The crystal structure of Ynk1, an NDPK from the yeast Saccharomyces cerevisiae, has been solved at 3.1 Å resolution.
Keywords: Ynk1, nucleoside diphosphate kinases, Saccharomyces cerevisiae
Abstract
Nucleoside diphosphate kinase (NDPK) catalyzes the transfer of the γ-phosphate from nucleoside triphosphates to nucleoside diphosphates. In addition to biochemical studies, a number of crystal structures of NDPK from various organisms, including both native proteins and complexes with nucleotides or nucleotide analogues, have been determined. Here, the crystal structure of Ynk1, an NDPK from the yeast Saccharomyces cerevisiae, has been solved at 3.1 Å resolution. Structural analysis strongly supports the oligomerization state of this protein being hexameric rather than tetrameric.
1. Introduction
Nucleoside diphosphate kinases (NDPKs) are highly conserved from prokaryotes to eukaryotes and play a critical role in nucleoside triphosphate synthesis, which maintains cellular homeostasis of nucleoside triphosphates (NTPs) and nucleoside diphosphates (NDPs). They catalyze the phosphorylation of nucleoside diphosphates to the corresponding triphosphorylated form in a ping-pong mechanism in which a conserved histidine at the active site is transiently phosphorylated and the high-energy phosphate is then transferred to a nucleoside diphosphate (Postel, 1998 ▶; Lascu et al., 2000 ▶). The high-energy phosphate is usually supplied by ATP and any nucleoside diphosphate except adenosine diphosphate can serve as the high-energy phosphate acceptor. In addition to their catalytic function, NDPKs are also involved in other cellular activities such as development, differentiation, cell proliferation, cell motility, tumour metastasis and apoptosis (Amutha & Pain, 2003 ▶). In addition to biochemical studies, a number of crystal structures of NDPKs from various species have been determined. To date, over 70 crystal structures of NDPKs have been solved, including those of point mutants and of complexes with nucleotides or analogues, which have greatly contributed to the establishment of a sound basis for the elucidation of their catalytic mechanism and stability against denaturation. All NDPKs of known structure are homo-oligomeric proteins, with the exception of human NDPK-A and NDPK-B, which can form hetero-oligomers; although most NDPKs form hexamers, some bacterial enzymes form tetramers (Lascu et al., 2000 ▶). Both hexamers and tetramers are constructed by the assembly of identical dimers. The hexameric structure is considered to be more stable than the tetrameric structure and also to have higher enzymatic activity.
The NDPK Ynk1 from the yeast Saccharomyces cerevisiae is encoded by open reading frame YNK1/YKL067W. Its NDPK activity is predominantly present in the cytosol, with a small amount in the intermembrane space of the mitochondria (Amutha & Pain, 2003 ▶), although there is not a cleavable mitochondrial targeting sequence at the N-terminus of protein as in human NDPK (Nm23-H4 and Nm23-H6; Milon et al., 2000 ▶) and pigeon NDPK (Lambeth et al., 1997 ▶). Ynk1 is also involved in both DNA and RNA metabolism and nucleoside-triphosphate synthesis, but YNK1 null mutants are viable and display normal sporulation, mating, morphology and growth rates (Fukuchi et al., 1993 ▶), which is uncommon in the NDPK family. Moreover, the quaternary structure of Ynk1 remains controversial, having first been identified as a hexamer (Palmieri et al., 1973 ▶) and subsequently demonstrated to be a tetramer (Jong & Ma, 1991 ▶).
Here, we present the X-ray crystal structure of S. cerevisiae Ynk1 at 3.1 Å resolution. The fold of the monomer and the strong dimer assembly are remarkably similar to all previously known structures. Furthermore, the dimers appear to be further assembled into hexamers during crystal packing. Most known bacterial NDPKs are tetrameric, whereas the known eukaryotic NDPKs are hexameric. However, the oligomeric state of NDPK in solution has been under debate. Even human NDPK-B and C were found to present a tetrameric structure in size-exclusion chromatography (Lascu et al., 2000 ▶). In this paper, the crystal structure of Ynk1 provided us with further insights into the interface and interactions between the dimers and the conserved residues involved in quaternary-structure formation.
