Abstract
The general receptor for phosphoinositides isoform 1 (GRP1) is recruited to the plasma membrane in response to activation of phosphoinositide 3-kinases and accumulation of phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3]. GRP1's pleckstrin homology (PH) domain recognizes PtdIns(3,4,5)P3 with high specificity and affinity, however, the precise mechanism of its association with membranes remains unclear. Here, we detail the molecular basis of membrane anchoring by the GRP1 PH domain. Our data reveal a multivalent membrane docking involving PtdIns(3,4,5)P3 binding, regulated by pH and facilitated by electrostatic interactions with other anionic lipids. The specific recognition of PtdIns(3,4,5)P3 triggers insertion of the GRP1 PH domain into membranes. An acidic environment enhances PtdIns(3,4,5)P3 binding and increases membrane penetration as demonstrated by NMR and monolayer surface tension and surface plasmon resonance experiments. The GRP1 PH domain displays a 28 nM affinity for POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine/PtdIns(3,4,5)P3 vesicles at pH 6.0, but binds 22-fold weaker at pH 8.0. The pH sensitivity is attributed in part to the His355 residue, protonation of which is required for the robust interaction with PtdIns(3,4,5)P3 and significant membrane penetration, as illustrated by mutagenesis data. The binding affinity of the GRP1 PH domain for PtdIns(3,4,5)P3-containing vesicles is further amplified (by ∼6-fold) by nonspecific electrostatic interactions with phosphatidylserine/phosphatidylinositol. Together, our results provide new insight into the multivalent mechanism of the membrane targeting and regulation of the GRP1 PH domain.
Keywords: general receptor for phosphoinositides isoform 1; pleckstrin homology domain; phosphoinositide; phosphatidylinositol 3,4,5-trisphosphate
The signaling lipid phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3] is produced in plasma membranes in response to stimulation of cell surface receptors by growth factors and hormones (1). Class I phosphoinositide (PI) 3-kinases phosphorylate the inositol headgroup of the relatively abundant phosphatidylinositol 4,5-bisphosphate [Ptdns(4,5)P2], transiently elevating the level of PtdIns(3,4,5)P3 from undetectable to nearly 10% of the PtdIns(4,5)P2 level (2–4). The concentration of PtdIns(3,4,5)P3 is tightly regulated by the activity of PI 5- and 3-phosphatases, such as SHIP1/2 and PTEN, which dephosphorylate the inositol ring generating PtdIns(3,4)P2 and PtdIns(4,5)P2 (5, 6). Despite the transitory accumulation and low concentrations in the plasma membrane, PtdIns(3,4,5)P3 is implicated in fundamental biological processes including growth, proliferation, migration, and survival of cells (1, 7). The PtdIns(3,4,5)P3-mediated signals are primarily recognized and transduced by pleckstrin homology (PH) domain-containing proteins that bind strongly and in some cases specifically to PtdIns(3,4,5)P3. Mutations in the PH domains that disrupt or promote PtdIns(3,4,5)P3 binding cause various signaling disarrays leading to severe immune system disorders and cancers (8, 9). Among the best characterized PtdIns(3,4,5)P3-recognizing protein effectors are Bruton's tyrosine kinase, ADP-ribosylation factor (Arf)-nucleotide binding site opener (ARNO), Cytohesin-1, and general receptor for phosphoinositides isoform 1 (GRP1) (10–12). GRP1 is a member of the GTP/GDP exchange factors family that catalyzes guanine nucleotide exchange of Arf proteins essential for actin rearrangement, endo- and exocytosis, membrane budding, and trafficking. Whereas the enzymatic activity of GRP1 is attributed to the central Sec7 homology domain, its membrane anchoring relies exclusively on the carboxy-terminal PH domain (13).
The PH domains comprise one of the largest families of signaling modules and are the most thoroughly characterized among PI binding domains (14, 15). The PH domain was identified within a set of human proteins in 1993 and named after the two homologous regions of pleckstrin, the major protein kinase C substrate of platelets (16, 17). Since then, it has been found in several hundred eukaryotic proteins involved in intracellular signaling, membrane trafficking, cytoskeletal dynamics, and lipid modifications. The PH domain contains ∼120 residues that are folded in a highly conserved three-dimensional structure despite little sequence similarity between the family members. The fold consists of a seven-stranded β-barrel, capped by an amphipathic α helix at one edge, whereas the opposite edge is framed by three variable loops. The β1-β2, β3-β4, and β6-β7 loops form a PI binding pocket, and their length and primary sequence define the PH domain specificity. Like other PH modules, the GRP1 PH domain is electrostatically polarized and has a strong positive electrostatic potential surrounding the binding site, which may contribute to both the specific PtdIns(3,4,5)P3 binding and the weak nonspecific electrostatic interactions with other acidic lipids in membranes (14, 18, 19). Although the majority of PH domains exhibit broad selectivity and associate with several PIs, recruiting their host proteins to different intracellular membranes, PtdIns(4,5)P2 and PtdIns(3,4,5)P3 remain the most common targets for this family of lipid binding effectors.
The details of how the PH domain recognizes the pattern of sequential 3-, 4-, and 5-phosphate groups in PtdIns(3,4,5)P3 is provided by the crystal structure of the GRP1 PH domain in complex with inositol 1,3,4,5-tetrakisphosphate (IP4), an isolated headgroup of PtdIns(3,4,5)P3 (20, 21). The tetrakisphosphate molecule lies in the center of a deep, positively charged pocket formed by the β1-β2, β3-β4, and β6-β7 loops and the strands they connect. The distal phosphates are the most buried in the pocket, whereas the 1-phosphate group is positioned near the tips of the loops. A network of hydrogen bonds, formed between conserved lysine and arginine residues of β2, β3, β4, and β7 and all four phosphate groups of IP4, efficiently restrains the inositol molecule. A unique β-hairpin in the long β6-β7 loop of GRP1 is involved in additional hydrogen bonding contacts with the 5-phosphate group of IP4. These additional interactions with the 5-phosphate account for the high specificity of the GRP1 PH domain toward PtdIns(3,4,5)P3.
