Allene oxide synthase, an atypical cytochrome P450 from Parthenium argentatum, was crystallized and diffraction data were collected to 2.4 Å resolution.
Keywords: allene oxide synthase, cytochrome P450 CYP74A2, oxylipin, jasmonate, guayule (Parthenium argentatum), crystallization
Abstract
Oxylipins are oxygenated derivatives of fatty acids and pivotal signaling molecules in plants and animals. Allene oxide synthase (AOS) is a key cytochrome P450 CYP74 enzyme involved in the biosynthesis of plant oxylipin jasmonates to convert 13(S)-hydroperoxide to allene oxide. Guayule (Parthenium argentatum) AOS, CYP74A2, was expressed in Escherichia coli. Protein was purified using affinity chromatography and size exclusion chromatography, and then crystallized. Two different crystal forms were obtained from 0.2 M (NH4)H2PO4, 50% MPD, 0.1 M Tris, pH 8.5 at 277 K using the hanging-drop vapor-diffusion method. Preliminary X-ray analysis was carried out, and the crystals were found to belong to the tetragonal space group I422 with cell parameters a = b = 126.5, c = 163.9 Å, and the monoclinic space group C2 with cell parameters a = 336.5, b = 184.2, c = 159.0 Å, β = 118.6°. Diffraction data were collected to 2.4 Å resolution from a tetragonal form of crystal using a home X-ray source.
1. Introduction
Oxylipins derived from oxygenated fatty acids are bioactive compounds involved in signal and defense reactions in mammals, higher plants and algae (Blee, 2002 ▶; Pohnert, 2005 ▶); for example, prostaglandins play essential roles in numerous physiological functions in mammals, and jasmonates act as ubiquitous plant growth regulators. Allene oxide synthase (AOS) is a key cytochrome P450 enzyme involved in the biosynthesis of jasmonates. It catalyzes a dehydration reaction to convert 13(S)-hydroperoxide to allene oxide.
AOS is atypical and belongs to the CYP74 family. The classic cytochrome P450s require electron-transfer partners, i.e. FAD-containing reductase and an iron–sulfur redoxin (for class I), and a P450 reductase (for class II). AOS and other CYP74 enzymes do not require O2 and NADPH-dependent cytochrome P450 reductase for their activities. They use hydroperoxides as the oxygen donors as well as sources of reducing equivalents. Sequence analysis showed that AOS contains an unusual heme-binding region and possesses a defective I-helix without the highly conserved threonine present in the classic P450s (Song et al., 1993 ▶).
There are different CYP74 enzymes involved in several distinct pathways for oxylipin biosynthesis, including AOS in the AOS branch for jasmonic acid biosynthesis, hydroperoxide lyase (HPL) in the HPL branch for the production of leaf aldehydes, and divinylether synthase (DES) in the DES branch for the formation of divinyl ethers (Hofmann et al., 2006 ▶). These CYP74 enzymes use fatty acid hydroperoxides as substrates, and are classified as four subfamilies (Stumpe & Feussner, 2006 ▶). AOSs specificially using 13(S)-hydroperoxides as substrates are named 13-AOSs and classified into the CYP74A subfamily. 9/13-AOS recognizes both 9(S)- and 13(S)-hydroperoxides, and 9-AOS has specificity for 9(S)-hydroperoxides. The 9/13- and 9-AOSs are grouped into the CYP74C subfamily. HPLs with specificity for 13-hydroperoxides fall into the CYP74B subfamily, and DESs form the CYP94D subfamily.
Guayule (Parthenium argentatum) plants accumulate large quantities of rubber in rubber particles which are a potential alternative source of latex products for medical applications (Siler et al., 1996 ▶). Rubber particle protein (RPP) is the most abundant protein in rubber particles (Backhaus et al., 1991 ▶); it has high amino-acid sequence identity to flax AOS, and was characterized as AOS and classified as CYP74A2 (Pan et al., 1995 ▶).
Crystallographic study of eukaryotic P450s is very challenging because they are usually membrane-associated proteins. To date, some eukaryotic P450 structures have been reported, including several microsomal P450s from rabbit and human (Williams et al., 2000 ▶, 2003 ▶, 2004 ▶; Scott et al., 2003 ▶; Schoch et al., 2004 ▶; Yano et al., 2005 ▶; Rowland et al., 2006 ▶), and endoperoxide metabolizing P450 prostacyclin synthases (PGIS) from human and zebrafish (CYP8A1) (Chiang et al., 2006 ▶; Li et al., 2008 ▶). AOSs are the same class of P450s as PGISs, but with very low sequence identity (~15%).