2. Experimental
2.1. Sample preparation
The open reading frame YNK1/YKL067W was amplified by PCR and cloned into a pET28a-derived expression vector with the coding sequence for a hexahistidine (6×His) tag immediately after the start codon. The plasmid containing YNK1 was transformed into Escherichia coli BL21 (DE3) for expression. The transformant cells were grown in 2×YT medium (16 g bactotryptone, 10 g yeast extract and 5 g NaCl) at 310 K until the OD600 reached 0.8. Expression was induced at 291 K with 0.2 mM IPTG for 20 h. The cells were collected by centrifugation and resuspended in lysis buffer (20 mM HEPES pH 7.0, 500 mM NaCl, 20 mM MgCl2 and 14 mM β-mercaptoethanol). After freeze–thawing followed by sonication, the soluble fraction was separated by centrifugation and passed through a 0.22 µm filter. The His-tagged proteins were purified using an Ni–NTA column (Amersham Biosciences) according to standard protocols. Fractions containing Ynk1 were pooled and purified by gel-filtration chromatography using a Hiload 16/60 Superdex 200 column (Amersham Biosciences) equilibrated with buffer containing 20 mM HEPES pH 7.0, 50 mM NaCl. The purity of the pooled fractions was ascertained by SDS–PAGE and the protein sample was concentrated to 19 mg ml−1 in the same buffer for further use.
2.2. Crystallization and data collection
Crystals of Ynk1 were grown by hanging-drop vapour diffusion at 291 K. 1 µl protein solution (19 mg ml−1 in 20 mM HEPES pH 7.0, 50 mM NaCl) was mixed with 2 µl reservoir solution containing 13% 2-propanol, 19% TBA (t-butyl alcohol), 0.1 M sodium citrate pH 5.6. The crystals grew to maximum dimensions of 150 × 150 × 100 µm in one month. Because the crystal was very fragile, it was directly flash-frozen with liquid nitrogen without cryoprotectant. Diffraction data were collected at 100 K on an in-house Rigaku rotating-anode generator (FR-E SuperBright) producing Cu Kα radiation and equipped with an R-AXIS IV++ image-plate detector at the Institute of Biochemistry and Cell Biology, Shanghai, China. A total of 70 images were collected with 1° oscillations at a wavelength of 1.54179 Å using an exposure time of 20 min per image. The data were integrated using MOSFLM (Leslie, 1999 ▶) and scaled using SCALA (Evans, 1997 ▶) from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). A summary of the statistics of the data used for structure determination is given in Table 1 ▶. The crystals belong to space group F23 with a dimer in the asymmetric unit. The unit-cell parameters are a = b = c = 185.92 Å, α = β = γ = 90.00°.
Table 1. Crystallographic data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Data processing | |
| Resolution range () | 29.403.10 (3.273.10) |
| Measured reflections | 63290 (6547) |
| Unique reflections | 9654 (1333) |
| B factor from Wilson plot (2) | 71 |
| Asymmetric unit contents | Dimer |
| Completeness (%) | 98.68 (94.76) |
| Multiplicity | 6.56 (4.91) |
| I/(I) | 5.78 (1.52) |
| R merge † | 0.12 (0.56) |
| Refinement statistics | |
| Resolution range () | 153.10 (3.183.10) |
| R cryst ‡ | 0.23 (0.28) |
| Free R factor§ | 0.21 (0.33) |
| No. of atoms (protein/ligand/water) | 229/10/07 |
| R.m.s.d. bond lengths () | 0.017 |
| R.m.s.d. bond angles () | 1.94 |
| Mean B value (2) | 60.000 |
| Ramachandran plot | |
| Residues in most favoured regions (%) | 89.4 |
| Residues in allowed regions (%) | 10.6 |
| Residues in disallowed regions (%) | 0 |
R
merge = 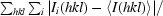
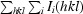 , where Ii(hkl) is the intensity of a reflection and I(hkl) is the average intensity of that reflection.
, where Ii(hkl) is the intensity of a reflection and I(hkl) is the average intensity of that reflection.
R
cryst = 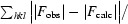
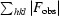 .
.
4.6% of the data were set aside for free R-factor calculation.
2.3. Structural determination and refinement
Molecular replacement was performed with MOLREP (Vagin & Teplyakov, 1997 ▶) from CCP4i using human nucleotide diphosphate kinase B (NDPK-B; PDB code 1nue; Moréra et al., 1995 ▶) as a search model. Model refinement was performed with REFMAC5 (Murshudov et al., 1999 ▶) and O (Jones et al., 1991 ▶), which was used for manual rebuilding. TLS refinement (using each chain as a separate body) was performed at the low resolution at 3.1 Å. Twofold noncrystallographic symmetry (NCS) restraints were applied to the homodimer in the asymmetric unit throughout refinement. The final model had an R factor of 23.1% and an R free of 26.1%. The structure was analyzed using PROCHECK (Laskowski et al., 1993 ▶). In addition to the His tag at the N-terminus, each chain lacks residues 1 and 55–61. The low resolution (3.1 Å) and high B factor (mean 60 Å2) of the crystal are a consequence of the large unit cell (a = b = c = 185.92 Å) and the high solvent content (70.6%). The final coordinates and structure factors have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with code 3b54.
