addition of TGFβ1 or vehicle. TGFβ1 treatments were generally for either 4 h for RNA analyses, or 6 h for protein analyses, based on previous studies (23). Cells were not passed beyond four passages.
Real Time PCR
Total RNA was isolated from fibroblast cell cultures and purified using RNeasy mini-RNA purification kits (Qiagen). RNA obtained was run on a 1% agarose gel to ensure RNA quality by analyzing for the presence of 18 and 28 S rRNA in proper relative proportions. RNA was quantified via spectrophotometry and 1 μg of RNA per treatment condition was added to 30 μl of reverse transcription reactions using random primers and the Applied Biosystems Reverse Transcription kit. Thermal cycler conditions for reverse transcription were 25 °C for 10 min, 37 °C for 60 min, and 95 °C for 5 min and cDNA was stored at -20 °C prior to use in real time PCR. Four microliters of each reverse transcription reaction was used for 50 μl of real time PCR using the traditional 96-well format. TaqMan probes for CCN2/CTGF (catalog number Hs00170014_m1) and glyceraldehyde-3-phosphate dehydrogenase (catalog number Hs99999905_m1) were used in real time PCR analyses. Real time PCR were run in an Applied Biosystems GeneAmp Prism 7700 System at thermal cycler conditions of 50 °C for 2 min, 95 °C for 10 min, and 60 °C for 1 min for 40 cycles. Data were analyzed using the 2-ΔΔct method and CCN2/CTGF mRNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA and no treatment controls (36).
Western Blot Analysis
Fibroblast cell cultures were grown as described above and total protein harvested by dissolving cell layers in 500 μl of sample buffer containing 62.5 mm Tris, 10% glycerol, 2% SDS, and 5% β-mercaptoethanol per 100-mm culture dish. Samples were then boiled for 5 min and stored at -80 °C. Samples were then subjected to 10% SDS-PAGE and Western blotting with antibodies specific for proteins of interest. Proteins were transferred to polyvinylidene difluoride membranes overnight at 4 °C in blotting buffer containing 0.025 m Tris, 0.192 m glycine, and 20% methanol. Membranes were blocked with blocking solution containing 5% dry milk in TBST (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.1% Tween) for 1 h in the presence of primary rabbit antibodies overnight at 4 °C with mild shaking. Membranes were then washed with TBST 5 times for 10 min each. Membranes were next subjected to incubation in the presence of secondary, anti-rabbit horse-radish peroxidase-conjugated antibodies in 5% milk and TBST for 2 h at room temperature with mild shaking. Membranes were washed with TBST 5 times for 5 min each. Chemiluminescent detection of bound horseradish peroxidase-conjugated secondary antibodies was determined using the LumiGlo/peroxide and exposed to film. Membranes were subsequently stripped using Restore Western Stripping Solution and re-probed for loading controls as required.
cAMP ELISA
ELISA kits for the detection of cAMP were obtained from Sigma (catalog number CA200) and experiments followed the manufacturer’s suggested guidelines. Briefly, fibroblasts were cultured in 6-well plates and grown until confluent and subsequently serum deprived for a minimum of 12 h. Cells were then challenged with a low (10 nm) or high (1 μm) concentration of PGE2 for the times indicated in the text. Culture lysates were then harvested using 150 μl of 0.1 n HCl per well and maintained on ice until the assay was conducted. Absorbance was measured at 550 nm and data were analyzed via interpolation with standards using semi-logarithmic paper. A minimum of n = 5 for each experimental condition was performed.
Analysis of TGFβ1-induced Smad Activation and Nuclear Localization
Gingival fibroblast cultures were grown to near confluence and serum starved for 12 h. Cultures were then pre-incubated with 10 μm MAPK inhibitors PD98059, U0126, SP600125, and SB203580 for 1 h. Media were then removed and replaced with identical concentrations of MAPK inhibitor in addition to 5 ng/ml TGFβ1 and incubated for an additional 30 min. Total cell layer protein was harvested to determine the effect of MAPK inhibition on Smad-3 phosphorylation/activation by TGFβ1 using Western blot analysis. Nuclear extracts were also obtained following the 30-min incubation in the presence of TGFβ1 with or without challenge with the JNK inhibitor SP600125 (10 μm). Cells were lysed and nuclei obtained free of cytosolic proteins according to the manufacturer’s instructions (Active Motif Nuclear Extraction Kit). Phosphorylated Smad-3 levels were then determined in nuclear extracts via Western blot normalized to lamin B.
Expression of Recombinant Adenovirus
Gingival fibroblasts were grown in 12-well plates to 80% confluence. Adenovirus expressing β-galactosidase was used as the control and assessment of the level of infection and gene expression in our cells. The dominant negative adenoviral construct for JNK1 (Cell Biolabs, Inc., ADV-115) was introduced at a concentration of 1010 infectious units/ml and cultures were grown for 48 h to permit expression. At this concentration of virus we achieved ∼100% infection of gingival fibroblasts as determined by β-galactosidase staining determined separately with a β-galactosidase-expressing viral construct (Cell Biolabs). Forty-eight hours post-infection, TGFβ1 was added and cultures were incubated for 6 h prior to harvest of total cell layer protein for Western blot analysis of CCN2/CTGF protein.
RESULTS
Tissue-specific Inhibition of the TGFβ1-stimulated Expression of CCN2/CTGF by PGE2
Human gingival tissues persistently contain relatively high levels of PGE2 as a consequence of normal levels of inflammation in the oral environment and normal masticatory function (33, 34). PGE2 has been shown to block the TGFβ1-stimulated expression of CCN2/CTGF in lung and kidney fibroblastic cells (31, 32). However, we have demonstrated that CCN2/CTGF is expressed at high levels in phenytoin-induced gingival overgrowth lesions in human patients (22, 23). We therefore investigated the ability of PGE2 to inhibit TGFβ1-stimulated CCN2/CTGF expression in gingival fibroblasts. Primary fibroblast cell cultures from human gingiva, and control lung (IMR90) and primary kidney mesangial fibroblastic cells were grown and pre-treated with PGE2 for 1 h followed by the addition of 5 ng/ml TGFβ1 as described under “Experimental Procedures” to determine the effects of PGE2 on the TGFβ1-induced expression of CCN2/CTGF as a function of the tissue of origin. Data demonstrate that TGFβ1 increases CCN2/CTGF expression in cultures of primary human gingival fibroblasts, renal mesangial cells, and lung fibroblasts. Pretreatment with 10 nm PGE2 completely blocks the expression of CCN2/CTGF mRNA (Fig. 1A) and protein levels (Fig. 1B) in human renal mesangial cells and IMR90 (lung) fibroblasts. In contrast, CCN2/CTGF mRNA expression induced by TGFβ1 in human gingival fibroblasts is not reduced by challenge with 10 nm PGE2 and only slightly reduced in response to 1 μm PGE2 (Fig. 1A). The receptor-independent activator of adenylate cyclase, forskolin, was next utilized to investigate a role for elevated cAMP in the absence of PGE2 receptor stimulation. Forskolin completely blocked CCN2/CTGF mRNA expression in response to TGFβ1 in human lung and renal mesangial cell cultures (Fig. 1A). Gingival fibroblast cultures, however, were found to be more resistant to the inhibitory effects of forskolin although forskolin did more potently reduce CCN2/CTGF mRNA expression than did 1 μm PGE2 (Fig. 1A). Western blot analysis for CCN2/CTGF protein in the different fibroblastic cultures confirmed that gingival fibroblasts are resistant to the inhibitory effects of PGE2 (Fig. 1B). The differing degrees of susceptibility, or resistance, to inhibition with PGE2 suggest that unique signaling mechanisms occur in human gingival fibroblasts. Thus, we focused our investigation on human gingival fibroblasts and the human lung fibroblasts to further explore these tissue-specific mechanisms of regulating the TGFβ1-induced expression of CCN2/CTGF by PGE2.
FIGURE 1. Gingival fibroblasts are resistant to PGE2 down-regulation of TGF β1-stimulated CCN2/CTGF levels.
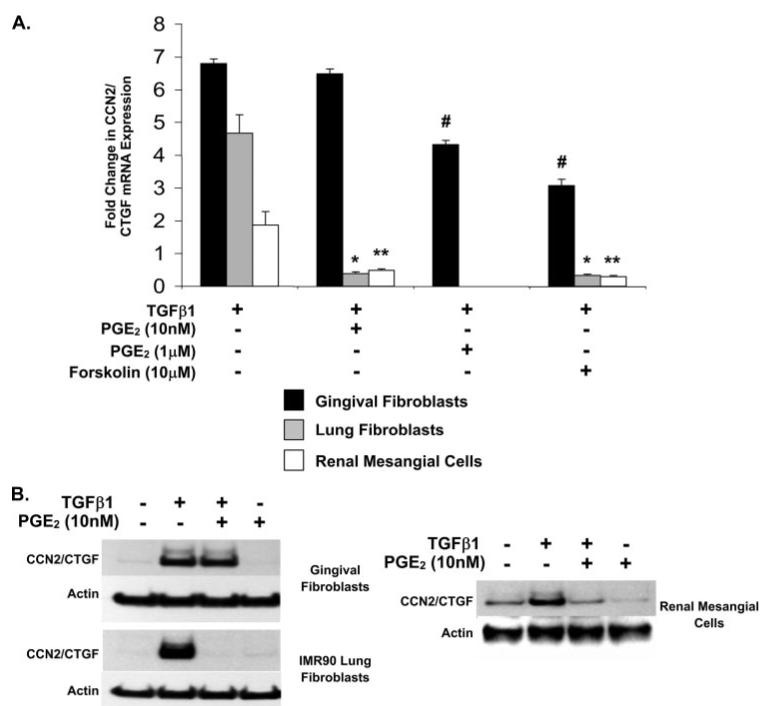
Cultured fibroblasts derived from different human tissues were treated with 10 nm PGE2 or 10 μm forskolin for 30 min prior to the addition of 5 ng/ml TGFβ1 and CCN2/CTGF mRNA and protein levels were determined by real time PCR or Western blot analysis following a 4- or 6-h incubation, respectively. CCN2/CTGF mRNA expression in response to 1 μm PGE2 was also evaluated in gingival fibroblasts. A, real time PCR analysis of RNA isolated from cultured human IMR90 lung, renal, and gingival fibroblastic cells. Data are measured as the mean ± S.D. Analyses were conducted using triplicate cultures and experiments were repeated at least three times with identical results. CCN2/CTGF mRNA expression was normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA and compared with no treatment control cultures for the evaluation of statistically significant changes (*, p < 0.0005, **, p < 0.005, # p < 0.0001). B, Western blot analysis of the inhibition of the TGFβ1-induced expression of CCN2/CTGF protein by PGE2 in human gingival fibroblasts, lung fibroblasts, and renal mesangial cells. Total β-actin was used as a gel-loading control. Each analysis was conducted at least twice with identical results.