Despite the critical role in GRP1 function, the precise mechanism by which the PH domain associates with PtdIns(3,4,5)P3-enriched membranes remains unclear. Because PtdIns(3,4,5)P3 is present at a very low level even in stimulated cells, elucidation of the overall membrane targeting is essential for our understanding of how the GRP1 PH domain distinguishes PtdIns(3,4,5)P3 and selectively binds this relatively rare lipid but does not recognize other, more-abundant PIs. Here we present the molecular basis of membrane docking, penetration, and pH regulation of the GRP1 PH domain based on structural and quantitative analysis of its interactions with lipids and membrane-mimicking monolayers and bilayers. Our results, derived from lipid binding measurements using NMR, surface plasmon resonance (SPR), liposome binding assays, and monolayer surface tension combined with mutagenesis data, provide novel insights into the mechanism of membrane recruitment and anchoring by GRP1.
MATERIALS AND METHODS
Subcloning, expression, and purification of GRP1 PH domain
The DNA fragment encoding residues 261–385 of the PH domain of human GRP1 was cloned into the pRSET A vector (Invitrogen). The unlabeled, 15N-labeled, and 15N/13C-labeled proteins were expressed in Escherichia coli Rosetta in Luria-Bertani or minimal media supplemented with 15NH4Cl and 13C6-glucose (Cambridge Isotope). Bacteria were harvested by centrifugation after induction with isopropyl-1-thio-β-D-galactopyranoside (0.1 mM) and lysed by French Press. The 6× His-fusion proteins were purified on a Talon-resin column (Clontech Laboratories, Inc.). The His tag was cleaved with EKMax (Invitrogen). The proteins were further purified by ion exchange chromatography on a HiTrap SP HP column (Amersham) in Bis-Tris buffer, pH 6.5, and concentrated in Millipore concentrators.
PCR mutagenesis of GRP1 PH
Site-directed mutagenesis of the GRP1 PH domain was performed using a QuikChange kit (Stratagene). The sequence of the H355A construct was confirmed by DNA sequencing.
NMR spectroscopy and sequence-specific assignments
Multidimensional homo- and heteronuclear NMR spectra were recorded at 25°C on Varian INOVA 500, 600, and 800 MHz spectrometers. The amino acid spin system and sequential assignments were made using three-dimensional 15N-edited NOESY-heteronuclear single quantum coherence (HSQC) (22, 23), 15N-edited TOCSY-HSQC (24) and triple-resonance experiments, such as HNCACB (25), CBCA(CO)NH (26), HNCO (26), C(CO)NH (27), and H(CCO)NH (27).
NMR titrations of lipids
The 1H,15N HSQC spectra of 0.2 mM uniformly 15N-labeled PH domain were collected using 1,024 t1 increments of 2,048 data points, 96 number of increments, and spectral widths of 7,500 and 1,367 Hz in the 1H and 15N dimensions, respectively. Lipid binding was characterized by monitoring chemical shift changes in the 1H,15N HSQC spectra of the PH domain as C4-PtdIns(3,4,5)P3, C4-PtdIns(4,5)P2, C4-PtdIns(4)P (Echelon Biosciences, Inc.), and inositol hexakisphosphate (IP6) (Aldrich) were added stepwise up to 4 mM. Significant changes in the resonances were judged to be greater than the average plus one standard deviation.
Monolayer measurements
The penetration of the wild-type and mutant GRP1 PH domains into the phospholipid monolayer was investigated by measuring the change in surface pressure (π) of invariable surface area during addition of the proteins. The experiments were performed using a 1 ml circular Teflon trough and wire probe connected to a Kibron MicroTrough X (Kibron, Inc., Helsinki). A lipid monolayer containing various combinations of phospholipids was spread onto the subphase composed of either 10 mM KH2PO4/0.16 M KCl (pH 6.0), 10 mM HEPES/0.16 M KCl (pH 7.4), or 10 mM HEPES/0.16 M KCl (pH 8.0) until the desired initial surface pressure (π0) was reached. After stabilization of the signal (∼5 min), 10 μg of protein was injected into the subphase through a hole in the wall of the trough. The surface pressure change (Δπ) was monitored for 45 min. The Δπ value reached a maximum after 30 min in all experiments.
Liposome binding
The liposome binding assays were performed as described in (28). Briefly, solutions of phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS) (Avanti), and C16- PtdIns(3,4,5)P3 (Echelon Biosciences, Inc.) dissolved in CHCl3:MeOH:H2O (65:25:4) were mixed and dried down under vacuum. The lipids were resuspended in 50 mM MOPS, 150 mM KCl, 1 mM DTT, pH 7.0, and incubated at 65°C for 1 h. The liposomes were then frozen in liquid nitrogen and thawed at 37°C for three cycles. The liposome solution was passed through an Avanti extruder to make 1.0 μm liposomes. Liposomes were collected by centrifugation at 25,000 g for 20 min and resuspended to a final concentration of 4 mM total lipids in 100 μl of binding buffers, containing 150 mM NaCl, 1 mM DTT, and 50 mM either NaAc/HAc (pH 5.0), MES (pH 6.0), MOPS (pH 7.0), or Tris (pH 8.0). Liposomes were incubated with the 6× His-fusion GRP1 PH domain (100–250 μg/ml final protein concentration) for 30 min at room temperature and then collected again by centrifugation. The liposome pellets were resuspended in 100 μl of buffer and analyzed using SDS-PAGE and Coomassie brilliant blue staining.
SPR measurements
All SPR experiments were performed at 25°C as described previously (29). Equilibrium SPR measurements were carried out at a flow rate of 5 μl/min. After injecting 85 μl of GRP1 PH domain, the protein-lipid association and dissociation were measured for 1,020 s and 500 s, respectively. The sensorgrams were obtained using five or more different concentrations of the protein (within a 10-fold range of Kd) for each condition tested. The sensorgrams were corrected for refractive index change by subtracting the control surface response. A total of three data sets were generated to obtain a standard deviation. The maximal response (Req, saturation value) was determined from the plots and plotted versus protein concentrations (C). The Kd value was determined by a nonlinear least-squares analysis of the binding isotherm using the equation Req = Rmax/(1 + Kd/C) (30).
