Abstract
The frizzled gene family of putative Wnt receptors encodes proteins that have a seven-transmembrane-spanning motif characteristic of G protein-linked receptors, though no loss-of-function studies have demonstrated a requirement for G proteins for Frizzled signaling. We engineered a Frizzled-2 chimera responsive to β-adrenergic agonist by using the ligand-binding domains of the β2-adrenergic receptor. The expectation was that the chimera would be sensitive both to drug-mediated activation and blockade, thereby circumventing the problem of purifying soluble and active Wnt ligand to activate Frizzled. Expression of the chimera in zebrafish embryos demonstrated isoproterenol (ISO)-stimulated, propranolol-sensitive calcium transients, thereby confirming the β-adrenergic nature of Wnt signaling by the chimeric receptor. Because F9 embryonic teratocarcinoma cells form primitive endoderm after stable transfection of Frizzled-2 chimera and stimulation with ISO, they were subject to depletion of G protein subunits. ISO stimulation of endoderm formation of F9 stem cells expressing the chimeric receptor was blocked by pertussis toxin and by oligodeoxynucleotide antisense to Gαo, Gαt2, and Gβ2. Our results demonstrate the requirement of two pertussis toxin-sensitive G proteins, Gαo and Gαt, for signaling by the Frizzled-2 receptor.
Wnt genes comprise a class of 18 vertebrate genes encoding 350- to 380-aa secreted signaling proteins, which are likely to be involved in modulating diverse processes in developing embryos and in a subset of adult tissues (1–4). Recent data support the hypothesis that Wnts activate the function of members of the frizzled gene family of prospective receptors (5–10). In the absence of a Wnt signal, active glycogen synthase kinase 3 (GSK3, zeste white 3/shaggy in Drosophila) phosphorylates β-catenin at an amino-terminal site (11), targeting it for ubiquitination and degradation through a proteosome pathway that also involves axin and the product of the adenomatous polyposis coli (APC) gene (12–21). Signaling by Wnt-1 and functionally related Wnts through Frizzled homologues then activates the function of Dishevelled, which represses the activity of GSK3 (3, 22). Inhibition of GSK3 through Wnt signaling (22, 23) or when somatic mutations occur in APC (24, 25) results in elevation of β-catenin levels. This leads to accumulation of β-catenin in nuclei (11, 26), where it interacts with members of the lymphoid enhancer factor (LEF)/T cell factor (TCF) classes of architectural high mobility group (HMG) box transcription factors (27–30) to regulate expression of target genes (24, 25, 27, 31–35).
There is also growing evidence that some Wnts work through a pathway that is distinct from the one described above (2, 36–38). For example, Xwnt-5a, unlike Wnt-1 and Xwnt-8, does not induce a duplication of the axis in Xenopus when ectopically expressed, but instead causes morphogenetic defects (36, 37). This is not just a peculiarity of Xwnt-5a, because Xwnt-1, 8, 8b, and 3a are functionally equivalent in axis induction assays, whereas Xwnt-5a, 4, and 11 are functionally equivalent in this distinct Wnt-signaling activity (39, 40). This grouping of Wnts closely resembles the grouping determined by the McMahon lab (41), who assayed based on the ability of Wnts to transform mouse mammary epithelial cells (for review, see ref. 2). Further suggesting that not all Wnts work through the β-catenin pathway, Wnt7a regulates dorsoventral polarity in the chick limb in a manner distinct from the function of β-catenin (38).
Does signaling by Wnt-5a differ from signaling by Wnt-1 in early embryos? We recently have found that Wnt-5a, but not Wnt-8 (a Wnt-1-like Wnt), elevates intracellular calcium release in early embryos (42). This can be mimicked by expression of rat Frizzled-2, but not Frizzled-1 (43). Reciprocally, rat Frizzled-1 but not Frizzled-2 couples to the Wnt-1 pathway (6). Thus, some Wnts and Frizzled-1 activate Wnt-1-like signaling but do not elevate intracellular calcium, whereas some Wnts and Frizzled-2 that do not activate Wnt-1 signaling nevertheless elevate levels of intracellular calcium. We also have provided evidence that this release of calcium occurs in a G protein-coupled manner (43). Is this release of calcium truly consistent with the existence of multiple Wnt-signaling pathways? We have investigated the Wnt and Frizzled responsiveness of calcium-sensitive enzymes such as CaMKII (calcium/calmodulin-dependent protein kinase II) and PKC (protein kinase C). We find that both CaMKII and PKC are activated by Frizzled-2 but not Frizzled-1 in multiple assays (L. Scheldahl, M. Kuehl, and R.T.M., unpublished observations), and in a manner dependent on pertussis toxin-sensitive G proteins. These data therefore are consistent with a separate Frizzled-signaling pathway mediated by calcium and G proteins. However, to date, no specific G protein subunits have been shown to be required for any cellular response to Frizzled signaling, an issue we address in the current study. Using a novel chimeric receptor molecule created from the rat frizzled-2 and hamster β-adrenergic receptor (βAR), we establish that the pertussis toxin-sensitive G proteins Go and Gt are required for signaling by Frizzled-2.