3. Results and discussion
3.1. Overall structure
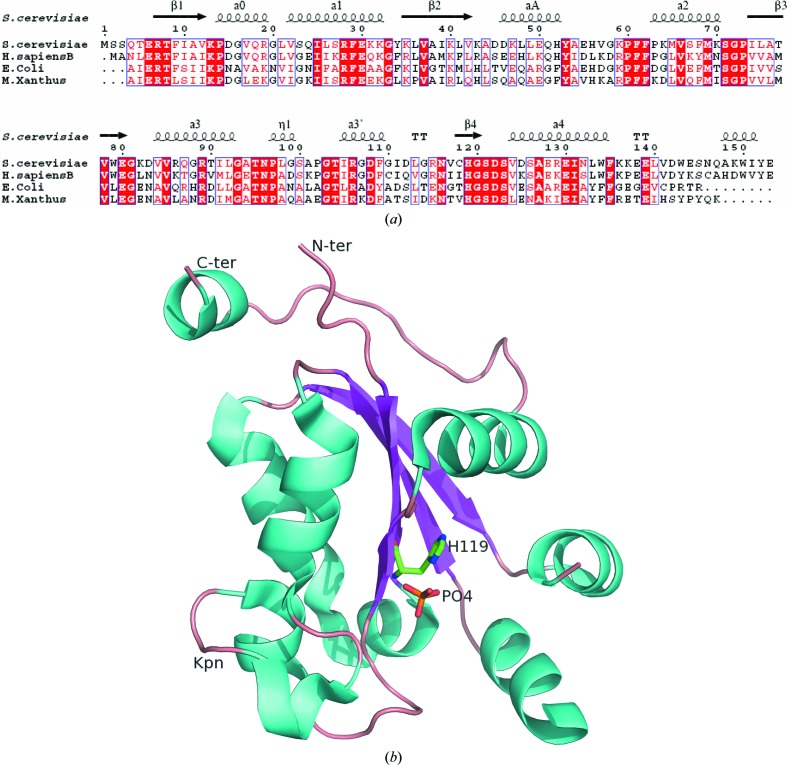
NDPKs are highly conserved from bacteria to humans (Fig. 1 ▶ a). The fold of Ynk1 resembles those of other NDPKs: the overall structure of one subunit is made up of seven α-helices, which partially cover the two faces of a central four-stranded antiparallel β-sheet (Fig. 1 ▶ b), which has been called the α/β-sandwich or ferredoxin fold as it was first observed in Pseudomonas aerogenes ferredoxin (Adman et al., 1973 ▶). In the crystal structure, the asymmetric unit is made up of two identical chains labelled A and B. Each chain of the final model includes residues 2–54 and 62–153 (Fig. 1 ▶ b). Although NDPK from Myxococcus xanthus is a tetramer (Williams et al., 1993 ▶), the r.m.s. deviation between superimposed Cα atoms is 0.962 Å, indicating that the two proteins share a very similar overall structure. The C-terminus and Kpn loop deviated much more than other parts of the structure. These two fragments are always flexible as they are involved in substrate binding and quaternary-structure formation. As expected from the high level of sequence identity (61%; Fig. 1 ▶ a), the structures of the Ynk1 and human NDPK-B (Webb et al., 1995 ▶) monomers are very similar, with a Cα r.m.s. deviation of 0.621 Å. The largest differences were found in the two disordered loops, one of which is at the link between αA and α2 and the other of which is at the N-terminus. The loop containing residues 54–61 that connects helices αA and α2 is highly solvent-exposed in each chain. As in all NDPK structures without nucleotide, this loop is quite flexible; it is required for nucleotide binding (Janin et al., 2000 ▶). In our structure, this loop could not be fitted into the electron-density map, so the corresponding residues were deleted from the final model. The first five residues at the N-terminus could also not be assigned correctly. As in other available NDPK structures, a β-branched residue at position 117 is the central residue of a classical γ-turn and has a positive ϕ angle (Janin et al., 2000 ▶) which deviates slightly from the allowed area of the Ramachandran plot in this low-resolution structure.
Figure 1.