PtdIns(3,4,5)P3 beads pull-down
The 2–5 μg of 6× His-GRP1 PH domain was incubated with 100 μl of PtdIns(3,4,5)P3 beads or control beads (Echelon Biosciences, Inc.) suspended in 150 mM NaCl, 2 mM DTT, 0.5% NP40, and 50 mM either MES (pH 6.0), MOPS (pH 7.0), Tris (pH 8.0), or CHES (pH 9.0) buffers at room temperature for 1 h. The beads fraction and supernatant were separated by centrifugation and analyzed by SDS-PAGE.
RESULTS AND DISCUSSION
The PtdIns(3,4,5)P3 lipid and its isolated headgroup occupy similar binding sites of the GRP1 PH domain
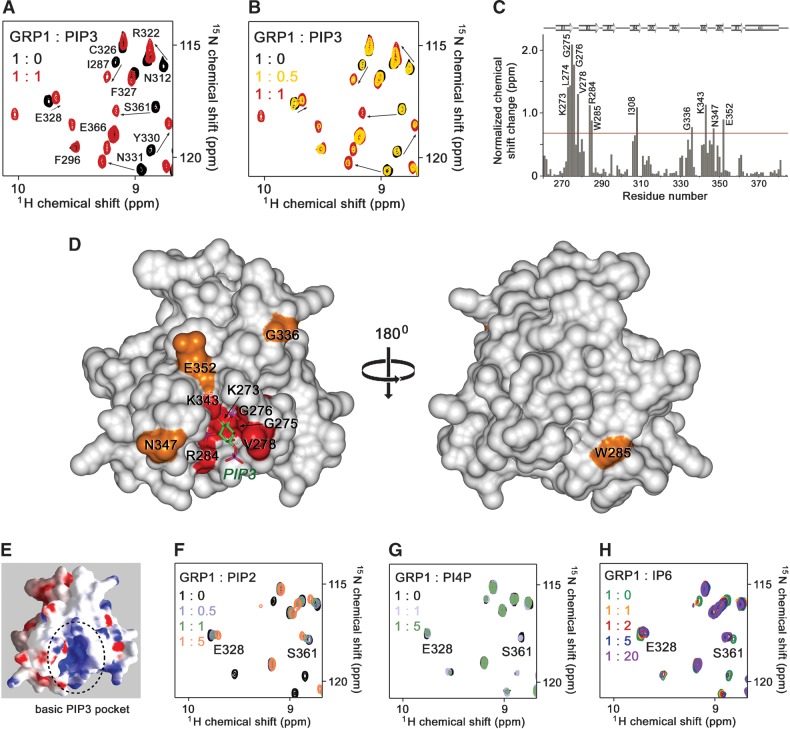
To establish the molecular basis of the lipid recognition, the interaction of the GRP1 PH domain with PtdIns(3,4,5)P3 was investigated by NMR. The 1H,15N HSQC spectra of uniformly 15N-labeled protein were collected while a soluble di-butanoyl (C4) form of PtdIns(3,4,5)P3 was gradually added into the NMR sample (Fig. 1A, B). As PtdIns(3,4,5)P3 was titrated in, the resonances of the ligand-free PH domain decreased in intensity and disappeared completely once the protein-lipid ratio had reached 1:1 (Fig. 1B). Concomitantly, another set of resonances corresponding to the lipid-bound state of the protein gradually appeared and replaced the initial set of resonances at equal molar concentrations of the PH domain and PtdIns(3,4,5)P3. This pattern of chemical shift changes is characteristic of slow exchange on the NMR time scale and indicates a robust interaction between the PH domain and PtdIns(3,4,5)P3. Almost identical resonance perturbations in the PH domain spectra were induced by IP4, an isolated headgroup of PtdIns(3,4,5)P3, suggesting that both molecules are bound in the same binding pocket, with the inositol ring involved in the most critical contacts. Only small differences were detected for three residues (K279 of the β1-β2 loop and T344 and A346 of the β6-β7 loop) (see supplementary Fig. I). These residues are located near the 1-phosphate group of IP4 and most likely are affected by the glycerol moiety, attached to this phosphate in the PtdIns(3,4,5)P3 lipid.
Fig. 1.
The phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3 or PIP3] binding site of the general receptor for phosphoinositides isoform 1 (GRP1) pleckstrin homology (PH) domain. A: The superimposed 1H,15N heteronuclear single quantum coherence (HSQC) spectra of the PtdIns(3,4,5)P3-bound (red) and the ligand-free (black) PH domain (0.2 mM). B: The 1H,15N HSQC spectra of the PH domain collected during titration with PtdIns(3,4,5)P3. The spectra are color-coded according to the concentration of the lipid. C: The histogram shows normalized (42) chemical shift changes induced in the backbone amides of the PH domain by addition of PtdIns(3,4,5)P3 at a 1:1 protein-to-lipid ratio. The horizontal line indicates the average change plus one standard deviation. The secondary structure is shown above the histogram. D: Residues that display significant chemical shift change in c are labeled on the GRP1 PH domain (Protein Data Bank ID: 1FGY) surface and colored in red and orange for large and medium changes, respectively. E: The electrostatic surface potential of the GRP1 PH domain, colored blue and red for positive and negative charges, respectively, reveals the basic pocket where the inositol headgroup is bound. F–H: The superimposed 1H,15N HSQC spectra of the 15N-labeled GRP1 PH domain collected during gradual addition of (F) PtdIns(4,5)P2 (or PIP2), (G) PtdIns(4)P (or PI4P), and (H) inositol hexakisphosphate. The spectra are color-coded according to the concentration of the ligands, as shown in the insets.