Forskolin and PGE2 Are Weak Inhibitors of CCN2/CTGF Protein Expression in Human Gingival Fibroblasts
Human gingival fibroblast cultures are resistant to the inhibitory effects of nanomolar concentrations of PGE2 on CCN2/CTGF protein expression (Fig. 1B), but 1 μm PGE2 and 10 μm forskolin resulted in a weak inhibition of CCN2/CTGF mRNA expression in gingival fibroblasts (Fig. 1A). We next determined if this inhibition also occurred at the level of CCN2/CTGF protein expression. Gingival fibroblast cultures were pretreated with 10 nm PGE2, 1 μm PGE2, or 10 μm forskolin for 30 min. Cells were then fed with media supplemented with 5 ng/ml TGFβ1 and PGE2, or forskolin, and cultures were incubated for another 6 h prior to harvesting total cell layer protein for Western blot analysis. A representative Western blot demonstrates that 10 nm PGE2 had no effect on the TGFβ1-stimulated expression of CCN2/CTGF protein in gingival fibroblast cultures as expected, whereas treatment with 1 μm PGE2 or forskolin resulted in a slight reduction of CCN2/CTGF protein (Fig. 2A). Densitometry analyses of three separate Western blots from different experiments demonstrate that 1 μm PGE2 results in a 10-13% reduction in the TGFβ1-induced expression of CCN2/CTGF protein, whereas forskolin more potently reduced CCN2/CTGF protein by 30-34% (Fig. 2B). Data confirm that gingival fibroblasts are resistant to down-regulation of TGFβ1-stimulated CCN2/CTGF protein levels by PGE2, and that forskolin is a weak inhibitor.
FIGURE 2. Weak down-regulation of CCN2/CTGF protein by 1 μm PGE2 and forskolin in human gingival cells.
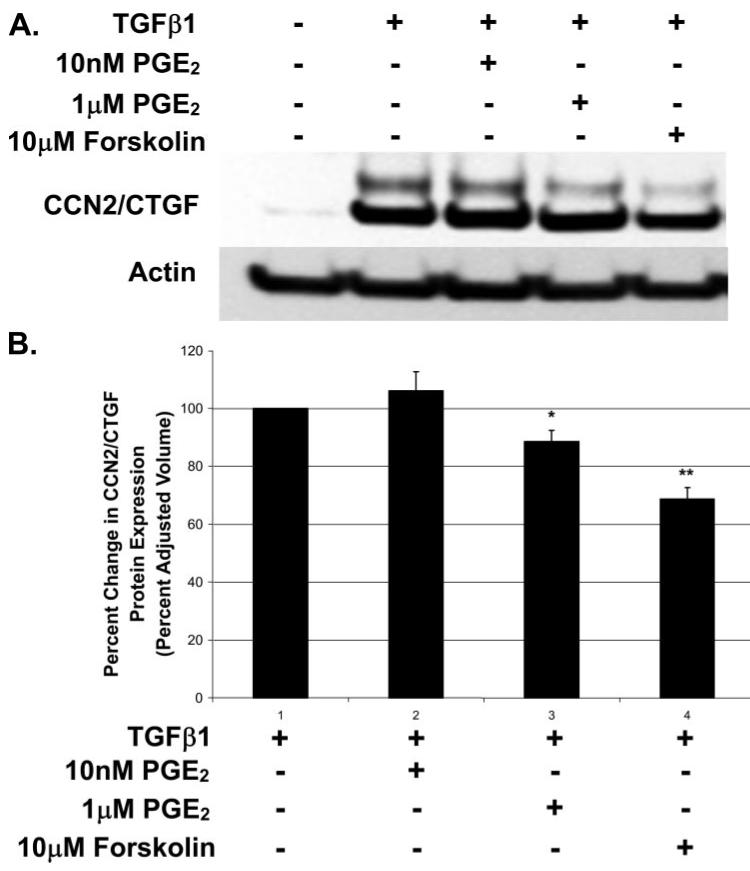
Human gingival fibroblast cultures were treated with 10 nm PGE2, 1 μm PGE2, or 10 μm forskolin 30 min prior to the addition of 5 ng/ml TGFβ1. Total protein was collected 6 h later and CCN2/CTGF protein expression was determined via Western blot analysis. A, Western blots showing CCN2/CTGF protein expression in human gingival fibroblasts in response to micromolar concentrations of PGE2 or forskolin and 5 ng/ml TGFβ1. B, densitometric analysis of CCN2/CTGF protein expression in gingival fibroblasts from three separate experiments. Data are represented as the mean ± S.D. and statistical evaluation per each experimental condition determined (*, p < 0.005; **, p < 0.0005) versus cultures stimulated with TGFβ1 alone.
Stimulation of the EP2 Prostanoid Receptor Blocks the Expression of CCN2/CTGF in Human Lung but Not in Gingival Fibroblasts
Published reports have shown that PGE2 blocks CCN2/CTGF expression in human lung (IMR90) and normal rat kidney fibroblasts by a cAMP-mediated mechanism (31, 32). Of four PGE2 receptors that have been characterized, EP1-EP4, EP2, and EP4 are Gαs-coupled receptors capable of directly stimulating adenylate cyclase activity and the accumulation of cAMP (37, 38). Direct evidence for EP2 receptor involvement in mediating the inhibitory effects of PGE2 have been reported (39). The EP3 prostanoid receptor is capable of either stimulating or inhibiting cAMP production depending on the predominant isoform expressed in a particular tissue or cell type (40). We therefore conducted experiments to determine the effects of the direct stimulation of specific PGE2 receptors EP2 and EP3, to determine which receptor isoforms were responsible for promoting the inhibitory effects of PGE2 on CCN2/CTGF expression. Cultures of human lung and gingival fibroblasts were pretreated with the EP2- or EP3-specific agonists, butaprost and sulprostone, respectively, for 30 min prior to stimulation with TGFβ1. The direct stimulation of the EP2 receptor with butaprost resulted in the complete inhibition of the TGFβ1-stimulated expression of CCN2/CTGF mRNA (Fig. 3A) and protein (Fig. 3B) in cultures of lung fibroblasts, as expected. In gingival fibroblasts, we observed only a decrease of 50% in CCN2/CTGF mRNA expression in response to EP2 stimulation (Fig. 3A) and no reduction in CCN2/CTGF protein levels (Fig. 3C). Stimulation of EP3 with sulprostone did not have any significant effect on CCN2/CTGF mRNA expression in lung fibroblasts, but appeared to slightly increase CCN2/CTGF protein in gingival fibroblasts (Fig. 3, A-C). These data suggest that cAMP accumulation in response to the stimulation of the EP2 prostanoid receptor is responsible for the specific inhibition of CCN2/CTGF expression in human lung fibroblasts and is consistent with published studies (32). By contrast, gingival fibroblasts are resistant to the inhibitory effects of EP2 activation (Fig. 3C).
FIGURE 3. EP2 and EP3 prostaglandin receptor agonist regulation of TGFβ1-stimulated CCN2/CTGF.
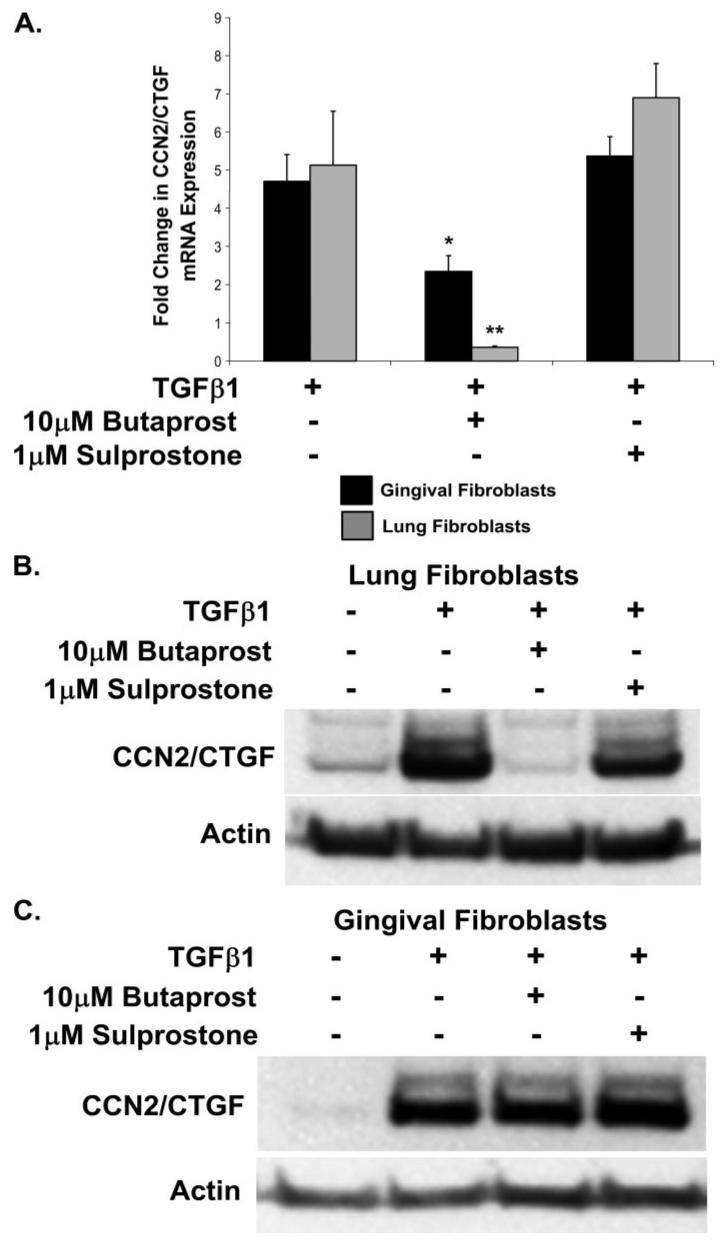
Fibroblast cultures were pretreated for 30 min with an agonist of either EP2 or EP3 prostanoid receptors, butaprost and sulprostone, respectively, prior to the addition of 5 ng/ml TGFβ1. Cultures were incubated for an additional 4 h prior to harvest of total RNA for real time PCR analysis or 6 h prior to harvest of total protein for Western blot. A, real time PCR analysis demonstrating that CCN2/CTGF mRNA expression is completely inhibited by EP2, but not EP3, receptor stimulation in human lung fibroblasts (**, p < 0.005). Stimulation of the EP2 prostanoid receptor with butaprost only partially reduced CCN2/CTGF mRNA in human gingival fibroblasts (*, p < 0.01). Data represent the mean ± S.D. from at least three separate experiments, each performed in triplicate. CCN2/CTGF mRNA was normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA expression. B, Western blot for CCN2/CTGF protein levels normalized to β-actin showing that the TGFβ1-stimulated expression of CCN2/CTGF protein was completely blocked by the activation of EP2 but not EP3 in human lung fibroblasts, and (C) the activation of the EP3, but not the EP2, prostanoid receptor slightly increased the TGFβ1-induced expression of CCN2/CTGF protein in human gingival fibroblasts. Western blot analyses were conducted at least twice with identical results.
