Abstract
We determined the 2.45 Å crystal structure of the nucleosome core particle from Drosophila melanogaster and compared it to that of Xenopus laevis bound to the identical 147 base-pair DNA fragment derived from human α-satellite DNA. Differences between the two structures primarily reflect 16 amino acid substitutions between species, 15 of which are in histones H2A and H2B. Four of these involve histone tail residues, resulting in subtly altered protein–DNA interactions that exemplify the structural plasticity of these tails. Of the 12 substitutions occurring within the histone core regions, five involve small, solvent-exposed residues not involved in intraparticle interactions. The remaining seven involve buried hydrophobic residues, and appear to have coevolved so as to preserve the volume of side chains within the H2A hydrophobic core and H2A-H2B dimer interface. Thus, apart from variations in the histone tails, amino acid substitutions that differentiate Drosophila from Xenopus histones occur in mutually compensatory combinations. This highlights the tight evolutionary constraints exerted on histones since the vertebrate and invertebrate lineages diverged.
Keywords: chromatin, nucleosome core particles, protein–DNA interaction, Drosophila, crystal structure
INTRODUCTION
Genomic DNA in the eukaryotic nucleus is compacted and organized in protein–DNA complexes called chromatin. The notion of a repeating unit of chromatin structure, composed of eight histone proteins and ∼200 base pairs of DNA, was proposed over 30 years ago.1,2 Within this unit, the first level of chromatin organization, revealed by micrococcal nuclease digestion, is the nucleosome core particle (NCP). The NCP is composed of an octamer containing two copies of each of the four histone proteins (H3, H4, H2A, and H2B), around which ∼146 base pairs of DNA are tightly wrapped in 1.65 turns of a left-handed superhelix (reviewed in Ref.3).
In addition to its structural role in genome organization, the nucleosome is the point of convergence for many DNA regulatory processes: recombination, repair, replication, and transcription. In particular, nucleosomes are highly dynamic and are directly involved in the regulation of transcription.4 ATP-dependent remodeling complexes physically modulate chromatin structure at the nucleosome level, actively altering the accessibility of specific sequences to transcription factors.5,6 Nucleosomes also carry information via changes in composition (histone variants) and posttranslational modifications (PTMs).7,8 The role of both types of modification on the regulation of genomic activity is currently the subject of intense research. Finally, the nucleosome is involved in major cellular regulatory mechanisms related to cell cycle and aging, cell differentiation, and cellular reprogramming,9 and plays a critical role in viral infection10 and cancer.11
Structural studies of the nucleosome over the last 20 years have painted an increasingly accurate picture of how the nucleosome accomplishes its packaging and regulatory roles. Initial low-resolution studies12–14 followed by the crystal structure of the histone octamer15 elucidated the overall architecture of the NCP. The structure of an NCP (Xla-NCP146) composed of recombinant Xenopus laevis histones and a 146-bp palindromic fragment of human α-satellite DNA revealed details of the DNA structure and its interactions with the histones.16 A higher resolution structure (Xla-NCP147) using a related 147-bp DNA fragment allowed for a detailed analysis of the DNA conformation, solvent structure, and interactions with ions.17–19 Structures of a Xenopus NCP containing the histone variant H2A.Z20 or macroH2A21 and of NCPs comprising chicken,22 yeast,23 and human histones24 have brought additional functional and evolutionary insights.
To extend this analysis, we determined the crystal structure of the NCP from Drosophila melanogaster, the first from an invertebrate species. Drosophila histones share a high degree of sequence identity with those of Xenopus, ranging from 83% and 89% identity for H2B and H2A, respectively, to 99% for H3 and H4. Most of these changes localize to histone tail residues that are disordered in the available NCP crystal structures. However, a substantial number involve structured histone residues. We compare the Drosophila and Xenopus NCP structures and focus particularly on histone residues that have diverged between these species.
MATERIALS AND METHODS
Crystallization
NCPs were prepared from recombinant D. melanogaster histones and a 147 bp palindromic DNA fragment derived from human α-satellite DNA, as described previously.25 Crystallization trials were carried out by the hanging drop vapor-diffusion technique at 4°C by equilibrating a droplet containing 3 mg/mL Dm-NCP147, 80–85 mM MnCl2, 50–80 mM KCl, and 20 mM potassium cacodylate (pH 6.0) against a reservoir solution containing of 40–42.5 mM MnCl2, 25–40 mM KCl, and 20 mM potassium cacodylate (pH 6.0). To improve diffraction quality, crystals were soaked overnight in the reservoir solution supplemented with 24% (v/v) 2-methyl-2,4-pentanediol as cryoprotectant and flash-cooled in liquid nitrogen.