PtdIns(3,4,5)P3 binding triggers large resonance perturbations in the binding site of the GRP1 PH domain but does not induce global conformational changes
To identify the most-perturbed residues, the resonances of the lipid-bound and apo-states of the PH domain were assigned using a standard set of three-dimensional NMR experiments, and compared. The normalized 1H and 15N chemical shift differences are shown as a histogram in Fig. 1C. The most-pronounced resonance changes upon PtdIns(3,4,5)P3 binding were observed for the K273, L274, G275, G276, V278, R284, and W285 residues of the β1 and β2 strands and the β1-β2 loop, I308 of β4, and G336, K343, N347, and E352 residues of the extended β6-β7 loop (Fig. 1C). All of these residues either directly interact with PtdIns(3,4,5)P3, creating a dense net of hydrogen bonding contacts, or are located near the interacting residues, as seen in the crystal structure of the IP4-bound GRP1 PH domain (20, 21). The amino group of K273 forms hydrogen bonds with the 3- and 4-phosphate groups of the inositol ring. The backbone amides of G276 and V278, in concert with the side chain of K343, restrain the 5-phosphate moiety, whereas R284 is critical for coordinating the 3-phosphate group. In addition, several amino acids were conformationally perturbed, being in close proximity to the residues involved in the hydrogen bonding contacts. Thus, L274 and G275 of the β1-β2 loop are flanked by the key residues, K273 and G276. The W285 residue is adjacent to R284, and I308 and E352 are near the R305 and N354 residues, which donate hydrogen bonds to the 3-phosphate and 5-phosphate, respectively.
Mapping the chemical shift changes onto the GRP1 PH domain surface revealed a well-defined, deep pocket where PtdIns(3,4,5)P3 is bound (Fig. 1D). Whereas the outer walls of the pocket are formed by neutral or hydrophobic residues, the inner region displays a strong positive potential (Fig. 1E), implying that electrostatic and hydrogen-bonding interactions play a major role in restraining the inositol ring. A unique set of these contacts is responsible for the strong preference of the GRP1 PH domain for PtdIns(3,4,5)P3 over other negatively charged lipids. For example, PtdIns(4,5)P2 was bound in the same binding pocket, but this interaction was much weaker, judging by significant line broadening and/or gradual movement of NMR resonances, indicative of an intermediate-to-fast exchange (Fig. 1F). This is in agreement with the reported affinities of the GRP1 PH domain for PtdIns(3,4,5)P3 (50 nM) and PtdIns(4,5)P2 (∼104-fold lower) (18), and for the headgroups of these lipids, IP4 [27 nM [31]; 35 nM (12)] and IP3 [>4.5 μM (31); 6 μM (12)]. Furthermore, no chemical shift changes were observed upon titration of PtdIns(4)P, demonstrating that this most-abundant in plasma membrane monophosphorylated PI does not bind at all (Fig. 1G). On the other hand, an increase of the net negative charge of the headgroup did not enhance the binding. A 20-fold excess of IP6 caused only minor changes in the NMR spectrum of the GRP1 PH domain (Fig. 1H), confirming a weak binding in the micromolar range (31). Taken together, these results demonstrate that the GRP1 PH domain specifically recognizes the pattern of sequential phosphate groups in the inositol ring of PtdIns(3,4,5)P3. The lack of significant resonance perturbations in other regions of the PH domain suggests that the PtdIns(3,4,5)P3 binding induces local but not global conformational changes in the protein.
PtdIns(3,4,5)P3 binding facilitates the membrane insertion of the GRP1 PH domain
A number of PI binding modules, including FYVE, PX, and ENTH domains have been shown to penetrate membranes following the recognition of PIs (30, 32–34). The PI coordination partially reduces positive potential surrounding the binding pocket, facilitating insertion of the adjacent hydrophobic residues into the bilayer. In general, PH domains harbor fewer hydrophobic residues near the PI binding site, however, recent studies have demonstrated that the PH domains of PLCδ1 (35, 36) and DAPP1 (37) penetrate membranes significantly. Initially, the ability of the GRP1 PH domain to penetrate membranes was examined by monitoring changes in surface tension of lipid monolayers in a neutral (pH 7.4) buffer. Phospholipid monolayers at the air-water interface serve as a highly sensitive tool for measuring the membrane penetration by peripheral proteins (30, 32–34). A POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) (80:20) or POPC/POPE/PtdIns(3,4,5)P3 (77:20:3) monolayer of initial surface pressure (π0) was spread at constant area, and the change in surface pressure (Δπ) after the injection of the protein was monitored (Fig. 2A, open and filled circles). Δπ is inversely proportional to π0, and an extrapolation of Δπ0 versus π0 yields the critical surface pressure πc, which specifies an upper limit of π0 that a protein can penetrate into (38). As shown in Fig. 2A, the GRP1 PH domain has low intrinsic membrane-penetrating ability. The πc value of a POPC/POPE monolayer was found to be ∼22 dyne/cm. Thus, in the absence of PtdIns(3,4,5)P3, the GRP1 PH domain does not significantly insert into membranes. However, incorporation of 3 mol% PtdIns(3,4,5)P3 into the monolayer substantially enhanced penetration, raising the πc value to ∼28 dyne/cm.
Fig. 2.
Binding of the GRP1 PH domain to PtdIns(3,4,5)P3-containing monolayers and bilayers is pH dependent. A: Insertion of the GRP1 PH domain into a POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) (80:20) monolayer (open symbols) and a POPC/POPE/PtdIns(3,4,5)P3 (77:20:3) monolayer (filled symbols) at pH 6.0 (triangle), 7.4 (circle), and 8.0 (square) monitored as a function of π. B,C: The SDS-PAGE gel and the histogram show partitioning of the PH domain between the supernatant (s) and liposome pellet (p) at indicated pHs in the liposome binding assays. The experimental points were averaged over at least three measurements. D: Affinity of the GRP1 PH domain for PtdIns(3,4,5)P3-containing membranes increases in an acidic environment. Surface plasmon resonance measurements used to determine Kds of the PH domain association with POPC/POPE/PtdIns(3,4,5)P3 vesicles. The solid line represents a theoretical curve obtained based on Rmax and Kd values determined by nonlinear least-squares analysis. Error bars indicate ± SD.