PGE2-induced cAMP Accumulation Is Greater in Human Lung Fibroblasts Than in Gingival Fibroblasts
Two of the four known PGE2 receptor subtypes are capable of stimulating adenylate cyclase activity and the production of cAMP upon binding PGE2 (37, 38). We hypothesized that cAMP production and/or accumulation could be higher in cells with a greater susceptibility to the inhibitory effects of PGE2 on CCN2/CTGF expression (lung fibroblasts versus gingival fibroblasts). Cultures of both human lung and gingival fibroblasts were treated for various times with either low (10 nm) or high (1 μm) concentrations of PGE2, and cAMP accumulation was determined by ELISA. Data demonstrate that 10 nm PGE2 was sufficient to produce a strong and transient increase of cAMP in lung fibroblast cultures with maximal production 5 min after treatment, with cAMP levels gradually returning to basal levels by 60 min (Fig. 4). The only statistically significant (*, p < 0.001) increase in cAMP in gingival fibroblast cultures in response to 10 nm PGE2 occurred 30 min after treatment (Fig. 4) and was less than 5% of the increase in cAMP observed in cultures of human lung fibroblasts. Lung fibroblast cultures treated with 1 μm PGE2 exhibited a sustained accumulation of cAMP beyond 60 min following treatment. In contrast, cAMP levels in gingival fibroblasts were increased only transiently in response to 1 μm PGE2 with maximal accumulation at 30 min with levels rapidly returning to basal levels by 60 min post-treatment (Fig. 4). The receptor-independent activator of adenylate cyclase, forskolin (10 μm), yielded a significant but somewhat less robust increase in cAMP than did 1 μm PGE2 in cultures of lung and gingival fibroblasts as measured at 30 min post-treatment (Fig. 4).
FIGURE 4. PGE2-stimulated cAMP production by human lung and gingival fibroblasts.
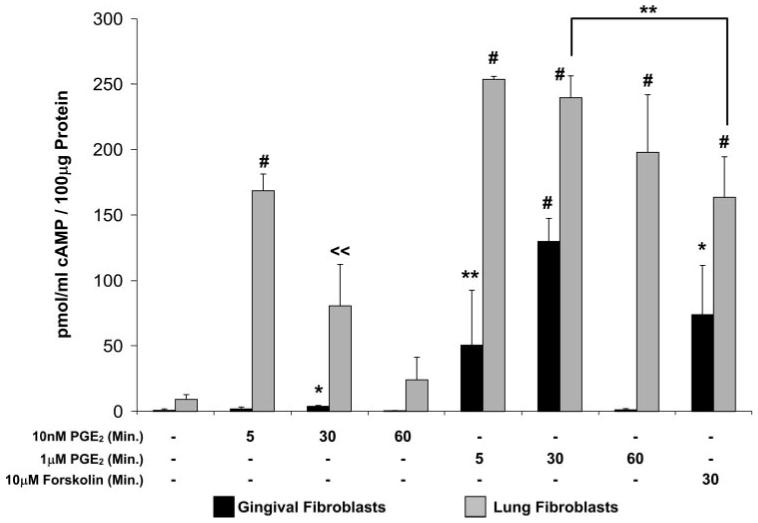
Fibroblast cultures were treated with 10 nm PGE2, 1 μm PGE2, or 10 μm forskolin for the times indicated. Cyclic AMP levels were assessed using cAMP ELISA as described under “Experimental Procedures” and normalized against total cell layer protein as determined by the Bradford assay. Each experimental condition was performed multiple times (n = 5). Data are expressed as the mean ± S.D. Statistically significant increases in cAMP are based on comparison with control versus treated cultures of the same cell type. In lung but not gingival fibroblast cultures, 1 μm PGE2 stimulated a significant increase compared with stimulation with 10 μm forskolin (*, p < 0.005; **, p < 0.02; # p < 0.00001; ≪ p < 0.001).
Tissue-specific MAPK Signaling Requirements for TGFβ1-stimulated CTGF Expression
TGFβ1 has been shown to stimulate the activation of MAPKs, p38, ERK, and JNK (35). We hypothesized that the observed tissue-specific regulation of the TGFβ1-mediated expression of CCN2/CTGF may involve the differential activation of MAPK signaling cascades. We therefore challenged cultures of both human lung and gingival fibroblasts with inhibitors of individual MAPKs to determine the MAPK signaling requirements for the TGFβ1-stimulated expression of CCN2/CTGF mRNA and protein. Cultures were pretreated with the indicated inhibitors for 60 min and media was replaced with fresh medium containing 5 ng/ml TGFβ1 and inhibitor, and then incubated for 4 or 6 h as indicated under “Experimental Procedures.” The inhibition of JNK with 10 μm SP600125 resulted in a dramatic decrease in CCN2/CTGF mRNA (Fig. 5A) and protein (Fig. 5B) expression in gingival fibroblasts, whereas challenge with 10 μm SB203580 (p38 inhibitor), U0126, or PD98059 (ERK1/2 inhibitors) was without significant effect. By contrast, the presence of any MAPK inhibitor in cultures of human lung fibroblasts significantly reduced CCN2/CTGF mRNA (Fig. 5C) and protein expression (Fig. 5D). Densitometric analysis from four separate experiments demonstrates that the inhibition of JNK with 10 μm SP600125 was significant and reduced CCN2/CTGF protein expression in response to TGFβ1 by 60% in gingival fibroblasts (Fig. 5E). Data demonstrate that different MAPK signaling pathways are required to promote optimal expression of CCN2/CTGF mRNA in response to TGFβ1 in human lung and gingival fibroblasts. Control experiments demonstrated that treatment of MAPK inhibitors without TGFβ1 did not elevate CCN2/CTGF expression (data not shown).
FIGURE 5. MAP kinase inhibitors and effects on TGFβ1-stimulated CCN2/CTGF production.
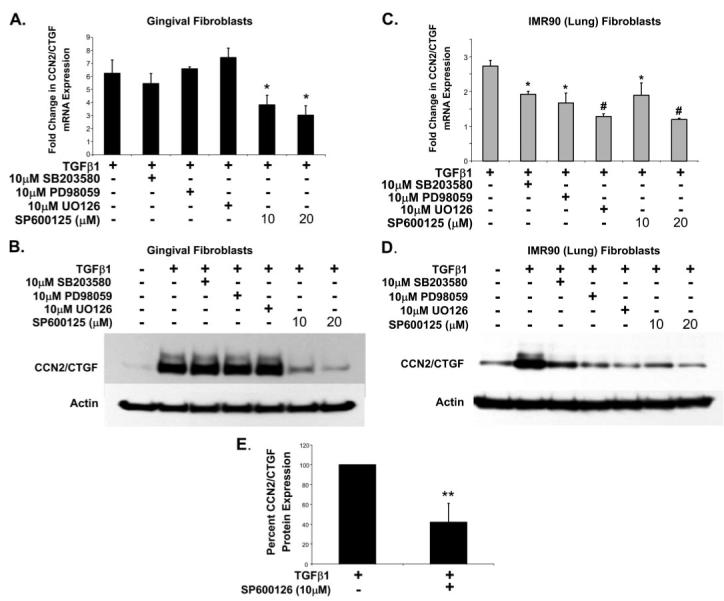
Cultured human lung and gingival fibroblasts were treated with MAPK inhibitors for 1 h and media was then replaced with fresh MAPK inhibitor and 5 ng/ml TGFβ1. CCN2/CTGF mRNA and protein expression levels were determined by real time PCR or Western blot analysis following a 4- or 6-h incubation, respectively, following the introduction of TGFβ1. Real time PCR and Western blot analyses in cultures of gingival fibroblasts (A and B), or IMR90 lung fibroblasts (C and D), are shown, respectively. Each experimental condition was conducted in triplicate and each experiment conducted twice with identical results. E, densitometric analysis from four separate experiments was conducted to determine the extent of inhibition on gingival CCN2/CTGF protein expression by the JNK inhibitor SP600125 (*, p < 0.05; #, p < 0.01; **, p < 0.005).