(a) Structure-based alignment of NDPK sequences. The secondary structure is that of Ynk1. The homologous proteins are from Saccharomyces cerevisiae, Homo sapiens, Escherichia coli and Myxococcus xanthus. The alignment was drawn using ESPript (Gouet et al., 1999 ▶). (b) Ribbon diagram of a monomer of Ynk1; secondary structures are coloured cyan (helices), magenta (strands) and wheat (loops). This figure was prepared using PyMOL (DeLano, 2002 ▶).
3.2. The active site and the quaternary structure
The NDPK active site comprises the nucleophilic histidine and the nucleotide-binding site. There is a single binding site per subunit which accepts two types of substrate: the nucleoside triphosphate that donates the phosphate group and the nucleoside diphosphate that receives the phosphate group. The nucleotide-binding site forms a cleft on the protein surface between the Kpn loop and the αA–α2 hairpin; the nucleophilic His119 resides at the bottom of the cleft (Fig. 1 ▶ b). The NDPK active sites are identical and independent within a tetramer or hexamer. They are also structurally identical in different enzymes and almost all residues involved in the active site are fully invariant from bacteria to human.
The residues contributing to dimerization are located in helix α1, strand β2 and the C-terminal segment. The dimer interface is 1013 Å2 in size, similar to those of other NDPKs (Janin et al., 2000 ▶), indicating that Ynk1 uses the same dimer to assemble the hexamer. A characteristic of Ynk1 is that an additional hydrogen bond is present in the interface between the side chains of residue Ser27 and the same residue of its counterpart subunit via a water molecule, which might stabilize the dimer.
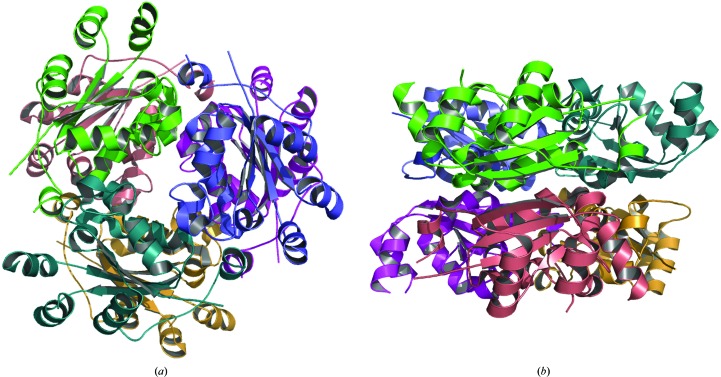
Although Ynk1 appears to contain a dimer in the asymmetric unit, in the crystal a hexameric structure is assembled by crystallographic symmetry and the hexameric structure is similar to the other known hexamers (Fig. 2 ▶). Ynk1 consists of 153 residues, which is similar to other eukaryotic NDPKs and longer than prokaryotic NDPKs. It has a longer C-terminus then eukaryotic NDPKs and this is critical for hexamer formation. The conserved C-terminal dipeptide Tyr-Glu is in the same position as in other NDKs from eukaryotes. However, Glu153 NH does not makes a hydrogen bond to the side chain of Asp112 of the neighbouring subunit as in the other enzymes. Instead, the side chain of Lys149, which is not conserved in other hexameric NDPKs, makes a hydrogen bond to the side chain of Asn146 from a neighbouring subunit; this is not found in other hexameric structures. Other key residues involved in hexamer formation, such as Arg19, Lys32, Pro97 and Pro102, are also conserved in Ynk1, contributing to the trimer interface through nonpolar interactions or hydrogen bonds to neighbouring subunits.
Figure 2.
Top view (a) and side view (b) of the hexameric structure of Ynk1 obtained by crystallographic symmetry operation.
The oligomeric structure of Ynk1 in solution has been reported as either a hexamer or a tetramer using size-exclusion chromatography and sucrose-gradient methods (Palmieri et al., 1973 ▶; Jong & Ma, 1991 ▶). In the present study, Ynk1 forms a hexamer in the crystal. Tetrameric forms have been classified into two types, type I from M. xanthus (Williams et al., 1993 ▶) and type II from E. coli (Moynié et al., 2007 ▶), which were assembled in different ways. The type II tetramer seems to be the most frequent assembly mode in bacterial tetrameric NDPKs (Moynié et al., 2007 ▶). A number of residues involved in tetramer formation are conserved in these NDPKs. The two critical residues (Ynk1 numbering) involved in tetramer formation are Lys32 and Lys107 in type I and Ala32 and Lys107 in type II; these residues are replaced by Lys32 and Gly107 in Ynk1 and other hexameric NDPKs. In addition, two aromatic residues at positions 52 and 133 (Ynk1 numbering) that are conserved in both type I and II tetramers are substituted by His52 and Leu133 in Ynk1. Therefore, it is unlikely that Ynk1 will form tetramers. This debate regarding the oligomeric structure also exists for other NDPKs (Hemmerich & Pecht, 1992 ▶; Kowluru & Metz, 1994 ▶). The most likely explanation for the debate is that ionic or hydrophobic interactions of some NDPKs with the gel matrix lead to protein retardation under the buffer conditions (Lascu et al., 2000 ▶). Recently, light-scattering and chemical cross-linking analyses of NDPK from the moderate halophile Halomonas sp. 593 (HaNDK) unambiguously demonstrated that this enzyme forms a dimeric structure (Yonezawa et al., 2007 ▶).