Most AOSs are membrane-associated proteins and target the chloroplast. Guayule AOS (CYP74A2) is an unusual P450 that does not possess the N-terminal membrane anchor and the organelle targeting sequences found in other CYP74A enzymes. This unique feature may be related to a special function in rubber particles, such as a possible involvement in rubber biosynthesis, which remains to be explored. Because of the lack of a membrane anchor, guayule AOS can be heterologously expressed in Escherichia coli in a water-soluble form (Pan et al., 1998 ▶). We chose guayule AOS as a model to study the structure of plant AOS and its related species. Here we report the crystallization of guayule AOS purified from overexpressing E. coli, and the preliminary X-ray diffraction analysis.
2. Materials and methods
2.1. Cloning and expression
The guayule (P. argentatum) AOS open-reading frame (ORF) was amplified from pRPP30 (Pan et al., 1995 ▶) by polymerase chain reaction (PCR) using Pfu thermostable DNA polymerase (Stratagene, La Jolla, CA). The following primers for the PCR amplification were used: 5′-CATGCCATGGACCCATCGTCTAAACC-3′ (forward) with an NcoI site (in bold) and 5′-CCGCTCGAGTATACTAGCTCTCTTCAGGAACGTAA-3′ (reverse) with an XhoI site (in bold). The PCR products were digested with NcoI and XhoI, purified, and ligated into NcoI/XhoI-digested pET28b vector (Novagen, Madison, WI) to produce the bacterial expression construct pETAOS. The construct codes for the entire native protein with a hexahistidine tag (i.e. LEHHHHHH) at the C-terminus. The insert was sequenced to ensure the authenticity of the ORF.
The pETAOS construct was transformed into E. coli BL21(DE3) cells (Invitrogen Carlsbad, CA, USA) according to the manufacturer’s specifications. Fresh colonies grown on LB plates containing kanamycin (50 µg ml−1) were inoculated into 25 ml LB medium containing kanamycin (50 µg ml−1). The overnight cultures were then inoculated into 1 l LB medium containing kanamycin (50 µg ml−1) and grown at 310 K. Induction was initiated when the absorbance at 600 nm (A 600nm) reached 0.4–0.6 by adding isopropyl-β-d-thiogalactoside (IPTG) to a final concentration of 0.5 mM. The heme precursor δ-aminolevulinic acid was added simultaneously at a final concentration of 0.5 mM. The culture was grown for an additional 48 h at 298 K. The cells were harvested by centrifugation at 6000g for 10 min at 277 K.
2.2. Purification
Harvested cells were suspended in lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, 1% TritonX-100, pH 8.0), and lysed by passing through a French press. Cell debris was removed by centrifugation at 29 000g for 1 h. The resulting supernatant was mixed gently with Ni–nitrilotriacetic acid (Ni2+–NTA) agarose (Qiagen, Valencia, CA, USA) for 40–60 min at 277 K. The mixture was then transferred into a disposable column and washed extensively with wash buffer (50 mM sodium phosphate, 300 mM NaCl, 20 mM imidazole, pH 8.0). His-tagged proteins were then eluted with elution buffer (50 mM sodium phosphate, 300 mM NaCl, 250 mM imidazole, pH 8.0). Further purification was accomplished with chromatography using a HiPrep Sephacryl S-200HR gel-filtration column (GE Healthcare BioSciences Corp., Piscataway, NJ, USA). The buffer for chromatography contained 50 mM potassium phosphate, pH 8.0, 150 mM NaCl, and 5% glycerol. The purity of AOS protein was judged by SDS–PAGE to be 90–95% (data not shown). The reduced-CO difference spectrum at 450 nm, which is a unique feature of P450s, was performed according to the method previously described (Omura & Sato, 1964 ▶). Similar to other P450s, the purified AOS protein showed a characteristic absorption peak at 450 nm. The catalytic activity of the purified AOS was determined according to the method previously described (Pan et al., 1998 ▶) to ensure the active form of purified AOS.
2.3. Crystallization
Initial crystallization screening was performed using the hanging-drop vapor-diffusion method and the commercial crystal screen kits, including Crystal Screen and Crystal Screen 2 from Hampton Research and Wizard I and Wizard II from Emerald Biosystems. Crystallization experiments were set up using 24-well VDX plates (Hampton Research). AOS protein at a concentration of ~15 mg ml−1 in 50 mM potassium phosphate, 150 mM NaCl, 5% glycerol, pH 8.0, was mixed with a reservoir solution in a drop size of 1+1 µl. The mixture was equilibrated over 300 µl reservoir solution at 277 K.