Dominant Negative JNK1 Inhibits TGFβ1-stimulated CCN2/CTGF Expression
Inhibition of JNK with SP600125 resulted in a significant reduction of CCN2/CTGF mRNA and protein expression induced by TGFβ1 in gingival fibroblasts. To independently confirm that these results were not due to nonspecific effects of this pharmacologic inhibitor, we infected gingival fibroblast cultures with recombinant adenovirus expression dominant negative JNK1 (DN-JNK1). Gingival fibroblast cultures were infected and treated as described under “Experimental Procedures.” Fig. 6A demonstrates that the overexpression of DN-JNK1 protein is achieved following infection with the recombinant adenovirus. Gingival fibroblast cultures expressing DN-JNK1 showed a significant reduction in CCN2/CTGF expression upon stimulation with TGFβ1 compared with control cultures infected with adenovirus expressing β-galactosidase (Fig. 6, B and C).
FIGURE 6. The overexpression of dominant-negative JNK1 blocks CCN2/CTGF expression.
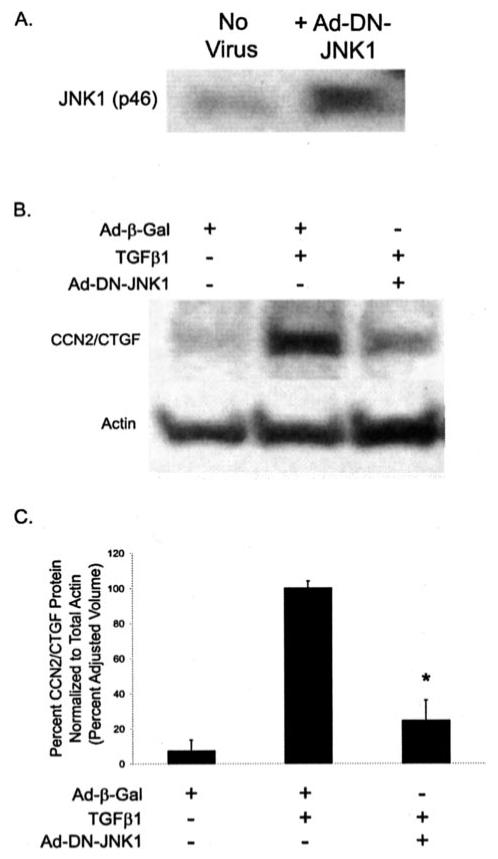
Gingival fibroblasts infected with recombinant adenovirus as described under “Experimental Procedures.” Infection with recombinant adenovirus resulted in nearly 100% infection efficiency and expression of β-galactosidase (data not shown). With an identical viral load of recombinant adenovirus expressing a dominant-negative (DN) form of JNK1 (p46), infected cultures showed an increase in the expression of JNK1 2-2.5-fold greater compared with uninfected cultures as determined using Western blot (A), normalized to total JNK2. Gingival fibroblast cultures infected with DN-JNK1 expressing adenovirus resulted in a significant decrease in the TGFβ1-induced expression of CCN2/CTGF (B and C) (*, p < 0.0005). No significant difference in CCN2/CTGF protein levels was determined between Ad-β-galactosidase-infected control cultures and cultures infected with adenovirus expressing DN-JNK1 and treated with TGFβ1. Three separate experiments are represented by these data.
TGFβ1 Activation and Nuclear Localization of Smad-3 Is Independent of JNK Activation
Smad-3 has been shown in Smad-3 knock-out studies to be a critical component of TGFβ1-stimulated CCN2/CTGF expression and the progression of fibrosis in the lungs, kidneys, and skin (41-43). Interestingly, one study has suggested that Smad-3 phosphorylation by JNK facilitates both its activation by the TGFβ receptor complex and its nuclear accumulation (44). We therefore wished to determine whether the decreased expression of CCN2/CTGF by JNK inhibition was the consequence of interference with TGFβ1-mediated inhibition of Smad-3 activation or nuclear accumulation. Our data suggest that the JNK inhibitor SP600125 (10 or 20 μm) caused no reduction in TGFβ1-mediated phosphorylation of Smad-3 (Fig. 7A) or the nuclear localization of phosphorylated Smad-3 (Fig. 7B).
FIGURE 7. JNK inhibition does not inhibit TGFβ-1-induced Smad-3 activation or nuclear localization.
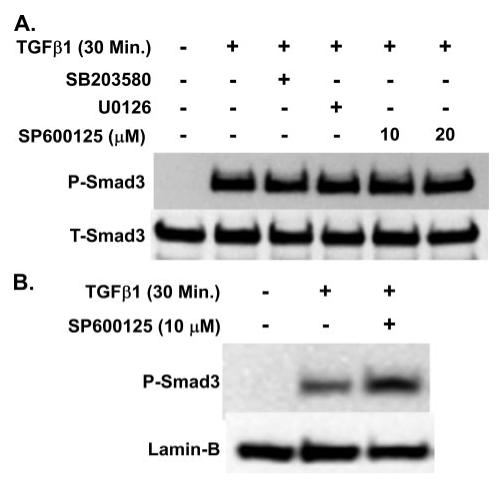
Treatment of gingival fibroblast cultures with 10 μm MAPK inhibitors, or 20 μm of the JNK inhibitor SP600125, had no effect on the TGFβ1-induced phosphorylation of Smad-3 (A). Inhibition of JNK activity with 10 μm SP600125 did not inhibit the nuclear localization of Smad-3 stimulated by TGFβ1 (B). Experiments were conducted twice with identical results.
cAMP Inhibition of MAPK Signaling Is Tissue-specific
Inhibition of any MAPK significantly reduces CCN2/CTGF mRNA and/or protein levels in response to TGFβ1 in human lung fibroblasts. We hypothesize that cAMP accumulation may inhibit one or more MAPKs in these cells and may be the mechanism through which lung cells exhibit CCN2/CTGF down-regulation in response to PGE2. We further suspected that PGE2 and cAMP would not affect MAPK signaling in gingival fibroblasts. The receptor-independent stimulator of adenylate cyclases, forskolin, was next used to examine the inhibitory effect(s) of cAMP on the TGFβ1-induced activation of MAPKs in the absence of PGE2 receptor activation. Pretreatment of fibroblast cultures for 30 min with 10 μm forskolin prior to the addition of TGFβ1 significantly reduced the TGFβ1-induced phosphorylation of JNK in gingival fibroblasts but had little effect on the phosphorylation of p38 or ERK1/2 (Fig. 8A). In human lung (IMR90) fibroblasts, however, forskolin greatly reduced JNK, p38, and ERK1/2 phosphorylation (Fig. 8B). These data suggest that the cAMP-mediated inhibition of all three MAPKs accounts for the observed sensitivity to the inhibitory effects of PGE2 on CCN2/CTGF expression in IMR90 (lung) fibroblasts. The small reduction of CCN2/CTGF observed at 1 μm PGE2 levels in gingival fibroblasts may depend on the cAMP-mediated inhibition of JNK.
FIGURE 8. Effect of forskolin on TGFβ1-stimulated MAPK activation.
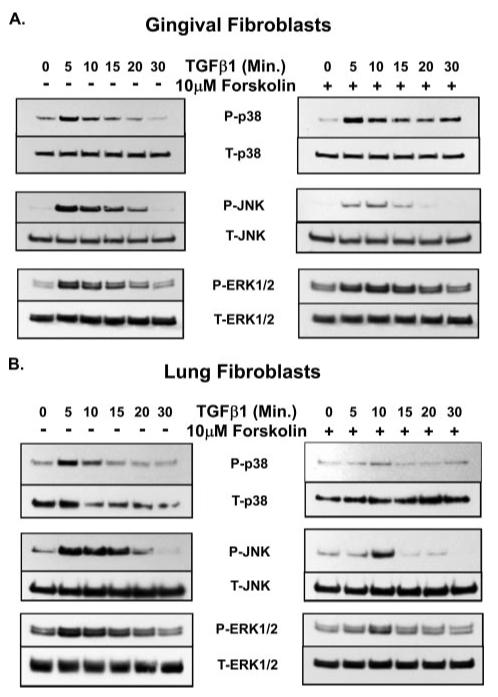
Fibroblast cultures were treated with 5 ng/ml TGFβ1 for the times indicated and total cellular protein was harvested for Western blot analysis of the phosphorylation status of MAPKs (JNK, ERK, and p38). Forskolin (10 μm) was added to some cultures for 30 min prior to the addition of TGFβ1 to determine the effect of forskolin on MAPK activation. Forskolin-mediated inhibition of MAPK activation in human gingival fibroblasts (A) and human lung fibroblasts (B) is shown. Each experiment was performed at least twice with identical results. P-MAPK, phosphorylated MAPK; T-MAPK, total MAPK.
JNK/p38 or JNK/ERK Inhibition Blocks CCN2/CTGF Protein Expression in Human Lung Fibroblasts
Forskolin, via the receptor-independent accumulation of cAMP, inhibits the activation of each major MAPK family member in IMR90 lung fibroblasts. Whereas the inhibition of any one MAPK family member results in a significant decrease in CCN2/CTGF expression induced by TGFβ1 in human lung fibroblasts, it is likely that the combined inhibition of MAPKs by cAMP could result in the complete inhibition observed upon challenge with low concentrations of PGE2. It follows, therefore, that the inhibition of MAPK family members with combinations of pharmacological inhibitors would completely block the TGFβ1-stimulated expression of CCN2/CTGF protein in cultures of lung fibroblasts. Cultures were pretreated for 1 h with different combinations of two MAPK inhibitors (10 μm). Media was subsequently replaced with media containing the identical composition and concentration of MAPK inhibitors and 5 ng/ml TGFβ1. Cultures were incubated for an additional 6 h prior to harvest of cell layer protein for Western blot analysis. Data show that the total inhibition of the TGFβ1-induced expression of the CCN2/CTGF protein resulted from the combination of the JNK inhibitor, SP600125, and either p38, with 10 μm SB203580, or ERK, with 10 μm U0126 (Fig. 9). These data confirm our findings that the activation of more than one MAPK is required in human lung fibroblasts for the maximal stimulation of CCN2/CTGF expression by TGFβ1 in IMR90 lung fibroblasts (Fig. 9), and that PGE2/cAMP inhibits CCN2/CTGF expression by inhibiting more than one MAPK pathway in these lung cells.