Materials and Methods
F9 Cell Culture and Transfection Studies.
Mouse F9 teratocarcinoma cells were obtained from the American Type Culture Collection, propagated, and stably transfected (44, 45).
Construction of the Rat Frizzled-2 Receptor (Rfz-2)/β2-Adrenergic Receptor (β2AR) Chimera.
Primers synthesized by Operon were as follows: (i) 5′-ATTTAGGTGACACTATAGAACTC, (ii) 5′-CTCTGGGTAGCGGAATCGCTGCATGGCAATGGCAATGGCTGTGATGACC, (iii) 5′-ATGCAGCGATTCCGCTACCCAGAGCGCATAACCTCCTTGGCGTGTGCT, (iv) 5′-ATTGGCCTCGATGGCCGCGTGGCCCCACTTATCCACTG- CTATCACGCACAGGG, (v) 5′-GGCCACGCGGCCATCGAGGCCAATTCGCAGCGAATGGTCATCCTAATGGTGTGG, (vi) 5′-GCTCCGTCTTGGTGCCGTCGTGCTTCATGATGGTGCGGATCCTGGAATAGACAAAGACC, (vii) 5′-GGCACCAAGACGGAGCCGCTGGAGAGGCTCATGGTGCGTTTAGGCATCATCATGGGCA, (viii) 5′-CCACGAGTGCAGCGTCTTGCCCTGGAAGGCAATCCTGAAATCTGG, (ix) 5′-GGCAAGACGCTGCACTCGTGG, and (x) 5′-TTGGTCTAGACACTGTTCTTTATTTAAAAAAATAAAATAGG.
The construction of the Rfz-2/β2AR chimera is as follows. Step I: Using hamster β2-AR cDNA in the pSP70 (Promega) plasmid as a template, four PCR fragments were obtained (fragments 1, 2, 3, and 4) when oligos 1 and 2; 3 and 4; 5 and 6; and 7 and 8 were used as pairs for primers, respectively. Using Rfz-2 as a template, fragment 5 was obtained when oligos 9 and 10 were employed as primers. Step II: Using fragments 2 and 3 as a template, fragment “2/3” was obtained when oligos 3 and 6 were employed as primers. Similarly, using fragment 3 and 4 as a template, fragment “3/4” was obtained when oligos 5 and 8 were employed as primers. Step III: Using fragments 1 and 23 as a template, fragment “1/23” was obtained when oligos 1 and 6 were employed as primers. Using fragments 34 and 5 as a template, fragment “34/5” was obtained when oligos 5 and 10 were employed as primers. Step IV: Fragment “1/23” was digested with HindIII and BamHI and then inserted into the HindIII-BamHI site of pBluescript SK (Stratagene). Fragment “34/5” was digested with BamHI and XbaI and inserted into the BamHI-XbaI site of pBluescript SK. The “1/23” and “34/5” recombinants were digested with HindIII and BamHI and BamHI and XbaI, respectively, and the small fragments were inserted into the HindIII-XbaI site of pCDNA3 (Invitrogen).
Zebrafish Studies.
Zebrafish eggs were microinjected with a pressure injector (PLI90; Harvard Apparatus) with ≈8–10 pg of synthetic Rfz-2/β2AR RNA mixed with Fura-2-conjugated dextran (10,000 Mr; Molecular Probes) at the one-cell stage. Injected embryos were manually dechorionated with fine forceps and cultured in Egg Medium (42, 43). The chamber was slowly perfused with media containing either ISO (100 μM) or propranolol (100 μM). Calcium release was monitored as described (42).
Antisense Oligodeoxynucleotide Treatment.
The clones expressing the Rfz-2/β2AR chimera were propagated on 12-well plates (3,000 cells per well) and allowed to attach overnight. The clones were treated with phosphorothioate oligodeoxynucleotide (ODN; cell culture-grade, HPLC-purified ODNs from Operon Technologies, Alameda, CA) antisense to specific G protein subunits at least 48 hr in advance of challenge with ISO (44, 46). Antisense ODNs (28 nt, including initiator codon at 3′) were complexed with the N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP) liposomal carrier (1 μg of oligomers complexed with 1 μl of DOTAP carrier) and employed at final concentrations of ≈1 μM. Antisense oligomers were designed against the 5′ untranslated regions of the G protein subunits and include the ATG start codon.
β-Adrenergic Antagonist [125I]Cyanopindolol (CYP) Binding.