Crystallography
Diffraction data were collected at ESRF beamline ID14-3 (λ = 0.931 Å) on a MAR CCD detector and processed with XDS26 and programs of the CCP4 suite.27 Crystals obtained using described conditions17 were isomorphous to the Xla-NCP147 crystal form (Table I). The Xla-NCP147 structure (pdb id 1KX5) minus the N-terminal histone tail residues was used as a starting model. Positioning this model into the Dm-NCP147 unit cell resulted in a crystallographic R-factor of 0.40, which dropped to 0.32 upon rigid body refinement using CNS.28 A further round of restrained coordinate and B-factor refinement reduced this to 0.276 (Rfree = 0.301). Differences between the Drosophila and Xenopus structures were readily apparent in a 2Fo − Fc map calculated using phase information from the Xla-NCP147 atomic coordinates. Iterative rounds of manual model building using O29 and CNS refinement were carried out to incorporate amino acid substitutions, ions, and water molecules, and to rebuild the histone tails. The structure was refined at 2.45 Å to a final crystallographic R-factor of 0.229 (Rfree = 0.262) and good geometry.
Table I.
Data and Refinement Statistics
| Data collection | |
| Space group | P212121 |
| Cell parameters (Å) | a = 106.0, b = 182.0, c = 109.4 |
| ESRF beamline | ID14-3 |
| Resolution | |
| Overall (Å) | 30–2.45 |
| Outer shell (Å) | 2.5–2.45 |
| Completeness (%)a | 94.6 (84.6) |
| No. reflections, total | 234,968 (10,078) |
| No. reflections, unique | 74,234 (3871) |
| Redundancy | 3.2 (2.6) |
| Rsym (%)b | 6.2 (48.1) |
| I/σ(I) | 12.4 (1.7) |
| Structure Refinement | |
| Rcryst/Rfree (%)c | 22.9/26.2 |
| No. of atoms | |
| Protein | 6103 |
| DNA | 6021 |
| Water | 88 |
| Ions | 18 |
| R.m.s.d.bond lengths (Å)/angles (°) | 0.009/1.2 |
| Residues in Ramachandran plot (%) | |
| Most favored/allowed | 94.1/5.9 |
| Mean B-factors (Å2) | |
| Protein/DNA | 51.9/102.2 |
Values in parentheses are those for the outer resolution shell.
Rsym = Σhkl|Ihkl − <I>|/ΣhklIhkl, where Ihkl is the measured intensity of reflections with indices hkl.
Rcryst = Σhkl|Fo − Fc|/ΣhklFo, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree is equal to Rcryst for a randomly selected 5% subset of reflections not used in the refinement.
RESULTS AND DISCUSSION
Structural conservation between the Xenopus and Drosophila NCPs
As expected, Dm-NCP147 and Xla-NCP147 share a high degree of structural similarity. The histone octamers of the two particles superimpose with an overall root-mean-squares deviation (rmsd) of 0.58 Å for backbone Cα atoms, and 1.00 Å for all atoms including side chains (Table II). These values are approximately half those obtained upon alignment of the yeast and Xenopus octamers23, consistent with the notion that structural divergence recapitulates phylogeny.30–33 The individual histone Cα backbones can be aligned with rmsd values of 0.15–0.93 Å. However, these values reduce to 0.15–0.35 Å upon exclusion of a small number of N- or C-terminal residues, where the most significant differences occur. These reduced values correlate well with degree of sequence conservation, the more divergent H2A and H2B histones showing larger rmsd values than the nearly invariant H3 and H4 histones (Table II).
Table II.
Comparison of Dm-NCP147 and Xla-NCP147 Structures
| RMSD (Å)a |
Seq. identity (%)b |
|||
|---|---|---|---|---|
| All structured residues | Excluding terminic | Terminal res. excludedd | Overall/structured | |
| H2A | 0.36 (0.65) | 0.33 (0.64) | 118–119 | 88.7/91.6 |
| H2A′ | 0.67 (1.18) | 0.35 (0.84) | 12–13 | |
| H2B | 0.48 (1.02) | 0.28 (0.88) | 28–29 | 82.7/93.6 |
| H2B′ | 0.49 (0.77) | 0.33 (0.68) | 29–29 | |
| H3 | 0.15 (0.64) | 0.15 (0.64) | –– | 99.2/99.0 |
| H3′ | 0.26 (0.65) | 0.17 (0.58) | 135 | |
| H4 | 0.78 (0.95) | 0.15 (0.43) | 23–24, 102 | 99.0/100 |
| H4′ | 0.93 (1.72) | 0.18 (0.57) | 15–19 | |
| Histone octamer | 0.58 (1.00) | 0.30 (0.70) | All above | |
| DNA | 0.37 (0.34) | |||
| NCP | 0.54 (0.83) | 0.32 (0.59) | All above | |
RMSD values for the backbone Cα and DNA phosphate atoms; values in parentheses are for all atoms including side chains.