FIGURE 9. Effect of pairs of MAP kinase inhibitors on TGFβ1-stimulated CCN2/CTGF levels.
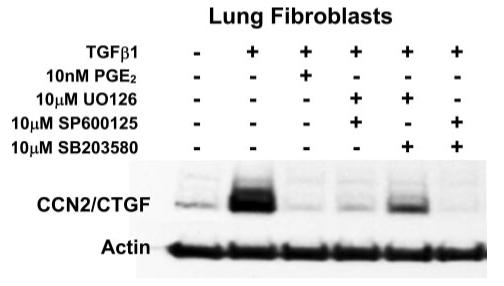
Cultures of IMR90 lung fibroblasts were pretreated with different combinations of MAPK inhibitors for 1 h, or 10 nm PGE2 for 30 min, prior to the addition of 5 ng/ml TGFβ1. Cultures were then incubated for an additional 6 h with fresh medium containing fresh MAPK inhibitor or PGE2 and TGFβ1 prior to harvest of total cellular protein for Western blot analysis of CCN2/CTGF protein. Experiments were conducted twice with identical results.
Inhibition of CCN2/CTGF Expression by PGE2/Forskolin Depends on PKA in Human Lung Fibroblasts
Because cAMP levels are dramatically increased in lung versus gingival fibroblasts, we suspected that the inhibition of CCN2/CTGF expression by PGE2 in lung cells may be due to high cAMP levels. A direct consequence of increased cAMP is the subsequent activation of protein kinase A (PKA). We therefore wished to determine whether the observed inhibitory effect of PGE2 and forskolin on CCN2/CTGF expression required PKA activity. Several inhibitors of PKA are available, but not all are entirely specific. For example, H89 inhibits ROCK even more potently than it inhibits PKA, whereas KT5720 has not been demonstrated to inhibit the activity of ROCK (45). ROCK has been shown to mediate TGFβ1 regulation of CCN2/CTGF in some cells (46). Thus, cultures of human lung and gingival fibroblasts were pretreated with 1 μm KT5720 for 1 h. Media were then removed and replaced with fresh media containing 1 μm KT5720 and concentrations of PGE2 as indicated. After a 30-min incubation, cells were refed with the indicated concentrations of KT5720, PGE2, and 5 ng/ml TGF-β1 and incubated for 6 h prior to harvest and analysis of cell layer protein. Data show that the inhibition of PKA by KT5720 prevents the complete inhibition of CCN2/CTGF expression by 10 nm PGE2 in cultures of human lung fibroblasts (Fig. 10A), and the modest inhibition by 1 μm PGE2 in gingival fibroblasts (Fig. 10B). Thus, in both lung and gingival fibroblasts, the PGE2-mediated inhibition of the TGFβ1-stimulated expression of CCN2/CTGF is mediated by activated PKA.
FIGURE 10. PKA dependence of PGE2 regulation of TGFβ1-stimulated CCN2/CTGF.
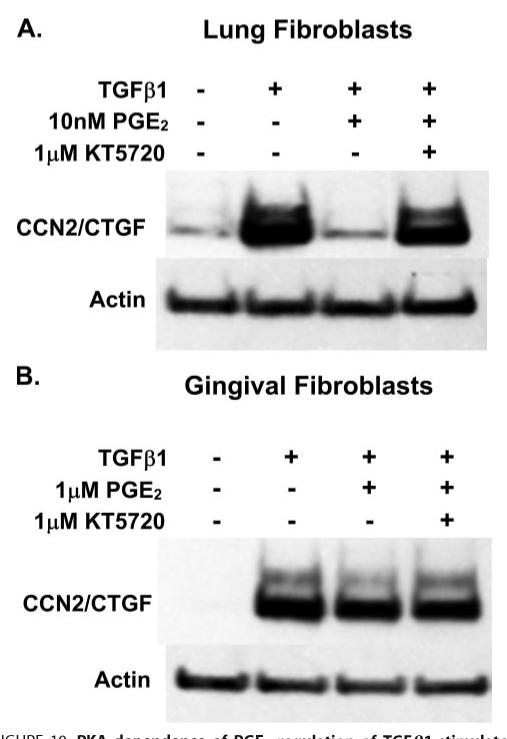
Cells were pretreated with 1 μm KT5720 for 1 h. Medium was aspirated and replaced with fresh medium containing KT5720 and the indicated concentrations of PGE2 and incubated for 30 min. Medium was again removed and replaced with medium containing KT5720, PGE2, and 5 ng/ml TGFβ1. After a 6-h incubation, total cell layer protein was harvested for Western blot analysis for CCN2/CTGF protein. A, the complete inhibition of CCN2/CTGF protein expression is rescued by PKA inhibition in human lung fibroblasts. B, the modest inhibition of CCN2/CTGF expression by PGE2 is relieved by the presence of the PKA inhibitor in human gingival fibroblasts. Experiments were conducted twice with similar results.
PGE2 Activates JNK via the EP3 Prostaglandin Receptor in Human Gingival Fibroblasts
Whereas we observed elevated cAMP levels in gingival fibroblasts in response to 1 μm PGE2 and 10 μm forskolin (Fig. 4), we found that the inhibition of the TGFβ1-induced expression of CCN2/CTGF protein was greater in response to forskolin (10 μm) than 1 μm PGE2 in these cells (Fig. 2B). We hypothesized that although increased cAMP can inhibit the activation of JNK, PGE2 may counteract this inhibition through the PGE2 receptor-stimulated phosphorylation of JNK. Cultures of gingival fibroblasts were treated with either TGFβ1 (control) or 1 μm PGE2 to determine the effect of PGE2 on the activation of JNK. We found that 1 μm PGE2 alone activates JNK in these cells but this stimulation was less than that induced by TGFβ1 alone (Fig. 11A). To further characterize the mechanism through which PGE2 stimulates the activation of JNK, we treated gingival fibroblast cultures with agonists for the EP2 or EP3 prostanoid receptor, butaprost or sulprostone, respectively. Stimulation of EP3 with sulprostone resulted in a robust increase in phosphorylated JNK (Fig. 11B). The stimulation of gingival fibroblasts with the EP2 agonist butaprost or forskolin, both of which are capable of stimulating the accumulation of cAMP, resulted in only a small increase in the phosphorylation of JNK (Fig. 11B). We conclude that a contributing mechanism by which gingival fibroblasts resist the inhibitory effects of PGE2, and cAMP, on the TGFβ1-induced expression of CCN2/CTGF is the stimulation of JNK activation through the activity of the EP3 receptor.
FIGURE 11. Regulation of JNK activation by TGFβ1, PGE2, EP-receptor agonists and forskolin.
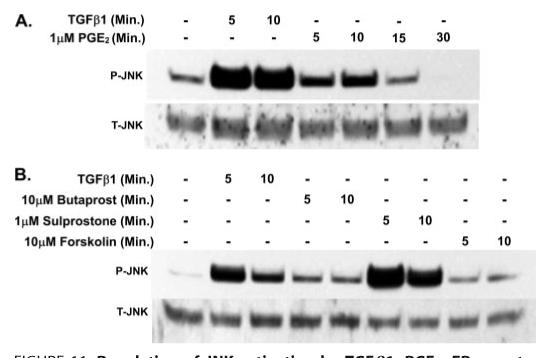
A, human gingival fibroblast cultures were treated with 1 μm PGE2 or 5 ng/ml TGFβ1 for the times indicated and total cellular protein harvested for Western blot analysis of JNK phosphorylation. B, Western blot for phosphorylated (P) and total (T) JNK showing that treatment of gingival fibroblasts with a specific agonist of the EP3 receptor (sulprostone) resulted in a strong stimulation of JNK. Experiments were conducted twice with identical results.
EP3 Prostanoid Receptor Stimulation Increases CCN2/CTGF Independent of TGFβ1
The dramatic increase in JNK activation induced by stimulation of the EP3 receptor led us to hypothesize that EP3 receptor activity could lead to CCN2/CTGF expression independent of TGFβ1. This would be significant in gingival tissues as these tissues are persistently in contact with elevated levels of PGE2 in vivo. In addition, the phenytoin-induced increase in PGE2 biosynthesis that occurs in gingival fibroblasts (57-59) may perpetuate the fibrotic response induced by CCN2/CTGF via EP3 stimulation. Cultures were serum starved, and treated with 1 μm sulprostone for 4 h prior to harvest of total RNA or 6 h prior to harvest of cell layer protein. Real time PCR and Western blot analysis of CCN2/CTGF mRNA and protein revealed that the stimulation of EP3 with its specific agonist sulprostone results in a ∼1.6-fold increase in CCN2/CTGF mRNA (Fig. 12A) and protein (Fig. 12, B and C) in gingival fibroblast cultures compared with no treatment control cultures.
FIGURE 12. EP3 stimulation increases CCN2/CTGF expression independent of TGFβ1.