The expression of the chimera in stably transfected F9 cells was quantified by using the radiolabeled, high-affinity β-adrenergic antagonist [125I]CYP to intact cells (47). Both Chinese hamster ovary (CHO) cells and mouse F9 teratocarcinoma cells were stably transfected with empty vector pCDNA3 (EV) or expression vector harboring the cDNA for the Rfz-2/β2AR chimera (pCDNA3 Rfz-2/β2AR).
Immunoblotting.
Aliquots of crude membrane fractions (0.1 mg protein per lane) from each F9 clone were subjected to SDS/PAGE, the separated proteins were transferred to nitrocellulose, and blots were stained with a rabbit polyclonal antibody against the first extracellular loop of β2-AR (CM2) (48) or with antipeptide antibodies to the indicated G protein subunits (Signal Transduction Laboratories, San Diego). The immune complexes were made visible by staining with a second antibody (goat anti-rabbit IgG) coupled to calf alkaline phosphatase.
Plasminogen Activator Assay.
The activity of tissue plasminogen activator (tPA) is the hallmark of the primitive endoderm (PE) phenotype. Stem cells induced to PE produce and secrete tPA. tPA secretion was monitored by the amidolytic assay (49).
Indirect Immunofluorescence Studies.
F9 cells were resuspended and spread in eight-well chamber slides (1,000 cells per well). For the studies of clones expressing the Rfz-2/β2AR chimera, cells attached in day 1 and were treated with or without ISO (0.01–10 μM). For some studies, cells were treated with 10 μM ISO with and without the β-adrenergic antagonist, propranolol (10 μM). After 4 days, the cells were fixed by 3% paraformaldehyde for 5 min and stained with the PE-specific marker TROMA-1 mAb for 30 min and, later, a Texas Red-labeled, goat anti-mouse IgG second antibody for 30 min.
Results and Discussion
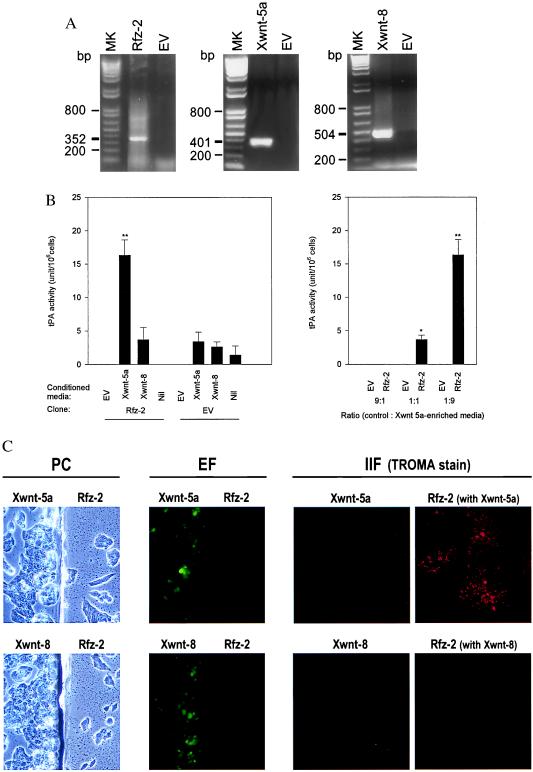
Because we have established F9 teratocarcinoma cells as a cultured cell system amenable to depletion of specific G proteins followed by analysis of differentiation phenotypes (44, 45, 50–52), we first needed to determine whether this was a suitable system for analysis of signaling by Frizzled homologues. F9 teratocarcinoma cells were stably transfected with Rfz-2 or the control vector, selected for use based on the expression of Rfz-2 mRNA (detected by reverse transcription–PCR, Fig. 1a), and then incubated with conditioned medium from other stable, transfectant F9 clones expressing either Xenopus Wnt5a (Xwnt-5a) or Wnt8 (Xwnt-8), also selected for use based on their expression of one Wnt mRNA (Fig. 1a). Application of the Xwnt 5a-containing medium to the Rfz-2-expressing cells induced differentiation of F9 cells to PE in a dose-dependent manner (Fig. 1b), as established by secretion of tPA, a hallmark for PE. In contrast, Rfz-2-expressing cells did not differentiate in response to conditioned media from cells transfected with either Xwnt-8 or the control vector, as evidenced by either tPA production (Fig. 1b) or positive staining by TROMA (Fig. 1c), a second hallmark of PE formation. Moreover, the formation of PE in response to Rfz-2 was abolished by prior treatment of the Rfz-2-expressing cells with pertussis toxin (not shown). These data are consistent with evidence that Rfz-2 acts with Xwnt-5a, but not Xwnt-8, to induce pertussis toxin-sensitive release of intracellular calcium (43). These data also establish the F9 cells as a viable read-out for signaling via Rfz-2.
Figure 1.