Percent sequence identity between Drosophila and Xenopus histones for all residues, and for residues present in the crystallographic model.
RMSD values in which structurally most divergent N- and/or C-terminal residues are excluded from the alignment.
N- and/or C-terminal residues excluded from the alignment.
The conformation of the DNA is essentially identical in the two structures (rmsd = 0.34 Å for all atoms). Unlike the structure of human NCP146, in which the DNA at three superhelix axis locations (SHLs) is shifted relative to Xla-NCP146,24 the DNA in Dm-NCP147 remains in register with that of Xla-NCP147.17 This is likely a reflection of the higher degree of order generally observed in NCP147 compared to NCP146, irrespective of the source of histones. As in previous NCP structures, a plot of B-factor versus base pair shows an oscillating pattern, with minima (40–80 Å2) where the DNA contacts histones, and maxima (80–160 Å) at intermediate positions. The manganese and chloride ions identified in Xla-NCP14718 are all preserved in our Drosophila structure. The entire structure can be superimposed onto that of Xla-NCP147 with an rmsd of 0.82 Å for all protein and DNA atoms, underscoring the high degree of tertiary and quaternary structure conservation.
Differences in the histone tails
A comparison of the Dm- and Xla-NCP147 structures reveals slight differences in the histone tail regions. These probably reflect the inherent structural disorder of the tails, but may also reflect sequence differences (Fig. 1, residues highlighted in pink. Sequence numbering throughout this paper is that of Xenopus, which is identical to the Drosophila numbering except for H2A). More specifically, in Xla-NCP147, H2A′ residue Lys13 inserts into the minor groove to hydrogen bond with Thy 45 in SHL 4 [Fig. 2(A,B)]. In the Drosophila structure, the corresponding Lys residue points more toward the solvent, interacting with the adjacent DNA phosphate group. A few residues away, the Drosophila and Xenopus H2A sequences diverge at two positions, with Drosophila residues Ser and Asn replacing Thr16 and Ser19, respectively. In the Xenopus NCP, Ser19 hydrogen bonds to the backbone amide of Thr16, one helical turn away, with the latter in van der Waals contact with the DNA phosphate backbone [Fig. 2(B)]. While the DNA contact is preserved in Dm-NCP147, the intrahelical hydrogen bond is not, its loss compensated by a hydrogen bond gained between the Ser16 and Asn19 side chains [Fig. 2(B)].
Figure 1.
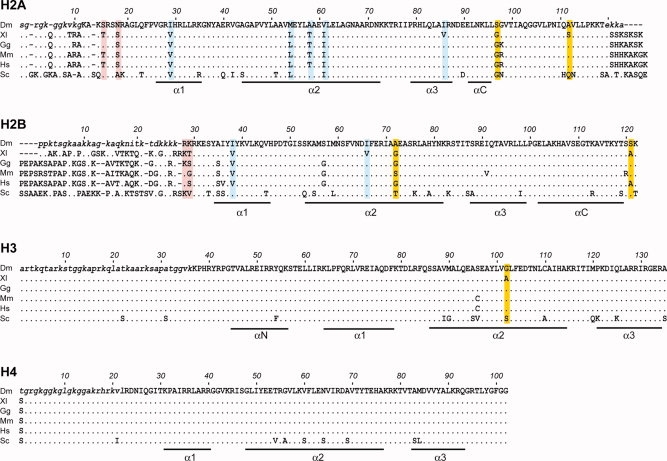
Sequence alignment of histones. Alignment of histones from Drosophila (Dm), Xenopus (Xl), chicken (Gg), mouse (Mm), human (Hs), and yeast (Sc). Drosophila H2A, H2B, H3, H4 sequences correspond to accession codes NP_724343, NP_724342, NP_724345, NP_724344, respectively. Only residues that differ from the Drosophila sequence are shown. Amino acid substitutions that differentiate the Drosophila and Xenopus histone core regions are highlighted in yellow and cyan; those in ordered tail residues are highlighted in pink. Unstructured residues are indicated in lower case.
Figure 2.