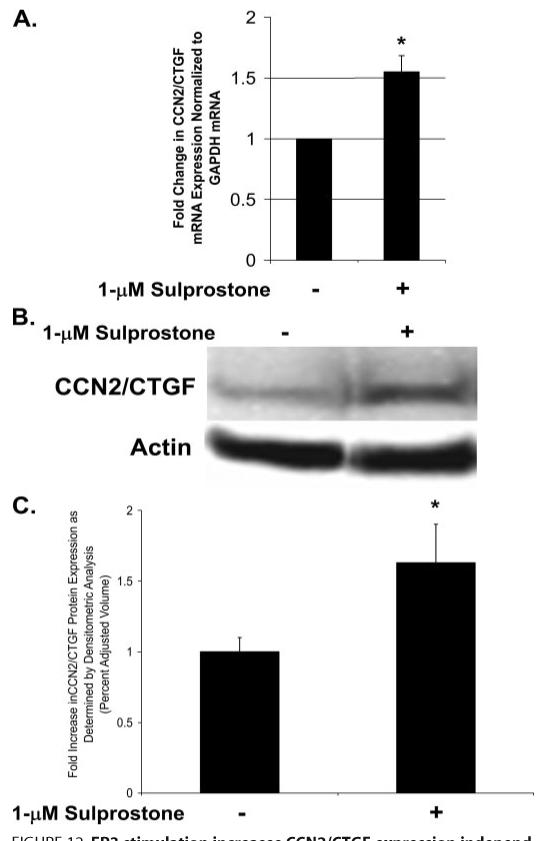
Cultures of gingival fibroblasts treated with the EP3 agonist sulprostone that stimulates CCN2/CTGF mRNA expression as determined by real time PCR analysis (A). Western blot analysis of CCN2/CTGF protein levels demonstrate a similar increase in CCN2/CTGF protein in response to stimulation with sulprostone (B and C) (*, p < 0.05). Data collected are representative from three separate experiments.
EP3 Prostanoid Receptor Stimulation Partially Rescues Forskolin-inhibited CCN2/CTGF Expression
We have shown that the receptor-independent activator of adenylate cyclase, forskolin, results in a more robust inhibition of CCN2/CTGF expression in contrast to PGE2 (Figs. 1A and 2) and that forskolin specifically interferes with the activation of JNK by TGFβ1 (Fig. 8A). Because the activation of the EP3 receptor of PGE2 stimulates JNK activation independent of TGFβ1, we surmise that this activation is a contributing mechanism by which gingival cells resist the inhibitory effects of PGE2 on CCN2/CTGF expression induced by TGFβ1. We wished to determine whether the targeted activation of EP3 could overcome the inhibitory effects of forskolin on CCN2/CTGF expression through the EP3-mediated activation of JNK. To test this, cultures were serum starved, and pretreated for 1 h with forskolin alone or in combination with sulprostone. Media was then replaced with fresh medium containing identical concentration(s) of forskolin/sulprostone and 5 ng/ml TGFβ1 and incubated for an additional 6 h prior to harvest of cell layer protein. Our data indicate that the stimulation of EP3 partially restored the TGFβ1-induced expression of CCN2/CTGF inhibited by forskolin (Fig. 13).
FIGURE 13. EP3 stimulation partially rescues CCN2/CTGF expression inhibited by forskolin.
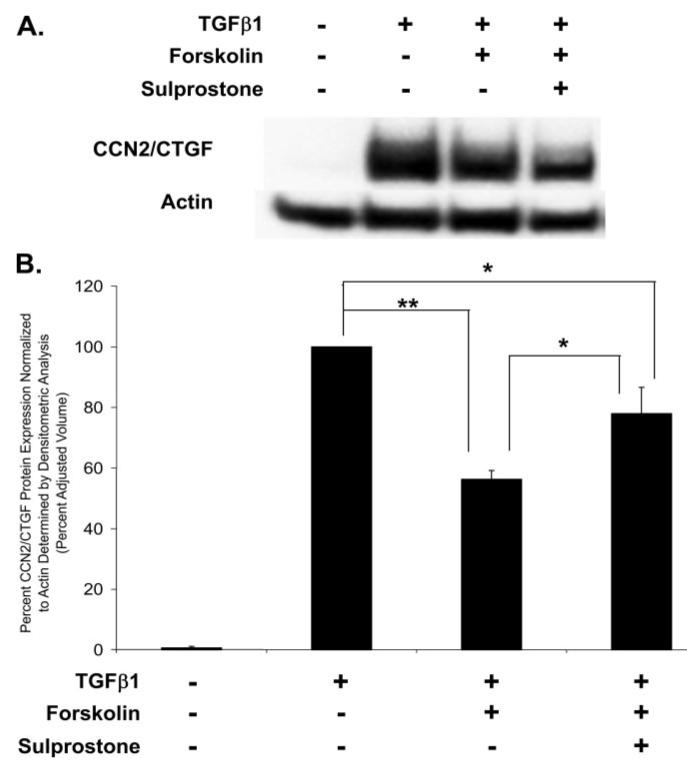
Gingival fibroblast cultures treated with forskolin, or a combination of forskolin and the EP3 agonist sulprostone, demonstrate that stimulation of the EP3 receptor partially rescues the TGFβ1-induced expression of CCN2/CTGF inhibited by forskolin, representative Western blot (A) and densitometric analysis from Western blots from three separate experiments (B) (*, p < 0.05; **, p < 0.00005).
The net result in gingival fibroblasts is that PGE2-dependent elevations of cAMP and subsequent activation of PKA have only a small inhibitory effect on TGFβ1-stimulated CCN2/CTGF levels compared with the effects of PGE2 on stimulating cAMP/PKA in lung fibroblasts. At the same time, PGE2 stimulates JNK and CCN2/CTGF expression in gingival fibroblasts via the EP3 prostaglandin receptor, further contributing to persistent expression of CCN2/CTGF (Fig. 14). By contrast, in lung cells, PGE2 causes a robust increase in cAMP that inhibits all three major MAP kinases and blocks CCN2/CTGF expression via a PKA-dependent mechanism.
FIGURE 14. Regulation of CCN2/CTGF expression in human gingival fibroblasts.
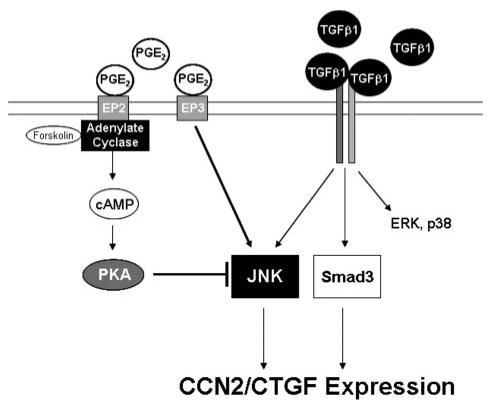
In human gingival fibroblasts, the sole MAPK requirement for TGFβ1 to promote optimal CCN2/CTGF expression is JNK. Whereas receptor-independent cAMP elevating agents reduce JNK activation induced by TGFβ1, the stimulation of the EP3 prostanoid receptor by PGE2 overcomes this inhibition by stimulating JNK activation to promote CCN2/CTGF expression in these cells.
DISCUSSION
This study provides direct evidence that human gingival fibroblasts are resistant to PGE2 down-regulation of TGFβ1-stimulated CCN2/CTGF levels, and that this resistance is tissue-specific. The mechanisms of resistance include a relative lack of inhibition of MAP kinase pathways by cAMP in gingival fibroblasts, and a simultaneous stimulation of JNK activation by PGE2/EP3 receptor activation. Moreover, in contrast to lung fibroblasts, we show that TGFβ1 stimulation of CCN2/CTGF levels in gingival cells is mediated primarily by JNK activation and not by p38 or ERK1/2. Moreover, neither the canonical TGFβ1 receptor-induced activation of Smad3 nor the nuclear localization of activated Smad3 are hindered by JNK MAPK. Elevated CCN2/CTGF levels contribute to fibrosis (47). These findings, therefore, are of potential clinical importance as they begin to identify pathways and mechanism(s) through which CCN2/CTGF levels are maintained in fibrotic gingival tissues in the presence of PGE2. The experimental approach of directly comparing the role of the three major MAP kinase pathways in mediating and maintaining CCN2/CTGF levels has, in addition, provided new insights into CCN2/CTGF regulation in lung fibroblasts. For example, we report that lung cells are highly efficient at increasing cAMP levels in response to PGE2, and that all three MAP kinases are inhibited in these cells in a cAMP/PKA-mediated mechanism. We show for the first time that the cAMP/PKA-dependent inhibition of all three MAP kinases, in turn, results in potent down-regulation of TGF-β1-stimulated CCN2/CTGF levels in lung fibroblasts.
It is well recognized that TGFβ1-dependent signal transduction pathways include activation of Smads (48, 49). Activation of Smad3 in particular is necessary but not sufficient for TGFβ1 stimulation of CCN2/CTGF production in normal fibroblasts (50, 51). It is understood that some effects of TGFβ1 are tissue or cell type-specific, and that TGFβ1-stimulated signal transduction pathways that are independent of primary Smad activation occur (52).
Specific and differential activation of MAP kinase pathways can mediate cell type or tissue-specific effects of TGFβ1 (35). Thus, the present study has focused on the presence or absence of cross-talk between PGE2-stimulated pathways and TGFβ1 stimulation of the three major MAP kinase pathways. As cAMP is a major mediator of PGE2 effects, we investigated the relationship between TGFβ1 stimulation of CCN2/CTGF production in the context of MAP kinase activation, and effects of PGE2 and cAMP production. Direct comparative analyses of lung and gingival cells were highly informative and confirmed that tissue-specific effects are in fact mediated by differences in the relationships between these pathways.
There are few previous examples of cAMP cross-talk with MAP kinase pathways. A consistent finding is that cAMP inhibits activation of ERK or p38 MAP kinases (53-55). No effect of cAMP on Smad activation has been observed so far (56). To our knowledge, the present study is the first report that cAMP can inhibit the activation of JNK. It is interesting that in gingival fibroblasts, JNK is the primary MAP kinase inhibited by forskolin/cAMP, and that ERK and p38 are largely unaffected, whereas in lung cells all three MAP kinases are inhibited by forskolin treatment. Moreover, this inhibition is functional with respect to CCN2/CTGF regulation, as we have shown by MAP kinase inhibitor studies in lung cells that all three MAP kinases contribute to TGFβ1-stimulated CCN2/CTGF production. Only the combined inhibition of JNK and either p38 or ERK results in the complete inhibition of the TGFβ1-induced expression of CCN2/CTGF in lung fibroblasts. This finding is somewhat similar to findings in a recent study using human lung HFL-1 fibroblasts in which JNK was found to be the primary MAP kinase mediating TGFβ1-stimulated increases in CCN2/CTGF (57). As TGFβ1 concentrations were not identical between the current study and the previous study, it is likely that this or other differences in treatment conditions could account for inconsistencies between the two studies. It is notable that our comparisons between lung, kidney, and gingival fibroblastic cells were all performed under identical experimental conditions, and, therefore, permit direct investigation of tissue-specific differences in signal transduction pathways.