Membrane insertion of the GRP1 PH domain is increased in the acidic pH
We next examined the extent of the membrane penetration of the GRP1 PH domain into PtdIns(3,4,5)P3-containing monolayers by varying the pH of the subphase buffer (Fig. 2B). When the pH was increased from 7.4 to 8.0, the monolayer surface pressure, πc, was reduced from 28 to 26 dyne/cm, indicating that basic conditions inhibit the membrane insertion of the GRP1 PH domain. Conversely, the πc value rose to 31 dyne/cm at pH 6.0, revealing that the membrane-penetrating ability is enhanced by an acidic environment. These results demonstrate that in low pH media, the GRP1 PH domain can easily penetrate physiological bilayers, because the surface pressure of cell membranes and large unilamellar vesicles is estimated to be in the range of 30–35 dyne/cm (30, 34, 38). In the absence of PtdIns(3,4,5)P3, the GRP1 PH domain did not insert, and the πc value of POPC/POPE monolayers remained unchanged for all pH conditions tested (∼22 dyne/cm). Thus, PtdIns(3,4,5)P3 is required for the strong anchoring and insertion of the protein into membranes. Likewise, PtdIns(3)P and PtdIns(4,5)P2 are necessary for the FYVE, PX, and ENTH domains to sufficiently penetrate phospholipid monolayers or micelles (30, 33, 34, 39).
Targeting of the GRP1 PH domain to PtdIns(3,4,5)P3-containing bilayers is pH dependent
To test whether the pH dependence is preserved for PtdIns(3,4,5)P3 embedded in bilayers, the protein-lipid interaction was investigated by liposome binding assays. The GRP1 PH domain was incubated with small unilamellar vesicles (SUVs) composed of PC, PE, and PtdIns(3,4,5)P3 at pH 5.0, 6.0, 7.0, or 8.0. Following centrifugation, the partitioning of the protein between the supernatant and pelleted fraction was examined. As shown in Fig. 2B, C, ∼95% of the GRP1 PH domain was retained in the pelleted liposome fraction at a low pH of 6.0. At each progressively higher pH value, the protein was increasingly redistributed to the supernatant. Densitometry analysis of the gel bands revealed that binding to PtdIns(3,4,5)P3-enriched vesicles was reduced from ∼95% to ∼25% as a result of alkalization of the buffer from pH 6.0 to pH 8.0 (Fig. 2C). In the absence of PtdIns(3,4,5)P3, the protein did not associate with SUVs, confirming that PtdIns(3,4,5)P3 is necessary for the membrane localization of GRP1.
Affinity of the GRP1 PH domain for PtdIns(3,4,5)P3-containing vesicles increases in an acidic environment
To quantitatively assess the effect of pH, equilibrium binding constants of the GRP1 PH domain's association with bilayers were measured by SPR (Fig. 2D and Table 1). Binding of the PH domain to POPC/POPE/PtdIns(3,4,5)P3 (78:20:2) vesicles was monitored at pH 6.0, 7.4, and 8.0. POPC/POPE (80:20) vesicles were used as a control, because it has been shown that the GRP1 PH domain is unable to localize with these vesicles even at high concentrations of protein. The obtained dissociation constants (Kds) demonstrate that the GRP1 PH domain associated with POPC/POPE/PtdIns(3,4,5)P3 vesicles, however, the binding affinity was significantly decreased when the pH of the buffer was raised from 6.0 to 8.0 (Table 1). At pH 6.0, the PH domain exhibited a 28 nM affinity for POPC/POPE/PtdIns(3,4,5)P3 vesicles, whereas this interaction was 22-fold weaker at pH 8.0 (Table 1). Thus, our data indicate that pH can mediate association of the GRP1 PH domain with PtdIns(3,4,5)P3-containing bilayers.
TABLE 1.
Lipid binding properties of the GRP1 PH domain and H355A mutant
| Protein | POPC/POPE/PtdIns(3,4,5)P3 (77:20:3) | POPC/POPE/POPS/PI/Cholesterol/PtdIns(3,4,5)P3 (9:35:22:9:22:3) |
|---|---|---|
| Kd (nM) | ||
| GRP1 wt pH 6.0 | 28 ± 2 | 4.8 ± 2 |
| H355A pH 6.0 | 120 ± 10 | 22 ± 5 |
| GRP1 wt pH 7.4 | 150 ± 10 | 28 ± 6 |
| H355A pH 7.4 | 480 ± 50 | 88 ± 9 |
| GRP1 wt pH 8.0 | 620 ± 40 | 110 ± 20 |
| H355A pH 8.0 | 970 ± 60 | 240 ± 30 |
POPE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine; PtdIns(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate; POPS, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine; PI, phosphatidylinositol; GRP1, general receptor for phosphoinositides isoform 1.
PtdIns(3,4,5)P3 binding of the GRP1 PH domain is pH sensitive
To examine whether the pH dependence is attributable to the direct interaction between GRP1 PH and PtdIns(3,4,5)P3, this binding was characterized in the absence of membrane mimetics by PtdIns(3,4,5)P3 pull-down assays and NMR spectroscopy. The His-tag fusion GRP1 PH domain was incubated with PtdIns(3,4,5)P3-immobilized beads at pH 6.0, 7.0, 8.0, and 9.0, and association with the lipid was examined by sedimentation and SDS-PAGE (Fig. 3A). The GRP1 PH domain showed a steady decrease in binding to PtdIns(3,4,5)P3 beads when pH was raised from 6.0 to 9.0, indicating that the PtdIns(3,4,5)P3 interaction is indeed pH sensitive and can be strengthened by the acidic media.
Fig. 3.