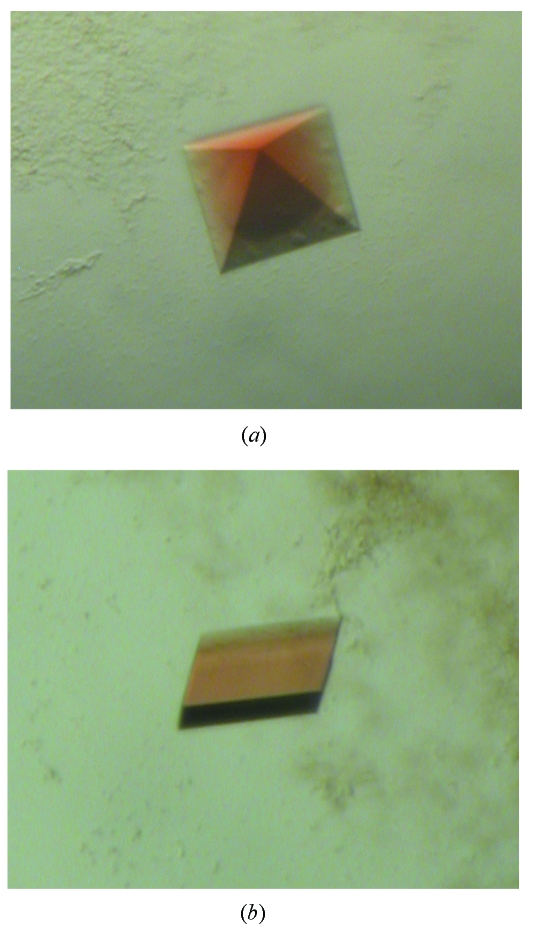
Crystals were obtained in several conditions from the initial screening. The AOS crystals are reddish due to the heme prosthetic group. To improve the quality and size of crystals, a large drop size of 2+2 µl was used, and optimization of crystallization conditions (e.g. precipitant concentration and pH) was carried out. The best crystals were grown from 0.2 M (NH4)H2PO4, 50% 2-methyl-2,4-pentanediol (MPD), 0.1 M Tris, pH 8.5. Interestingly, two different forms of AOS crystals were obtained from the same condition. One has a pyramid-like shape, and another is plate-shaped (Fig. 1 ▶). The crystals were grown in about 7–10 days to the dimensions of 0.2 × 0.2 × 0.1 mm and 0.2 × 0.1 × 0.05 mm for the pyramid- and plate-shaped crystals, respectively.
Figure 1.
Crystals of guayule AOS. (a) A tetragonal crystal form; (b) a monoclinic crystal form.
2.4. Diffraction data collection and processing
Crystals of AOS were picked up directly at 277 K from the crystallization droplets, mounted in nylon loops, and flash-frozen in liquid nitrogen, since the 50% MPD in the mother liquor may be used as cryoprotectant. Data collection was carried out using an R-AXIS IV++ image-plate detector and RU3H X-ray source. Diffraction images were indexed, integrated and scaled using the HKL2000 program suite (Otwinowski & Minor, 1997 ▶).
Both crystal forms of AOS were analyzed, and X-ray diffraction data sets were obtained from a pyramid-shaped crystal. Details of the data-collection statistics are summarized in Table 1 ▶.
Table 1. X-ray diffraction data statistics for AOS.
Numbers in parentheses are for the highest-resolution shell.
| Space group | I422 |
| Unit-cell parameters (Å, °) | a = b = 126.5, c = 163.9 |
| α = β = γ = 90 | |
| Resolution (Å) | 28.4–2.4 (2.49–2.4) |
| Total reflections | 127004 |
| Unique reflections | 26120 (2603) |
| Average redundancy | 4.9 (4.7) |
| Completeness (%) | 98.6 (100) |
| Rmerge (%) | 8.0 (59.4) |
| I/σ(I) | 15.2 (2.4) |
| Solvent content (%) | 59.1 |
| Matthews coefficient (Å3 Da−1) | 3.0 |
| Molecules per ASU | 1 |
3. Results and discussion
Analysis of the X-ray diffraction data indicates that the pyramid-shaped crystal belongs to the tetragonal space group I422, with unit-cell parameters a = b = 126.5, c = 163.9 Å, α = β = γ = 90°. The calculated Matthews coefficient (V M = 3.0 Å3 Da−1), corresponding to a solvent content of 59.1%, indicates the presence of one monomer of 55 kDa protein in the asymmetric unit. A complete data set to 2.4 Å resolution has been obtained with an R merge of 8.0%.
The plate-shaped crystal belongs to the monoclinic space group of C2, with unit-cell parameters a = 336.5, b = 184.2, c = 159.0 Å, β = 118.6°. This crystal form diffracted weakly, only to ∼4 Å. There would be eight molecules per crystallographic asymmetric unit with 75.2% solvent content if a V M of 4.9 Å3 Da−1 is assumed. This is consistent with the result of gel-filtration analysis, that AOS is present in solution as an octamer.