Structural differences within the H2A and H2B histone tails between Dm-NCP147 and Xla-NCP147. (a) Dm-NCP viewed along the superhelix showing the location of the N-terminal tail of H2A. (b) The N-terminal histone tail of H2A′, showing close-up of boxed region in a. Side chains from Drosophila are in light gray; from Xenopus in dark Grey. Hydrogen bonds unique to Drosophila are in black; those unique to Xenopus are in red. Residue substitutions are labelled in the direction from Xenopus to Drosophila. DNA bases are shown as sticks, except for Thy45. The view is slightly rotated relative to that in a. (c) Edge view of the NCP showing location of the Nterminal tail of H2B. (d) Histone tail of H2B′ showing close-up view of boxed region in c. Base atoms is shown for Thy50 and Cyt-49.
In the H2B′ chain of Xla-NCP147 (and -NCP146), residue Thr29 hydrogen bonds with the DNA phosphate backbone (at nucleotide position 30 of chain J) [Fig. 2(C,D)]. In Dm-NCP147, the corresponding Lys29 residue interacts with a phosphate group on the complementary strand (position 26 of chain I). The preceding Arg28 residue adopts a similar orientation as the Xenopus Lys28 residue, but inserts more deeply into the minor groove, interacting with the Cyt-49 (chain J) and Thy-50 (chain I). An Arg side chain in a minor groove is a recurrent motif, observed in both the tail and core regions of the various histone chains.16 Residues 28 and 29 in the Dm-NCP147 H2B chain are located approximately as in H2B′, but are considerably more disordered. Such variations between otherwise identical chains highlight the structural plasticity of the histone tails.
Amino acid substitutions in the histone cores
Twelve amino acid substitutions differentiate Drosophila from Xenopus within the histone core regions. Five substitutions are highly conservative replacements involving solvent-exposed, small (Gly, Ala, or Ser) residues which do not interact with other residues nor with the DNA (Fig. 1, highlighted in yellow). Three of these five substitutions [Gly98 → Ser in H2A; Gly72 → Ala in H2B; and Ala102 → Gly in H3 (written as Xenopus → Drosophila)] are unlikely to modify interactions between NCP particles, as the residues concerned face solvent regions internal to the octamer core. The other two (Ser113 → Ala in H2A and Ala121 → Ser in H2B) are converse substitutions which localize to the outer face of the NCP; their net effect is the displacement of a single hydroxyl group across the face of the NCP by 45 Å (or 75% of the octamer's diameter), which probably has no more than a modest effect on inter-NCP interactions.
The remaining seven substitutions differentiating Drosophila from Xenopus localize to the hydrophobic core of H2A and to the H2A-H2B dimer interface (Fig. 1, highlighted in cyan). The residues cluster into two groups, located on opposite sides of the pseudodyad [Fig. 3(A)]. Remarkably, all the substitutions are of a mutually compensatory nature. Two substitutions are juxtaposed in histone H2A at positions mediating interactions between the α2 and α3 helices. The converse nature of these substitutions, Ile62 → Val and Val87 → Ile, allows for the net translocation of a methyl group without perturbing the spatial coordinates of the protein backbone [Fig. 3(B)]. A similar phenomenon is observed for Xenopus residues H2A-Thr59 and H2B-Val38, which in Drosophila are Ala and Ile residues, respectively [Fig. 3(C)]. These two positions are juxtaposed in the dimer interface, such that the gain and loss of a methly group are mutually offset. The Thr59 hydroxyl group forms an intrachain hydrogen bond with the backbone helix α2, and so its loss in Drosophila is unlikely to affect dimer stability. Finally, in Xenopus, H2A residue Leu55 is sandwiched between Val30 of the same chain and H2B residue Val66. All three positions are substituted in Drosophila in such a way as to preserve the volume occupied by side chains in the hydrophobic core: replacement of Leu55 by the more slender Met is countered by replacement of the two valines by bulkier isoleucines [Fig. 3(D)].
Figure 3.

Structural differences in the H2A-H2B dimer between Dm-NCP147 and Xla-NCP147. (a) Overview of structured residues in the hydrophobic core of the H2A-H2B dimer that diverge between Xenopus and Drosophila. View is approximately along the pseudodyad. (b) Compensatory changes within the hydrophobic core of H2A. Residue substitutions are labeled in the direction from Xenopus to Drosophila. (c) Compensatory changes involving two residues in the H2A-H2B dimer interface. The hydrogen bond missing from the Dm structure is in red. (d) Compensatory changes involving three residues in the H2A-H2B dimer interface.
Their compensatory nature suggests that these substitutions are unlikely to influence the kinetics or stability of H2A-H2B dimer formation, and hence the dynamics of nucleosome assembly/disassembly. More generally, the phenomenon of counterbalanced substitutions may partly account for the poor correlation observed between sequence conservation and the number of intrachain contacts mediated by histone residues,34 because two or more poorly conserved residues may coevolve to preserve inter-residue contacts.