The direct interaction of the GRP1 PH domain with PtdIns(3,4,5)P3 is enhanced by lowering the pH. A: The SDS-PAGE gel displays the distribution of the His-tag fusion PH domain between the supernatant (s) and pelleted PtdIns(3,4,5)P3 beads or control beads (p) at indicated pHs. B: Superimposed 1H,15N HSQC spectra of the 15N-labeled GRP1 PH domain (0.2 mM) recorded in the absence (black) and presence of C4-PtdIns(3,4,5)P3 at pH 6.5 (green) and 8.0 (orange). C: His355 of the GRP1 PH domain mediates PtdIns(3,4,5)P3 binding. A schematic diagram showing the PtdIns(3,4,5)P3 headgroup coordination by the GRP1 PH domain (Protein Data Bank ID: 1FGY). Only charged residues of the proteins are depicted for clarity. D: Insertion of the wild-type GRP1 PH domain (filled symbols) and H355A mutant (open symbols) into a POPC/POPE (80:20) monolayer at pH 6.0 (diamond) and a POPC/POPE/PtdIns(3,4,5)P3 (77:20:3) monolayer at pH 6.0 (triangle), 7.4 (circle), and 8.0 (square) monitored as a function of π. E: Alignment of the PH domain sequences: absolutely, moderately, and weakly conserved residues are colored orange, green, and yellow, respectively. The secondary structure is shown above the sequences. The conserved His355 residue of the GRP1 PH domain is indicated by the orange circle.
The enhancement of PtdIns(3,4,5)P3 binding in a low pH environment was substantiated by NMR resonance perturbations observed in the PtdIns(3,4,5)P3-bound GRP1 PH domain upon varying the solution pH (Fig. 3B). The 1H,15N HSQC spectra of the PH domain were recorded in the absence (black) and presence (green) of PtdIns(3,4,5)P3 at a constant pH of 6.5. Binding of PtdIns(3,4,5)P3 caused large chemical shift changes in the protein (Fig. 3B). As the pH of the sample was increased to 8.0, the crosspeaks corresponding to the PtdIns(3,4,5)P3-bound PH domain (orange) shifted toward their positions in the ligand-free protein, thus indicating a weaker interaction under basic conditions. Consequently, the lipid-bound and -free states of the GRP1 PH domain appear to be stabilized by lowering and raising the pH, respectively. Taken together, our data demonstrate that the direct interaction of the GRP1 PH domain with PtdIns(3,4,5)P3 is pH dependent and becomes stronger in acidic conditions.
His355 of the GRP1 PH domain mediates PtdIns(3,4,5)P3 binding
The PtdIns(3,4,5)P3 binding pocket of the GRP1 PH domain contains the His355 residue, which forms a hydrogen bond with the 4-phosphate group of PtdIns(3,4,5)P3 (Fig. 3C). To determine whether His355 plays a role in the pH sensitivity, it was replaced with Ala, and binding of the H355A mutant to POPC/POPE/PtdIns(3,4,5)P3 vesicles was tested at different pH values. We found that the H355A mutant associates with vesicles ∼4-fold weaker than the wild-type protein at pH 6.0, 3-fold weaker at pH 7.4, and 1.6-fold weaker at pH 8.0 (Table 1). Overall, the binding affinity of H355A dropped by 8-fold when pH was changed from 6.0 to 8.0. In contrast, the wild-type GRP1 PH domain showed a 22-fold decrease in affinity. The H355A mutant did not entirely abolish this interaction, suggesting that lysine and arginine residues in the PtdIns(3,4,5)P3 binding site play critical roles in coordinating the lipid (Fig. 3C). Unlike the wild-type protein, the H355A mutant was unable to effectively insert into the POPC/POPE/PtdIns(3,4,5)P3 monolayer at any pH (Fig. 3D). In fact, it had lower penetrating power at pH 6.0, than did the wild-type GRP1 PH domain at pH 7.4. Thus, His355 is necessary for the strong membrane anchoring and insertion, and its protonation increases the binding affinity. These results also support the idea that membrane penetration by the GPR1 PH domain requires efficient PtdIns(3,4,5)P3 binding, which in turn can be promoted by acidic media.
Nonspecific electrostatic interactions with PS/PI enhance the GRP1 PH domain binding to PtdIns(3,4,5)P3-containing vesicles
Recent studies have shown that nonspecific electrostatic interactions with anionic phospholipids often stimulate membrane association of peripheral proteins. This can be particularly important in the case of the inner leaflet of the plasma membrane, where the anionic lipid concentration is high (40). To determine whether the interactions with acidic lipids other than PtdIns(3,4,5)P3 contribute to the membrane targeting by the GRP1 PH domain, we examined its association with plasma membrane mimetic liposomes containing PC, PE, PS, PI, cholesterol, and PtdIns(3,4,5)P3 (Table 1). The wild-type GRP1 PH domain bound the membrane-mimicking vesicles with a Kd of 4.8 nM at pH 6.0. Thus, the binding affinity increased nearly 6-fold for the vesicles containing anionic PS/PI/PtdIns(3,4,5)P3 lipids in comparison to the POPC/POPE/PtdIns(3,4,5)P3 vesicles. A comparable ∼6-fold amplification of the affinity was observed for the H355A mutant. This points to the significant contribution of the nonspecific electrostatic interactions to the membrane docking of the GRP1 PH domain and is in line with the recent report showing a 12-fold increase in the membrane affinity of GRP1 due to the presence of acidic lipids (18). Similarly, both the wild-type GRP1 PH domain and its H355A mutant exhibited an ∼5- to 6-fold increase in the affinity for the plasma membrane mimetic at pH 7.4 and pH 8.0, indicating comparable contributions of nonspecific electrostatic interactions at any pH. As in other PI binding modules (19, 30, 32, 41), a strong positive potential surrounding the PtdIns(3,4,5)P3 binding pocket of the GRP1 PH domain may facilitate the initial membrane association, reinforcing PtdIns(3,4,5)P3 binding and alleviating penetration.
The pH sensitivity of the GRP1 PH domain was maintained in the interaction with the plasma membrane-mimicking liposomes. The binding affinity was reduced by ∼23-fold upon alkalization of the buffer from pH 6.0 to pH 8.0 (Table 1). This decrease was similar to the 22-fold drop in binding affinity toward POPC/POPE/PtdIns(3,4,5)P3 vesicles, suggesting that the pH dependence is primarily due to PtdIns(3,4,5)P3 binding rather than to nonspecific electrostatic interactions with other acidic lipids.