Conditioned medium from cells expressing Xwnt-5a, but not Xwnt-8, stimulates differentiation of mouse F9 teratocarcinoma cells expressing Rfz-2 to PE, as established by the expression of the PE marker, tPA. (A) F9 clones stably transfected with the vector expressing Rfz-2 receptor, Xwnt-5a, Xwnt-8, or empty vector (EV) were selected on the basis of expression of target mRNA, as measured by reverse transcription–PCR. The molecular markers (MK) indicate relative size in bp of the amplified product. Boldface tick marks identify expected product. (B) Conditioned medium was collected from F9 clones stably transfected with Xwnt-5a, Xwnt-8, or the empty vector and used to supplement 1:9 (ratio of conditioned medium of target Rfz-2-expressing clones to that of the Wnt-expressing clones) the medium of the F9 cells stably expressing either Rfz-2 or the empty vector (EV; Left). Nil denotes the clones to which no conditioned medium was added. The ratio of conditioned medium of the target Rfz-2-expressing clones to that of the Wnt-expressing clones was varied from 9:1 to 1:9, and the PE formation was followed by tPA activity (Right). The tPA activity is calculated as described (47). The results are presented as the mean values ± SEM of at least four separate experiments. *, P < 0.05 for difference from the mean. (C) Coculture of Rfz-2 receptor-expressing F9 clones with clones expressing green fluorescent protein (GFP) and either Xwnt-5a or Xwnt-8. Coverslips on which Rfz-2-expressing cells had been cultured were transferred to Petri dishes seeded with clones expressing a single Wnt ligand (phase contrast, PC). Epifluorescence (EF) analysis reveals the GFP/Wnt-producing clones; the Rfz-2-bearing clones are negative. Indirect immunofluorescence (IIF) analysis of coculture partners reveals positive TROMA staining (indicative of PE formation) only for cocultures of Rfz-2 cells with Xwnt-5a- but not Xwnt-8-producing clones.
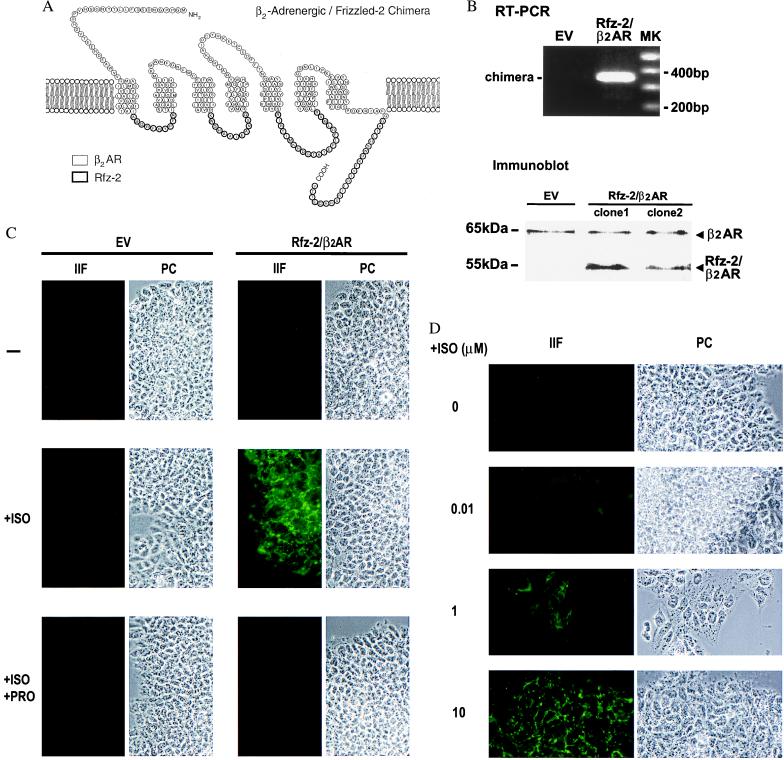
To facilitate the study of downstream-signaling events by Frizzled homologues, a chimera (Rfz-2/β2AR) was constructed by using the exofacial and transmembrane-spanning regions of the hamster β2-AR (48) with the predicted, cytoplasmic loops of the Rfz-2 gene product (Fig. 2a). This design was predicated on the hypothesis that Rfz-2 is a G protein-linked receptor (53, 54) in which its cytoplasmic signaling is defined by its cytoplasmic domains and not the putative ligand-binding and membrane-spanning domains. The prior creation of chimeric G protein-linked receptors between and within subfamilies provides ample precedence for this strategy (55–62), though these are the most distant relatives of this superfamily to be engineered as chimeras. After transfection of Rfz-2/β2AR into CHO and F9 cells, we monitored its expression by using radioiodinated β2-adrenergic antagonist CYP. As shown in Table 1, transfection of Rfz-2/β2AR elevated CYP binding in both cell types, though to a lesser extent in F9 cells, which have a full complement of endogenous βARs (47). Confirming expression of Rfz-2/β2AR, reverse transcription–PCR amplification of chimera mRNA and immunoblotting of crude membranes with antibody directed to an exofacial sequence of the β2AR in Rfz-2/β2AR revealed a unique, 55-kDa product (Fig. 2b). The next question to be addressed was whether Rfz-2/β2AR was functional.