4. Conclusion
The crystal structure of yeast NDPK strongly supports the oligomeric state of this protein being hexameric rather than tetrameric and also confirms that all NDPKs use dimers as a basic assembly unit regardless of the quaternary structure. However, whether or not this assembly unit is itself biologically significant remains unclear.
Supplementary Material
PDB reference: Ynk1, 3b54, r3b54sf
Acknowledgments
This work was supported by the Ministry of Education of China (Talents Project of New Century NCET-06-0374 and Program PRA B07-02 to YC), the National Natural Science Foundation of China (30670461 to YC, 30470366 to CZZ) and the Pujiang Talents project (06PJ14087 to YC).
References
- Adman, E. T., Sieker, L. C. & Jensen, L. H. (1973). J. Biol. Chem. 248, 3987–3996. [PubMed]
- Amutha, B. & Pain, D. (2003). Biochem. J. 370, 805–815. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. http://www.pymol.org.
- Evans, P. R. (1997). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 33, 22–24.
- Fukuchi, T., Nikawa, J., Kimura, N. & Watanabe, K. (1993). Gene, 129, 141–146. [DOI] [PubMed]
- Gouet, P., Courcelle, E., Stuart, D. I. & Metoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Hemmerich, S. & Pecht, I. (1992). Biochemistry, 31, 4580–4587. [DOI] [PubMed]
- Janin, J., Dumas, C., Moréra, S., Xu, Y., Meyer, P., Chiadmi, M. & Cherfils, J. (2000). J. Bioenerg. Biomembr. 32, 215–325. [DOI] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Jong, A. Y. & Ma, J. J. (1991). Arch. Biochem. Biophys. 291, 241–246. [DOI] [PubMed]
- Kowluru, A. & Metz, S. A. (1994). Biochemistry, 33, 12495–12503. [DOI] [PubMed]
- Lambeth, D. O., Mehus, J. G., Ivey, M. A. & Milavetz, B. I. (1997). J. Biol. Chem. 272, 24604–24611. [DOI] [PubMed]
- Lascu, L., Giartosio, A., Ransac, S. & Erent, M. (2000). J. Bioenerg. Biomembr. 32, 227–236. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Leslie, A. G. W. (1999). Acta Cryst. D55, 1696–1702. [DOI] [PubMed]
- Milon, L., Meyer, P., Chiadmi, M., Munier, A., Johansson, M., Karlsson, A., Lascu, I., Capeau, J., Janin, J. & Lacombe, M.-L. (2000). J. Biol. Chem. 275, 14264–14272. [DOI] [PubMed]
- Moréra, S., Chiadmi, M., LeBras, G., Lascu, I. & Janin, J. (1995). Biochemistry, 34, 11062–11070. [PubMed]
- Moynié, L., Giraud, M. F., Georgescauld, F., Lascu, L. & Dautant, A. (2007). Proteins, 67, 755–765. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. (1999). Acta Cryst. D55, 247–255. [DOI] [PubMed]
- Palmieri, R., Yue, R. H., Jacobs, H. K., Maland, L., Wu, L. & Kuby, S. A. (1973). J. Biol. Chem. 248, 4486–4499. [PubMed]
- Postel, E. H. (1998). Int. J. Biochem. Cell Biol. 30, 1291–1295. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst. 30, 1022–1025.
- Webb, P. A., Perisic, O., Mendola, C. E., Backer, J. M. & Williams, R. L. (1995). J. Mol. Biol. 251, 574–587. [DOI] [PubMed]
- Williams, R. L., Oren, D. A., Muñoz-Dorado, J., Inouye, S., Inouye, M. & Arnold, E. (1993). J. Mol. Biol. 234, 1230–1247. [DOI] [PubMed]
- Yonezawa, Y., Izutsu, K., Tokunaga, H., Maeda, H., Arakawa, T. & Tokunaga, M. (2007). FEMS Microbiol. Lett. 268, 52–58. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Ynk1, 3b54, r3b54sf