In conclusion, our results reveal the multifaceted membrane docking by the GRP1 PH domain involving PtdIns(3,4,5)P3 recognition that is i) facilitated by stabilizing electrostatic interactions with other acidic lipids, ii) regulated by pH, and iii) accompanied by insertion into the membrane. The His355 residue in the PI binding site may undergo reversible protonation/deprotonation as cytosolic pH fluctuations occur (28). This could modulate the affinity of the GRP1 PH domain for transiently accumulated PtdIns(3,4,5)P3 in the plasma membrane. Future research is necessary to establish the physiological significance of pH sensing by the GRP1 PH domain. Alignment of the PH domain sequences shows conservation of His355 in ARNO, Cyto, GAP1, and SecG (Fig. 3E), suggesting a general regulatory mode for this subset of PH domains.
Supplementary Material
Acknowledgments
The authors thank C. Burd, M. Lemmon, and A. Sorkin for discussions; M. Lemmon for providing cDNA of the GRP1 PH domain; and S. Lee, O. Subach, and A. Ogorodnikova for helping with experiments.
Abbreviations
GRP1, general receptor for phosphoinositides isoform 1
HSQC, heteronuclear single quantum coherence
IP4, inositol 1,3,4,5-tetrakisphosphate
PH, pleckstrin homology
PI, phosphoinositide
POPE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
PS, phosphatidylserine
PtdIns(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate
PtdIns(4,5)P2, phosphatidylinositol 4,5-bisphosphate
SPR, surface plasmon resonance
Published, JLR Papers in Press, May 9, 2008.
Footnotes
This research was supported by an Indiana University Biomedical Research grant; American Cancer Society Grant IRG-84-002-22 (R.V.S.); and National Institutes of Health Grants GM-070358, GM-073913 (V.V.V.), and GM-071424 and CA-95144 (T.G.K.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
References
- 1.Vanhaesebroeck B., S. J. Leevers, K. Ahmadi, J. Timms, R. Katso, P. C. Driscoll, R. Woscholski, P. J. Parker, and M. D. Waterfield. 2001. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 70 535–602. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B., and M. D. Waterfield. 1999. Signaling by distinct classes of phosphoinositide 3-kinases. Exp. Cell Res. 253 239–254. [DOI] [PubMed] [Google Scholar]
- 3.Czech M. P. 2000. PIP2 and PIP3: complex roles at the cell surface. Cell. 100 603–606. [DOI] [PubMed] [Google Scholar]
- 4.Pendaries C., H. Tronchere, M. Plantavid, and B. Payrastre. 2003. Phosphoinositide signaling disorders in human diseases. FEBS Lett. 546 25–31. [DOI] [PubMed] [Google Scholar]
- 5.Sly L. M., M. J. Rauh, J. Kalesnikoff, T. Buchse, and G. Krystal. 2003. SHIP, SHIP2, and PTEN activities are regulated in vivo by modulation of their protein levels: SHIP is up-regulated in macrophages and mast cells by lipopolysaccharide. Exp. Hematol. 31 1170–1181. [DOI] [PubMed] [Google Scholar]
- 6.Maehama T., and J. E. Dixon. 1998. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273 13375–13378. [DOI] [PubMed] [Google Scholar]
- 7.Cantley L. C. 2002. The phosphoinositide 3-kinase pathway. Science. 296 1655–1657. [DOI] [PubMed] [Google Scholar]
- 8.Lindvall J. M., K. E. Blomberg, J. Valiaho, L. Vargas, J. E. Heinonen, A. Berglof, A. J. Mohamed, B. F. Nore, M. Vihinen, and C. I. Smith. 2005. Bruton's tyrosine kinase: cell biology, sequence conservation, mutation spectrum, siRNA modifications, and expression profiling. Immunol. Rev. 203 200–215. [DOI] [PubMed] [Google Scholar]
- 9.Carpten J. D., A. L. Faber, C. Horn, G. P. Donoho, S. L. Briggs, C. M. Robbins, G. Hostetter, S. Boguslawski, T. Y. Moses, S. Savage, et al. 2007. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 448 439–444. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda M., T. Kojima, H. Kabayama, and K. Mikoshiba. 1996. Mutation of the pleckstrin homology domain of Bruton's tyrosine kinase in immunodeficiency impaired inositol 1,3,4,5-tetrakisphosphate binding capacity. J. Biol. Chem. 271 30303–30306. [DOI] [PubMed] [Google Scholar]
- 11.Klarlund J. K., A. Guilherme, J. J. Holik, J. V. Virbasius, A. Chawla, and M. P. Czech. 1997. Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science. 275 1927–1930. [DOI] [PubMed] [Google Scholar]
- 12.Cronin T. C., J. P. DiNitto, M. P. Czech, and D. G. Lambright. 2004. Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains. EMBO J. 23 3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiNitto J. P., A. Delprato, M. T. Gabe Lee, T. C. Cronin, S. Huang, A. Guilherme, M. P. Czech, and D. G. Lambright. 2007. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol. Cell. 28 569–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinitto J. P., and D. G. Lambright. 2006. Membrane and juxtamembrane targeting by PH and PTB domains. Biochim. Biophys. Acta. 1761 850–867. [DOI] [PubMed] [Google Scholar]
- 15.Lemmon M. A., and K. M. Ferguson. 2000. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 350 1–18. [PMC free article] [PubMed] [Google Scholar]
- 16.Haslam R. J., H. B. Koide, and B. A. Hemmings. 1993. Pleckstrin domain homology. Nature. 363 309–310. [DOI] [PubMed] [Google Scholar]
- 17.Mayer B. J., R. Ren, K. L. Clark, and D. Baltimore. 1993. A putative modular domain present in diverse signaling proteins. Cell. 73 629–630. [DOI] [PubMed] [Google Scholar]