Figure 2.
Expression of a chimera of the exofacial and transmembrane domains of the β2AR with the cytoplasmic domains of Rfz-2 gene products enables ISO-stimulated differentiation of F9 cells to PE. (A) Schematic representation of the Rfz-2/β2AR construct. (B) Reverse transcription–PCR and immunoblotting of Rfz-2/β2AR stably expressed in F9 cells. The RNA of F9 clones harboring either the empty expression vector (EV) or the vector expressing the Rfz-2/β2AR chimera was reverse-transcribed and amplified. The molecular markers (MK) indicate the relative size in bp of the amplified products. Note the identification of the 55-kDa chimeric receptor species in the immunoblots of clones 1 and 2 when stained with anti-β2AR antibodies. (C) The β-adrenergic agonist ISO (10 μM) stimulates whereas the β-adrenergic antagonist propranolol (10 μM) blocks differentiation to PE in F9 clones expressing the Rfz-2/β2AR chimera. Clones were treated with ligand for 4 days and then fixed and stained with TROMA-1 antibody to reveal PE formation. Indirect immunofluorescence (IIF) and phase-contrast (PC) images are displayed. (D) Dose response for ISO-stimulated differentiation to PE in F9 clones expressing the Rfz-2/β2AR chimera. Formation of PE was revealed by positive staining with the TROMA-1 antibody, shown by IIF. Based on multiple dose responses, the Kd for ISO-induced PE formation appears to be ≈2 μM in the F9 clones stably expressing the chimeric receptor.
Table 1.
Expression of the Rfz-2/β2AR chimera in Chinese hamster ovary (CHO-K) cells deficient in β-ARs and in F9 cells yields increased expression of β-AR binding, as measured by the binding of the radiolabeled, high-affinity β-adrenergic antagonist [125I]CYP
| Cell line | Vector | [125I]CYP binding, fmol/105 cells |
|---|---|---|
| CHO-K | EV | 0.6 ± 0.2 |
| Rfz-2/β2AR chimera | 18.0 ± 2.5 | |
| F9 | EV | 10.3 ± 0.9 |
| Rfz-2/β2AR chimera | 20.6 ± 4.4 |
Data are expressed as the mean values (±SEM) of at least four different clones and two independent experiments. EV, empty vector control.
We next investigated whether Rfz-2/β2AR elicited the same responses in F9 cells as we had observed with Rfz-2. Treating the Rfz-2/β2AR-expressing cells with the β2-adrenergic agonist ISO (10 μM) for 4 days induced differentiation to PE, demonstrated by strong, positive staining of the cells by the TROMA-1 antibody (Fig. 2c). The ability of ISO to induce PE could be blocked by the β-adrenergic antagonist propranolol (10 μM). Additionally, the formation of PE was found to be dose-dependent with respect to ISO (Fig. 2d). These results establish that Rfz-2/β2ARs induce PE in F9 cells in a β-agonist-stimulatable manner and reflect the normal signaling activity of wild-type Rfz-2. The data demonstrating that it is the Rfz-2 cytoplasmic domains of the chimera responsible for the signaling and not the β-adrenergic transmembrane and/or extracellular domains are as follows: F9 cells already display a complement of wild-type β2ARs; treatment of wild-type F9 cells with ISO, thereby elevating intracellular cyclic AMP, does not induce PE formation; stable transfection of F9 cells with wild-type βARs does not induce PE formation (not shown); and elevation of intracellular cyclic AMP by either cholera toxin or pertussis toxin, or the addition of dibutyryl cyclic AMP, does not provoke PE formation (47).