Although many structures have been reported for P450s, sequence identity is very low between AOS and the P450s of known structure (only ~10–16%). A molecular replacement study, using the 2.4 Å data set from the tetragonal crystal form, indicated that it is difficult to determine the structure of AOS from these P450 structures including the same class P450 prostacyclin synthase (PDB ID: 2iag). Heavy-atom derivatives are in preparation for structure determination using MIR or MAD methods.
A very large number of P450s are present in plants, and many are involved in the biosynthesis of natural products for catalyzing various chemical reactions. However, to our knowledge, no crystal structure for plant P450s has been reported. We have successfully crystallized this unique P450 enzyme AOS from guayule (P. argentatum). The high quality of crystals will lead to the elucidation of the structure of this important P450 enzyme, which will provide a structural insight to AOSs and related P450 enzymes, and facilitate the study of enzyme–substrate interactions and catalytic mechanisms.
Acknowledgments
We thank Drs Z. Fu and H. Pan for critical reading of the manuscript. This work was supported by the Samuel Roberts Noble Foundation.
References
- Backhaus, R. A., Cornish, K., Chen, S.-F., Huang, D.-S. & Bess, V. H. (1991). Phytochemistry, 30, 2493–2497.
- Blee, E. (2002). Trends Plant Sci.7, 315–322. [DOI] [PubMed]
- Chiang, C. W., Yeh, H. C., Wang, L. H. & Chan, N. L. (2006). J. Mol. Biol.364, 266–274. [DOI] [PMC free article] [PubMed]
- Hofmann, E., Zerbe, P. & Schaller, F. (2006). Plant Cell, 18, 3201–3217. [DOI] [PMC free article] [PubMed]
- Li, Y. C., Chiang, C. W., Yeh, H. C., Hsu, P. Y., Whitby, F. G., Wang, L. H. & Chan, N. L. (2008). J. Biol. Chem.283, 2917–2926. [DOI] [PMC free article] [PubMed]
- Omura, T. & Sato, R. (1964). J. Biol. Chem.239, 2370–2378. [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pan, Z., Camara, B., Gardner, H. W. & Backhaus, R. A. (1998). J. Biol. Chem.273, 18139–18145. [DOI] [PubMed]
- Pan, Z., Durst, F., Werck-Reichhart, D., Gardner, H. W., Camara, B., Cornish, K. & Backhaus, R. A. (1995). J. Biol. Chem.270, 8487–8494. [DOI] [PubMed]
- Pohnert, G. (2005). Chembiochem.6, 1–14. [DOI] [PubMed]
- Rowland, P., Blaney, F. E., Smyth, M. G., Jones, J. J., Leydon, V. R., Oxbrow, A. K., Lewis, C. J., Tennant, M. G., Modi, S., Eggleston, D. S., Chenery, R. J. & Bridges, A. M. (2006). J. Biol. Chem.281, 7614–7622. [DOI] [PubMed]
- Schoch, G. A., Yano, J. K., Wester, M. R., Griffin, K. J., Stout, C. D. & Johnson, E. F. (2004). J. Biol. Chem.279, 9497–9503. [DOI] [PubMed]
- Scott, E. E., He, Y. A., Wester, M. R., White, M. A., Chin, C. C., Halpert, J. R., Johnson, E. F. & Stout, C. D. (2003). Proc. Natl Acad. Sci. USA, 100, 13196–13201. [DOI] [PMC free article] [PubMed]
- Siler, D. J., Cornish, K. & Hamilton, R. G. (1996). J. Allergy Clin. Immunol.98, 895–902. [DOI] [PubMed]
- Song, W.-C., Funk, C. D. & Brash, A. R. (1993). Proc. Natl Acad. Sci. USA, 90, 8519–8523. [DOI] [PMC free article] [PubMed]
- Stumpe, M. & Feussner, I. (2006). Phytochem. Rev.5, 347–357.
- Williams, P. A., Cosme, J., Sridhar, V., Johnson, E. F. & McRee, D. E. (2000). Mol. Cell, 5, 121–131. [DOI] [PubMed]
- Williams, P. A., Cosme, J., Vinkovic, D. M., Ward, A., Angove, H. C., Day, P. J., Vonrhein, C., Tickle, I. J. & Jhoti, H. (2004). Science, 305, 683–686. [DOI] [PubMed]
- Williams, P. A., Cosme, J., Ward, A., Angove, H. C., Matak Vinkovic, D. & Jhoti, H. (2003). Nature (London), 424, 464–468. [DOI] [PubMed]
- Yano, J. K., Hsu, M. H., Griffin, K. J., Stout, C. D. & Johnson, E. F. (2005). Nat. Struct. Mol. Biol.12, 822–823. [DOI] [PubMed]