Patterns in histone evolution
The core histones are among the best-conserved proteins known. The sequence conservation is notably greater in structured (histone-fold) regions than in the histone tails (Fig. 1). Our analysis of Drosophila and Xenopus NCPs suggests that the sequence divergence in structured residues should have little impact on histone octamer assembly, histone–DNA interactions, or inter-NCP interactions. Clearly, NCP evolution has been tightly constrained since the speciation event that separated the vertebrate and invertebrate clades.
In contrast, yeast and higher eukaryotes exhibit considerably more differences in their histone-fold sequences (Fig. 1), suggesting that histone evolution underwent a burst prior to the appearance of metazoa, only to stagnate thereafter. Although yeast and metazoan NCP structures differ little at the mononucleosomal level, substantial differences in crystal packing interactions suggest that they may exhibit different internucleosomal interactions in vivo.23 This may be a reflection of the significantly lower requirements for DNA compaction of the much smaller yeast genome compared to that of metazoa.
Mutational studies of histones (both in vivo and invitro) have made it clear that maintaining nucleosome structure cannot entirely account for the extreme degree of histone sequence conservation. By corollary, sequence changes in histone mutants or variants are of little structural, but of decisive functional consequence. Histones account for a large percentage of the nucleosome's exposed surface–a highly sculpted, differentially charged landscape that interacts with many nuclear factors35 and that likely mediates nucleosome–nucleosome interactions to form chromatin higher order structure. Thus, unlike globular proteins, exposed surface residues are exceptionally constrained, and can only mutate if compensatory changes minimize the effects.
Acknowledgments
We thank Peter B. Becker for plasmids leading to the expression of the recombinant D. melanogaster histones; and ESRF/EMBL colleagues for access and support to the ESRF beamlines. C.R.C. acknowledges support from an EMBO Long Term Post-Doctoral Fellowship. Coordinates have been deposited in the Protein Data Bank (accession code 2PYO).
REFERENCES
- 1.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 4.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 5.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 6.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 7.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 8.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 9.Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Lieberman PM. Chromatin regulation of virus infection. Trends Microbiol. 2006;14:132–140. doi: 10.1016/j.tim.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 12.Finch JT, Lutter LC, Rhodes D, Brown RS, Rushton B, Levitt M, Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- 13.Bentley GA, Lewit-Bentley A, Finch JT, Podjarny AD, Roth M. Crystal structure of the nucleosome core particle at 16 A resolution. J Mol Biol. 1984;176:55–75. doi: 10.1016/0022-2836(84)90382-6. [DOI] [PubMed] [Google Scholar]
- 14.Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 15.Arents G, Burlingame RW, Wang B-C, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 17.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 18.Davey CA, Richmond TJ. DNA-dependent divalent cation binding in the nucleosome core particle. Proc Natl Acad Sci USA. 2002;99:11169–11174. doi: 10.1073/pnas.172271399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 20.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 21.Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S, Luger K. Structural characterization of the histone variant macroH2A. Mol Cell Biol. 2005;25:7616–7624. doi: 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harp JM, Hanson BL, Timm DE, Bunick GJ. Asymmetries in the nucleosome core particle at 2.5 Å resolution. Acta Crystallogr D Biol Crystallogr. 2000;56:1513–1534. doi: 10.1107/s0907444900011847. [DOI] [PubMed] [Google Scholar]
- 23.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. Embo J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsunaka Y, Kajimura N, Tate S, Morikawa K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005;33:3424–3434. doi: 10.1093/nar/gki663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 27.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. Embo J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbard TJ, Blundell TL. Comparison of solvent-inaccessible cores of homologous proteins: definitions useful for protein modelling. Protein Eng. 1987;1:159–171. doi: 10.1093/protein/1.3.159. [DOI] [PubMed] [Google Scholar]
- 32.Flores TP, Orengo CA, Moss DS, Thornton JM. Comparison of conformational characteristics in structurally similar protein pairs. Protein Sci. 1993;2:1811–1826. doi: 10.1002/pro.5560021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell RB, Saqi MA, Sayle RA, Bates PA, Sternberg MJ. Recognition of analogous and homologous protein folds: analysis of sequence and structure conservation. J Mol Biol. 1997;269:423–439. doi: 10.1006/jmbi.1997.1019. [DOI] [PubMed] [Google Scholar]
- 34.Sullivan SA, Landsman D. Characterization of sequence variability in nucleosome core histone folds. Proteins. 2003;52:454–465. doi: 10.1002/prot.10441. [DOI] [PubMed] [Google Scholar]
- 35.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]