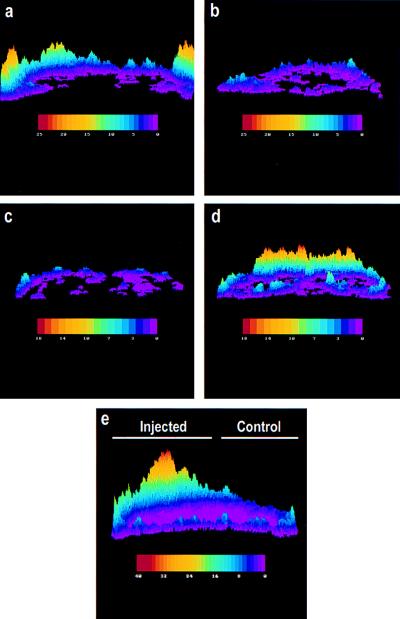
Because expression of Xwnt-5a (42) or Rfz-2 (43) in zebrafish embryos activates calcium transients, we next used this assay to ensure that the Rfz-2/β2AR functioned in a manner resembling that of wild-type Rfz-2. Microinjection of RNA encoding Rfz-2/β2AR into sibling zebrafish embryos led to the activation of calcium transients within 20 min after stimulation with ISO (Fig. 3a), but not with the β-adrenergic antagonist propranolol (Fig. 3b). Monitoring individual embryos injected with Rfz-2/β2AR RNA revealed that treatment with or without β-antagonist did not induce calcium transients (Fig. 3c), but switching the same embryo to agonist induced calcium transients within 20 min (Fig. 3d). As a control used in our previous studies (42, 43), we showed that injection of Rfz-2/β2AR RNA into only half the embryo resulted in inducible calcium transients only in the injected half (Fig. 3e). These data provide compelling evidence that the Rfz-2/β2AR chimera elicits an ISO-stimulated downstream-signaling response resembling that of Xwnt-5a and Rfz-2. The next question to be addressed was whether this signaling involved known signal transduction components.
Figure 3.
Expression of the Rfz-2/β2AR chimera in zebrafish embryos results in appearance of ISO-stimulated calcium transients. The composites of fura-2-dextran fluorescence ratio–image analysis represent the frequency of intracellular Ca2+ transients in embryos injected with RNA encoding Rfz-2/β2AR and then treated with drugs for 20 min before imaging. (a) A representative embryo perfused with a solution of β-adrenergic agonist (ISO, 100 μM) to stimulate the receptor. (b) A sibling representative embryo perfused with a solution of β-adrenergic antagonist (propranolol, 100 μM). (c and d) A representative embryo was treated with β-adrenergic antagonist (propranolol, 100 μM) for 20 min, imaged for 20 min (c), and then switched to agonist (ISO, 100 μM) for 20 min and imaged (d). (e) As a control, one-half of a zebrafish embryo was injected with Rfz-2/β2AR RNA in Texas Red dextran, and the embryo was challenged with ISO. Note that only the half of the embryo microinjected with Rfz-2/β2AR RNA (determined by Texas Red fluorescence; not shown) displays a calcium transient in response to challenge with ISO. Data were collected during the time of mesoderm induction between the 32- and 64-cell stages and the 1,000-cell stage. The pseudocolor ratio images represented in this set are a compilation of a 20-min time course in which the color bar represents the number of transients: red denotes high numbers, violet represents lower numbers, and the peaks represent more active regions.
The downstream factors required for signaling by Rfz-2 remain largely a mystery. We first used transfected CHO cells to test whether Rfz-2/β2AR stimulated adenylylcyclase activity in response to the β-AR agonist ISO, an assay made possible by the lack of β-adrenergic response in nontransfected CHO cells. Although CHO cells expressing the chimera display CYP binding, they exhibit no adenylylcyclase response to ISO (not shown), suggesting that adenylylcyclase is not activated in response to Frizzled-2 signaling in these cells.
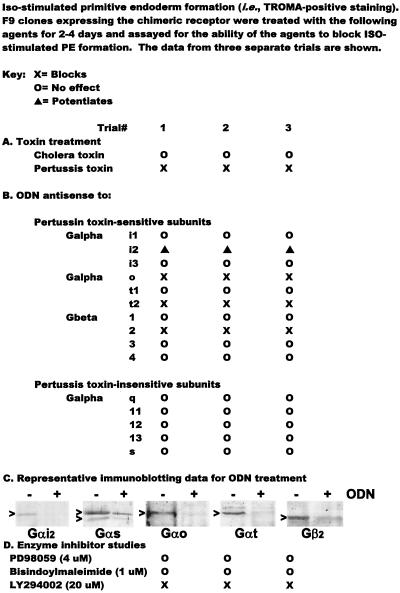
We next investigated whether pertussis toxin and other inhibitors of G protein signaling would block formation of PE in F9 cells in response to activating Rfz-2 signaling with Rfz-2/β2AR. Pertussis toxin treatment was tested in F9 clones expressing Rfz-2/β2AR and the clones then stimulated with ISO. The read-out in these experiments was TROMA staining (Fig. 4), a binary read-out that enables a cell-based assay for the expression of the TROMA antigen, a hallmark for PE formation. Pertussis intoxication abolished the ISO-stimulated formation of PE (Fig. 4), in agreement with the pertussis toxin sensitivity of induction of calcium transients by Rfz-2 (43).
Figure 4.