PGE2 exerts its effects on cells through binding and activating receptors EP1-4. EP2 and EP4 have been most often associated with elevating cAMP levels in target cells (37). The role of EP2 appears to be more essential in maintaining cAMP production because it has been suggested that EP4 may be desensitized through its subsequent phosphorylation by PKA activated in response to elevated cAMP, whereas EP2 is not (58, 59). Our studies in lung cells indicate that the direct stimulation of the EP2 receptor with its specific agonist butaprost completely blocked CCN2/CTGF expression and mimics the effects of forskolin and PGE2 itself. In gingival fibroblasts, however, EP2 stimulation resulted in only a small reduction of CCN2/CTGF expression, as did forskolin. Interestingly, stimulation of the EP3 receptor with its agonist, sulprostone, in combination with TGFβ1 resulted in a slight increase in CCN2/CTGF protein in gingival fibroblasts compared with cultures treated with TGFβ1 alone. Stimulation of EP3 receptor present at trace levels in Rat-1 cells could possibly also account for the increased level of CCN2/CTGF seen previously in Rat-1 fibroblasts (32).
It is interesting that gingival fibroblasts and lung fibroblasts differ so markedly in the ability to generate cAMP in response to PGE2. This diminished ability of gingival fibroblasts to produce cAMP in response to PGE2 is likely to contribute to the observed resistance of gingival fibroblasts to the effects of PGE2 because many of these effects are mediated by cAMP activation of PKA. At this point it is unclear by which molecular mechanisms the difference in cAMP accumulation occur. Preliminary analyses of EP1-4 receptor protein levels in gingival fibroblasts and lung fibroblasts do not show significant differences (data not shown), suggesting that differential expression of these receptors does not account for the difference in sensitivity to PGE2. It is more likely, therefore, that differences exist in the relative activity or levels of adenylyl cyclases, phosphodiesterases, or that a tissue-specific feedback mechanism is involved in regulating the activity of adenylyl cyclases or phosphodiesterases, respectively. As forskolin, a stimulator of adenylyl cyclase activity, does stimulate significant levels of cAMP in gingival fibroblasts, it seems likely that these cells may limit cAMP accumulation by producing elevated levels of phosphodiesterases. These questions are currently under investigation. It is important to recognize that data presented indicates that gingival fibroblast CCN2/CTGF is relatively insensitive to cAMP elevations. Treatment of gingival fibroblasts with 1 μm PGE2, which resulted in elevated cAMP levels, did not result in the potent down-regulation of TGFβ1-stimulated CCN2/CTGF expression that was observed in lung cell cultures. This suggests that additional mechanisms occur in gingival fibroblasts that confer resistance to the inhibitory effects of elevated cAMP levels in gingival fibroblasts, and we have identified these pathways in the present study.
Pro-inflammatory processes exacerbate phenytoin-induced gingival overgrowth (60). Pro-inflammatory cytokines including tumor necrosis factor α and interleukin-1β increase PGE2 production in gingival fibroblasts, and these effects are further stimulated by phenytoin (61-63). We suspect that in the presence of phenytoin and inflammatory cytokines, increased PGE2 production by gingival fibroblasts could prime cells for the increased expression of CCN2/CTGF by activating JNK via the EP3 prostanoid receptor. Exposure to TGFβ1 would then further induce CCN2/CTGF expression and promote the progression of fibrosis as is observed in patients with phenytoin-induced gingival overgrowth (64). The reliance on one MAPK signaling pathway (JNK) in gingival fibroblasts in promoting CCN2/CTGF expression by TGFβ1 and the ability of PGE2 to stimulate JNK activation, in combination with the limited cAMP production in these cells, appear to provide gingival cells a tissue-specific mechanism by which they can maintain CCN2/CTGF expression and balance of extracellular matrix accumulation with turnover under normal physiologic conditions. These same mechanisms may also serve to allow the gingiva-specific, pathologic accumulation of excess extracellular matrix in the presence of phenytoin by conferring resistance to down-regulation of CCN2/CTGF by inflammatory mediators including PGE2. Moreover, excess PGE2 may serve to perpetuate the fibrotic response in gingival cells to phenytoin because we have demonstrated that the targeted stimulation of the EP3 receptor alone stimulates CCN2/CTGF expression. In summary, our data identify two mechanisms by which the TGFβ1-stimulated expression of CCN2/CTGF in human gingival fibroblasts are resistant to the down-regulation by PGE2; (i) cAMP cross-talk with MAPK pathways is limited in gingival fibroblasts and (ii) PGE2 activation of the EP3 prostanoid receptor stimulates the activation of JNK and limits cAMP-dependent down-regulation of the TGFβ1-induced, JNK-dependent expression of CCN2/CTGF.
Footnotes
- CTGF
- connective tissue growth factor
- DN-JNK1
- dominant negative JNK1
- TGFβ
- transforming growth factor β
- MAPK
- mitogen-activated protein kinase
- JNK
- c-Jun NH2-terminal kinase
- SAPK
- stress-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- PGE2
- prostaglandin E2
- ELISA
- enzyme-linked immunosorbent assay
- PKA
- protein kinase A
This work was supported by National Institutes of Health NIDCR Grants R01 DE11004, K08 DE016609, and M01 RR00533. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Oemar BS, Luscher TF. Arterioscler. Thromb. Vasc. Biol. 1997;17:1483–1489. doi: 10.1161/01.atv.17.8.1483. [DOI] [PubMed] [Google Scholar]
- 2.Brigstock DR. Endocr. Rev. 1999;20:189–206. doi: 10.1210/edrv.20.2.0360. [DOI] [PubMed] [Google Scholar]
- 3.Bork P. FEBS Lett. 1993;327:125–130. doi: 10.1016/0014-5793(93)80155-n. [DOI] [PubMed] [Google Scholar]
- 4.Moussad EE, Brigstock DR. Mol. Genet. Metab. 2000;71:276–292. doi: 10.1006/mgme.2000.3059. [DOI] [PubMed] [Google Scholar]
- 5.Chujo S, Shirasaki F, Kawara S, Inagaki Y, Kinbara T, Inaoki M, Takigawa M, Takehara K. J. Cell. Physiol. 2005;203:447–456. doi: 10.1002/jcp.20251. [DOI] [PubMed] [Google Scholar]
- 6.Yokoi H, Mukoyama M, Sugawara A, Mori K, Nagae T, Makino H, Suganami T, Yahata K, Fujinaga Y, Tanaka I, Nakao K. Am. J. Physiol. 2002;282:F933–F942. doi: 10.1152/ajprenal.00122.2001. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh AK. Exp. Biol. Med. (Maywood) 2002;227:301–314. doi: 10.1177/153537020222700502. [DOI] [PubMed] [Google Scholar]
- 8.Duncan MR, Frazier KS, Abramson S, Williams S, Klapper H, Huang X, Grotendorst GR. FASEB J. 1999;13:1774–1786. [PubMed] [Google Scholar]
- 9.Frazier K, Williams S, Kothapalli D, Klapper H, Grotendorst GR. J. Investig. Dermatol. 1996;107:404–411. doi: 10.1111/1523-1747.ep12363389. [DOI] [PubMed] [Google Scholar]
- 10.Leask A, Sa S, Holmes A, Shiwen X, Black CM, Abraham DJ. Mol. Pathol. 2001;54:180–183. doi: 10.1136/mp.54.3.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abraham DJ, Shiwen X, Black CM, Sa S, Xu Y, Leask A. J. Biol. Chem. 2000;275:15220–15225. doi: 10.1074/jbc.275.20.15220. [DOI] [PubMed] [Google Scholar]
- 12.Ito Y, Aten J, Bende RJ, Oemar BS, Rabelink TJ, Weening JJ, Goldschmeding R. Kidney Int. 1998;53:853–861. doi: 10.1111/j.1523-1755.1998.00820.x. [DOI] [PubMed] [Google Scholar]
- 13.Riser BL, Denichilo M, Cortes P, Baker C, Grondin JM, Yee J, Narins RG. J. Am. Soc. Nephrol. 2000;11:25–38. doi: 10.1681/ASN.V11125. [DOI] [PubMed] [Google Scholar]
- 14.Paradis V, Dargere D, Vidaud M, De Gouville AC, Huet S, Martinez V, Gauthier JM, Ba N, Sobesky R, Ratziu V, Bedossa P. Hepatology. 1999;30:968–976. doi: 10.1002/hep.510300425. [DOI] [PubMed] [Google Scholar]
- 15.Tamatani T, Kobayashi H, Tezuka K, Sakamoto S, Suzuki K, Nakanishi T, Takigawa M, Miyano T. Biochem. Biophys. Res. Commun. 1998;251:748–752. doi: 10.1006/bbrc.1998.9543. [DOI] [PubMed] [Google Scholar]