- 18.Corbin J. A., R. A. Dirkx, and J. J. Falke. 2004. GRP1 pleckstrin homology domain: activation parameters and novel search mechanism for rare target lipid. Biochemistry. 43 16161–16173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh S. M., and D. Murray. 2003. Molecular modeling of the membrane targeting of phospholipase C pleckstrin homology domains. Protein Sci. 12 1934–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferguson K. M., J. M. Kavran, V. G. Sankaran, E. Fournier, S. J. Isakoff, E. Y. Skolnik, and M. A. Lemmon. 2000. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell. 6 373–384. [DOI] [PubMed] [Google Scholar]
- 21.Lietzke S. E., S. Bose, T. Cronin, J. Klarlund, A. Chawla, M. P. Czech, and D. G. Lambright. 2000. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol. Cell. 6 385–394. [DOI] [PubMed] [Google Scholar]
- 22.Zuiderweg E. R., and S. W. Fesik. 1989. Heteronuclear three-dimensional NMR spectroscopy of the inflammatory protein C5a. Biochemistry. 28 2387–2391. [DOI] [PubMed] [Google Scholar]
- 23.Marion D., L. E. Kay, S. W. Sparks, D. A. Torchia, and A. Bax. 1989. Three-dimensional heteronuclear NMR of nitrogen-15 labeled proteins. J. Am. Chem. Soc. 111 1515–1517. [Google Scholar]
- 24.Marion D., P. C. Driscoll, L. E. Kay, P. T. Wingfield, A. Bax, A. M. Gronenborn, and G. M. Clore. 1989. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1 beta. Biochemistry. 28 6150–6156. [DOI] [PubMed] [Google Scholar]
- 25.Wittekind M., and L. Mueller. 1993. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha-carbon and beta-carbon resonances in proteins. J. Magn. Reson. 101 201–205. [Google Scholar]
- 26.Grzesiek S., and A. Bax. 1992. Improved 3D triple-resonance NMR techniques applied to a 31-kDa protein. J. Magn. Reson. 96 432–440. [Google Scholar]
- 27.Grzesiek S., J. Anglister, and A. Bax. 1993. Correlation of backbone amide and aliphatic side-chain resonances in C-13/N-15-enriched proteins by isotropic mixing of C-13 magnetization. J. Magn. Reson. 101 114–119. [Google Scholar]
- 28.Lee S. A., R. Eyeson, M. L. Cheever, J. Geng, V. V. Verkhusha, C. Burd, M. Overduin, and T. G. Kutateladze. 2005. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. USA. 102 13052–13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stahelin R. V., and W. Cho. 2001. Differential role of ionic, aliphatic, and aromatic residues in membrane-protein interactions: a surface plasmon resonance study on phospholipases A2. Biochemistry. 40 4672–4678. [DOI] [PubMed] [Google Scholar]
- 30.Stahelin R. V., F. Long, K. Diraviyam, K. S. Bruzik, D. Murray, and W. Cho. 2002. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J. Biol. Chem. 277 26379–26388. [DOI] [PubMed] [Google Scholar]
- 31.Kavran J. M., D. E. Klein, A. Lee, M. Falasca, S. J. Isakoff, E. Y. Skolnik, and M. A. Lemmon. 1998. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 273 30497–30508. [DOI] [PubMed] [Google Scholar]
- 32.Stahelin R. V., A. Burian, K. S. Bruzik, D. Murray, and W. Cho. 2003. Membrane binding mechanisms of the PX domains of NADPH oxidase p40phox and p47phox. J. Biol. Chem. 278 14469–14479. [DOI] [PubMed] [Google Scholar]
- 33.Stahelin R. V., F. Long, B. J. Peter, D. Murray, P. De Camilli, H. T. McMahon, and W. Cho. 2003. Contrasting membrane interaction mechanism of AP180 N-terminal homology (ANTH) and Epsin N-terminal homology (ENTH) domains. J. Biol. Chem. 278 28993–28999. [DOI] [PubMed] [Google Scholar]
- 34.Kutateladze T. G., D. G. S. Capelluto, C. G. Ferguson, M. L. Cheever, A. G. Kutateladze, G. D. Prestwich, and M. Overduin. 2004. Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 279 3050–3057. [DOI] [PubMed] [Google Scholar]
- 35.Flesch F. M., J. W. Yu, M. A. Lemmon, and K. N. Burger. 2005. Membrane activity of the phospholipase C-delta1 pleckstrin homology (PH) domain. Biochem. J. 389 435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuzi S., N. Uekama, M. Okada, S. Yamaguchi, H. Saito, and H. Yagisawa. 2003. Structure and dynamics of the phospholipase C-delta1 pleckstrin homology domain located at the lipid bilayer surface. J. Biol. Chem. 278 28019–28025. [DOI] [PubMed] [Google Scholar]
- 37.Manna D., A. Albanese, W. S. Park, and W. Cho. 2007. Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J. Biol. Chem. 282 32093–32105. [DOI] [PubMed] [Google Scholar]
- 38.Marsh D. 1996. Lateral pressure in membranes. Biochim. Biophys. Acta. 1286 183–223. [DOI] [PubMed] [Google Scholar]
- 39.Lee S. A., J. Kovacs, R. V. Stahelin, M. L. Cheever, M. Overduin, T. G. Setty, C. Burd, W. Cho, and T. G. Kutateladze. 2006. Molecular mechanism of membrane docking by the VAM7P PX domain. J. Biol. Chem. 281 37091–37101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hildebrand J., D. Marique, and J. Vanhouche. 1975. Lipid composition of plasma membranes from human leukemic lymphocytes. J. Lipid Res. 16 195–199. [PubMed] [Google Scholar]
- 41.Karathanassis D., R. V. Stahelin, J. Bravo, O. Perisic, C. M. Pacold, W. Cho, and R. L. Williams. 2002. Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 21 5057–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grzesiek S., S. J. Stahl, P. T. Wingfield, and A. Bax. 1996. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 35 10256–10261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.