Analysis of downstream signaling of Rfz-2/β2AR chimera by using pertussis toxin treatment as well as treatment with ODN antisense to specific G protein subunits. Cells stably transfected with Rfz-2/β2AR chimera were treated either with ODN antisense to the specific G protein subunits indicated 48 hr in advance or with pertussis toxin 12 hr in advance of challenge with ISO (10 μM). For the control situation, only the clones expressing the Rfz-2/β2AR chimera and then treated with ISO differentiated to PE within 4 days. For pertussis toxin, the cells were intoxicated (10 ng toxin/ml) for 4 hr before the challenge with ISO. The PE phenotype was established by positive staining of the cells by TROMA antibody. The data are compiled from results of three separate trials for each ODN. The extent of subunit suppression in response to the treatment with specific ODNs was quantified from the immunoblots and ranged between 85% (Gαs) and 95% (Gαi2).
To ascertain the role of specific subunits of the heterotrimeric G proteins in mediating the cellular responses to Rfz-2/β2AR chimera, F9 clones stably expressing Rfz-2/β2AR were treated with phosphorothioate ODN antisense to specific G protein subunits, as employed previously to suppress expression of these subunits (44, 46, 51, 63, 64). ODN antisense to either Gαo or Gαt2 selectively blocked the ability of ISO to stimulate PE formation in cells expressing Rfz-2/β2AR (Fig. 4). As noted previously (51), suppression of Gαi2 by antisense RNA/DNA mimics the action of retinoic acid, leading to the PE phenotype. Clones treated with ODN antisense to Gαi1, Gαi3, and Gαt1, in contrast, displayed an ISO-induced PE formation (Fig. 4). Treatment with ODN antisense to Gα12, 13, s, and q resulted in no change in the ability of the clone expressing the chimeric receptor to differentiate to PE in response to ISO. Antisense ODNs to Gβ1, 3, and 4 also were without effect, whereas those to Gβ2 proved effective in the suppression of the ISO-induced PE response in the chimera-expressing cells. In all cases in which the antisense ODN blocks ISO-induced formation of PE in cells expressing Rfz-2/β2AR, analysis of the protein reveals that the expression of the targeted G protein subunit is reduced (Fig. 4c).
Inhibitors of phosphoinositide 3-kinase (PI3K), protein kinase C, mitogen-activated protein kinases ERK1 and 2, and cyclic nucleotide phosphodiesterases were tested in F9 clones expressing Rfz-2/β2AR and stimulated with ISO. The PD98059 inhibitor (4 μM) of mitogen-activated kinase kinase signaling to ERK1 and 2 displayed no effect, in spite of the observation that the ability of the morphogen retinoic acid to induced PE in F9 cells requires ERK1 and 2 (44). Likewise, treatment of the chimera-expressing clones with bisindolylmaleimide (1 μM), a selective protein kinase C inhibitor, displayed normal, ISO-stimulated PE. In contrast, inhibition of PI3K activity with the LY294002 compound (20 μM) abolished the ISO-stimulated PE response. These data implicate PI3K activity as participating in cellular responses to Rfz-2. Taken together, the inhibitor and antisense oligodeoxynucleotide data (Fig. 4) provide new insights into signaling components required for cellular responsiveness to Rfz-2.
Pertussis toxin effectively blocks ISO-stimulated differentiation of the F9 teratocarcinoma cells, directly implicating substrate G protein α-subunits for toxin-catalyzed ADP ribosylation and inactivation, such as members of the Gi family. However, the effects of pertussis toxin also could involve Gβ/γ subunits that also would be attenuated in toxin-treated cells, as observed previously in the studies of Wnt signaling in zebrafish embryos (43). Because of this uncertainty with the use of drugs to investigate signaling by Frizzled homologues, we turned to specific depletion of G protein subunits by antisense ODNs. The results of these studies indicate that signaling through Frizzled-2 in F9 cells to form PE requires Gαo and Gαt2, as well as Gβ2. Both Gαo and Gαt2 are substrates for pertussis toxin-catalyzed ADP ribosylation, and, hence, the drug studies and antisense oligonucleotide studies are in good agreement. Our data support the hypothesis that PI3K activity and three specific G protein subunits are required for Frizzled 2 to induce PE formation in F9 cells. Our studies do establish a novel in vitro system with which to address this issue, as well as the separate question of whether distinct Frizzled homologues signal through separate pathways. Finally, the construction of a novel chimeric Frizzled receptor that is amenable to soluble drug-induced activation and repression should enable further analysis of Frizzled signaling and cellular responses within minutes of receptor activation. The historic problem of lacking purified, soluble Wnt ligand thus has been circumvented, and the door is open for analysis of ligand-induced Frizzled signaling.