- 16.Williams EJ, Gaca MD, Brigstock DR, Arthur MJ, Benyon RC. J. Hepatol. 2000;32:754–761. doi: 10.1016/s0168-8278(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 17.Rachfal AW, Brigstock DR. Hepatol. Res. 2003;26:1–9. doi: 10.1016/s1386-6346(03)00115-3. [DOI] [PubMed] [Google Scholar]
- 18.Sedlaczek N, Jia JD, Bauer M, Herbst H, Ruehl M, Hahn EG, Schuppan D. Am. J. Pathol. 2001;158:1239–1244. doi: 10.1016/S0002-9440(10)64074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueberham U, Ueberham E, Gruschka H, Arendt T. Neuroscience. 2003;116:1–6. doi: 10.1016/s0306-4522(02)00670-x. [DOI] [PubMed] [Google Scholar]
- 20.Lasky JA, Ortiz LA, Tonthat B, Hoyle GW, Corti M, Athas G, Lungarella G, Brody A, Friedman M. Am. J. Physiol. 1998;275:L365–L371. doi: 10.1152/ajplung.1998.275.2.L365. [DOI] [PubMed] [Google Scholar]
- 21.Allen JT, Knight RA, Bloor CA, Spiteri MA. Am. J. Respir. Cell Mol. Biol. 1999;21:693–700. doi: 10.1165/ajrcmb.21.6.3719. [DOI] [PubMed] [Google Scholar]
- 22.Uzel MI, Kantarci A, Hong HH, Uygur C, Sheff MC, Firatli E, Trackman PC. J. Periodontol. 2001;72:921–931. doi: 10.1902/jop.2001.72.7.921. [DOI] [PubMed] [Google Scholar]
- 23.Hong HH, Uzel MI, Duan C, Sheff MC, Trackman PC. Lab. Investig. 1999;79:1655–1667. [PubMed] [Google Scholar]
- 24.Kantarci A, Black S, Xydas C, Murawel P, Uchida Y, Yucekal-Tuncer B, Atilla G, Emingil G, Uzel M, Lee A, Firatli E, Sheff M, Hasturk H, Van Dyke T, Trackman P. J. Pathol. 2006;210:59–66. doi: 10.1002/path.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oemar BS, Werner A, Garnier JM, Do DD, Godoy N, Nauck M, Marz W, Rupp J, Pech M, Luscher TF. Circulation. 1997;95:831–839. doi: 10.1161/01.cir.95.4.831. [DOI] [PubMed] [Google Scholar]
- 26.Ohnishi H, Oka T, Kusachi S, Nakanishi T, Takeda K, Nakahama M, Doi M, Murakami T, Ninomiya Y, Takigawa M, Tsuji T. J. Mol. Cell Cardiol. 1998;30:2411–2422. doi: 10.1006/jmcc.1998.0799. [DOI] [PubMed] [Google Scholar]
- 27.Schober JM, Chen N, Grzeszkiewicz TM, Jovanovic I, Emeson EE, Ugarova TP, Ye RD, Lau LF, Lam SC. Blood. 2002;99:4457–4465. doi: 10.1182/blood.v99.12.4457. [DOI] [PubMed] [Google Scholar]
- 28.di Mola FF, Friess H, Martignoni ME, Di Sebastiano P, Zimmermann A, Innocenti P, Graber H, Gold LI, Korc M, Buchler MW. Ann. Surg. 1999;230:63–71. doi: 10.1097/00000658-199907000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dammeier J, Brauchle M, Falk W, Grotendorst GR, Werner S. Int. J. Biochem. Cell Biol. 1998;30:909–922. doi: 10.1016/s1357-2725(98)00046-6. [DOI] [PubMed] [Google Scholar]
- 30.Lee EH, Joo CK. Investig. Ophthalmol. Vis. Sci. 1999;40:2025–2032. [PubMed] [Google Scholar]
- 31.Ricupero DA, Rishikof DC, Kuang PP, Poliks CF, Goldstein RH. Am. J. Physiol. 1999;277:L1165–L1171. doi: 10.1152/ajplung.1999.277.6.L1165. [DOI] [PubMed] [Google Scholar]
- 32.Yu J, Prado GN, Schreiber B, Polgar P, Taylor L. Arch. Biochem. Biophys. 2002;404:302–308. doi: 10.1016/s0003-9861(02)00276-x. [DOI] [PubMed] [Google Scholar]
- 33.Preshaw PM, Heasman PA. J. Clin. Periodontol. 2002;29:15–20. doi: 10.1034/j.1600-051x.2002.290103.x. [DOI] [PubMed] [Google Scholar]
- 34.van der Pauw MT, Klein Nulend J, van den Bos T, Burger EH, Everts V, Beertsen W. J. Periodontal Res. 2000;35:335–343. doi: 10.1034/j.1600-0765.2000.035006335.x. [DOI] [PubMed] [Google Scholar]
- 35.Javelaud D, Mauviel A. Oncogene. 2005;24:5742–5750. doi: 10.1038/sj.onc.1208928. [DOI] [PubMed] [Google Scholar]
- 36.Huggett J, Dheda K, Bustin S, Zumla A. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 37.Bastien L, Sawyer N, Grygorczyk R, Metters KM, Adam M. J. Biol. Chem. 1994;269:11873–11877. [PubMed] [Google Scholar]
- 38.Regan JW, Bailey TJ, Pepperl DJ, Pierce KL, Bogardus AM, Donello JE, Fairbairn CE, Kedzie KM, Woodward DF, Gil DW. Mol. Pharmacol. 1994;46:213–220. [PubMed] [Google Scholar]
- 39.Choung J, Taylor L, Thomas K, Zhou X, Kagan H, Yang X, Polgar P. J. Cell. Biochem. 1998;71:254–263. [PubMed] [Google Scholar]
- 40.Narumiya S, Sugimoto Y, Ushikubi F. Physiol. Rev. 1999;79:1193–1226. doi: 10.1152/physrev.1999.79.4.1193. [DOI] [PubMed] [Google Scholar]
- 41.Bonniaud P, Margetts PJ, Ask K, Flanders K, Gauldie J, Kolb M. J. Immunol. 2005;175:5390–5395. doi: 10.4049/jimmunol.175.8.5390. [DOI] [PubMed] [Google Scholar]
- 42.Lakos G, Takagawa S, Chen SJ, Ferreira AM, Han G, Masuda K, Wang XJ, DiPietro LA, Varga J. Am. J. Pathol. 2004;165:203–217. doi: 10.1016/s0002-9440(10)63289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato M, Muragaki Y, Saika S, Roberts AB, Ooshima A. J. Clin. Investig. 2003;112:1486–1494. doi: 10.1172/JCI19270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engel ME, McDonnell MA, Law BK, Moses HL. J. Biol. Chem. 1999;274:37413–37420. doi: 10.1074/jbc.274.52.37413. [DOI] [PubMed] [Google Scholar]
- 45.Davies SP, Reddy H, Caivano M, Cohen P. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haydont V, Mathe D, Bourgier C, Abdelali J, Aigueperse J, Bourhis J, Vozenin-Brotons MC. Radiother. Oncol. 2005;76:219–225. doi: 10.1016/j.radonc.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 47.Mori T, Kawara S, Shinozaki M, Hayashi N, Kakinuma T, Igarashi A, Takigawa M, Nakanishi T, Takehara K. J. Cell. Physiol. 1999;181:153–159. doi: 10.1002/(SICI)1097-4652(199910)181:1<153::AID-JCP16>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 48.Kretschmer A, Moepert K, Dames S, Sternberger M, Kaufmann J, Klippel A. Oncogene. 2003;22:6748–6763. doi: 10.1038/sj.onc.1206791. [DOI] [PubMed] [Google Scholar]
- 49.Massague J, Chen YG. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 50.Holmes A, Abraham DJ, Sa S, Shiwen X, Black CM, Leask A. J. Biol. Chem. 2001;276:10594–10601. doi: 10.1074/jbc.M010149200. [DOI] [PubMed] [Google Scholar]
- 51.Leask A, Holmes A, Black CM, Abraham DJ. J. Biol. Chem. 2003;278:13008–13015. doi: 10.1074/jbc.M210366200. [DOI] [PubMed] [Google Scholar]
- 52.Hartsough MT, Mulder KM. Pharmacol. Ther. 1997;75:21–41. doi: 10.1016/s0163-7258(97)00020-x. [DOI] [PubMed] [Google Scholar]
- 53.Fassett J, Tobolt D, Hansen LK. Mol. Biol. Cell. 2006;17:345–356. doi: 10.1091/mbc.E05-09-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dhillon AS, Pollock C, Steen H, Shaw PE, Mischak H, Kolch W. Mol. Cell. Biol. 2002;22:3237–3246. doi: 10.1128/MCB.22.10.3237-3246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng J, Diaz Encarnacion MM, Warner GM, Gray CE, Nath KA, Grande JP. Am. J. Physiol. 2005;289:C959–C970. doi: 10.1152/ajpcell.00153.2005. [DOI] [PubMed] [Google Scholar]
- 56.del Valle-Perez B, Martinez-Estrada OM, Vilaro S, Ventura F, Vinals F. FEBS Lett. 2004;569:105–111. doi: 10.1016/j.febslet.2004.05.058. [DOI] [PubMed] [Google Scholar]
- 57.Utsugi M, Dobashi K, Ishizuka T, Masubuchi K, Shimizu Y, Nakazawa T, Mori M. Am. J. Respir. Cell Mol. Biol. 2003;28:754–761. doi: 10.1165/rcmb.4892. [DOI] [PubMed] [Google Scholar]
- 58.Liggett SB, Freedman NJ, Schwinn DA, Lefkowitz RJ. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3665–3669. doi: 10.1073/pnas.90.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishigaki N, Negishi M, Ichikawa A. Mol. Pharmacol. 1996;50:1031–1037. [PubMed] [Google Scholar]
- 60.Penarrocha-Diago M, Bagan-Sebastian JV, Vera-Sempere F. J. Periodontol. 1990;61:571–574. doi: 10.1902/jop.1990.61.9.571. [DOI] [PubMed] [Google Scholar]
- 61.Modeer T, Brunius G, Iinuma M, Lerner UH. Br. J. Pharmacol. 1992;106:574–578. doi: 10.1111/j.1476-5381.1992.tb14377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brunius G, Yucel-Lindberg T, Shinoda K, Modeer T. Eur. J. Oral Sci. 1996;104:27–33. doi: 10.1111/j.1600-0722.1996.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 63.Modeer T, Anduren I, Lerner UH. J. Oral Pathol. Med. 1992;21:251–255. doi: 10.1111/j.1600-0714.1992.tb01005.x. [DOI] [PubMed] [Google Scholar]
- 64.Trackman PC, Kantarci A. Crit. Rev. Oral Biol. Med. 2004;15:165–175. doi: 10.1177/154411130401500305. [DOI] [PubMed] [Google Scholar]