Abbreviations
- β2AR
β2-adrenergic receptor
- ISO
isoproterenol
- ODN
oligodeoxynucleotide
- CYP
cyanopindolol
- tPA
tissue plasminogen activator
- PE
primitive endoderm
- CHO
Chinese hamster ovary
- PI3K
phosphoinositide 3-kinase
References
- 1.Cox R T, Peifer M. Curr Biol. 1998;8:R140–R144. doi: 10.1016/s0960-9822(98)70081-8. [DOI] [PubMed] [Google Scholar]
- 2.Moon R T, Brown J D, Torres M. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 3.Wodarz A, Nusse R. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 4.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 5.Bhanot P, Brink M, Samos C H, Hsieh J C, Wang Y, Macke J P, Andrew D, Nathans J, Nusse R. Nature (London) 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 6.Yang-Snyder J, Miller J R, Brown J D, Lai C J, Moon R T. Curr Biol. 1996;6:1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- 7.He X, Saint-Jeannet J P, Wang Y, Nathans J, Dawid I, Varmus H. Science. 1997;275:1652–1654. doi: 10.1126/science.275.5306.1652. [DOI] [PubMed] [Google Scholar]
- 8.Bhat K M. Cell. 1998;95:1027–1036. doi: 10.1016/s0092-8674(00)81726-2. [DOI] [PubMed] [Google Scholar]
- 9.Kennerdell J R, Carthew R W. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 10.Muller H A, Samanta R, Wieschaus E. Development. 1999;126:577–586. doi: 10.1242/dev.126.3.577. [DOI] [PubMed] [Google Scholar]
- 11.Yost C, Torres M, Miller J R, Huang E, Kimelman D, Moon R T. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 12.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orford K, Crockett C, Jensen J P, Weissman A M, Byers S W. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 14.Maniatis T. Genes Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 15.Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek T J, Perry W L, 3rd, Lee J J, Tilghman S M, Gumbiner B M, Costantini F. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 16.Hamada F, Tonoyasu Y, Takatsu Y, Nakamura M, Nagai S, Suzuki A, Fujita F, Shibuya H, Toyoshima K, Ueno N, Akiyama T. Science. 1999;283:173–174. doi: 10.1126/science.283.5408.1739. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakanaka C, Weiss J B, Williams L T. Proc Natl Acad Sci USA. 1998;95:3020–3023. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behrens J, Jerchow B A, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 20.Hart M J, de los S, Albert I N, Rubinfeld B, Polakis P. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto H, Kishida S, Uochi T, Ikeda S, Koyama S, Asashima M, Kikuchi A. Mol Cell Biol. 1998;18:2867–2875. doi: 10.1128/mcb.18.5.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papkoff J, Aikawa M. Biochem Biophys Res Commun. 1998;247:851–858. doi: 10.1006/bbrc.1998.8888. [DOI] [PubMed] [Google Scholar]
- 23.Larabell C A, Torres M, Rowning B A, Yost C, Miller J R, Wu M, Kimelman D, Moon R T. J Cell Biol. 1997;136:1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 25.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 26.Fagotto F, Gluck U, Gumbiner B M. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- 27.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 28.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 29.Clevers H, van de Wetering M. Trends Genet. 1997;13:485–489. doi: 10.1016/s0168-9525(97)01305-x. [DOI] [PubMed] [Google Scholar]
- 30.Eastman Q, Grosschedl R. Curr Opin Cell Biol. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 31.Cavallo R A, Cox R T, Moline M M, Roose J, Polevoy G A, Clevers H, Peifer M, Bejsovec A. Nature (London) 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 32.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes, Moerer P, van de Wetering M, Destree O, Clevers H. Nature (London) 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 33.Brannon M, Gomperts M, Sumoy L, Moon R T, Kimelman D. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKendry R, Hsu S C, Harland R M, Grosschedl R. Dev Biol. 1997;192:420–431. doi: 10.1006/dbio.1997.8797. [DOI] [PubMed] [Google Scholar]
- 35.Young C S, Kitamura M, Hardy S, Kitajewski J. Mol Cell Biol. 1998;18:2474–2485. doi: 10.1128/mcb.18.5.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moon R T. BioEssays. 1993;15:91–97. doi: 10.1002/bies.950150204. [DOI] [PubMed] [Google Scholar]
- 37.Torres M A, Yang-Snyder J A, Purcell S M, DeMarais A A, McGrew L L, Moon R T. J Cell Biol. 1996;133:1123–1137. doi: 10.1083/jcb.133.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kengaku M, Capdevila J, Rodriguez-Esteban C, De La P, Johnson R L, Belmonte J C I, Tabin C J. Science. 1998;280:1274–1277. doi: 10.1126/science.280.5367.1274. [DOI] [PubMed] [Google Scholar]
- 39.Cui Y, Brown J D, Moon R T, Christian J L. Development. 1995;121:2177–2186. doi: 10.1242/dev.121.7.2177. [DOI] [PubMed] [Google Scholar]
- 40.Du S J, Purcell S M, Christian J L, McGrew L L, Moon R T. Mol Cell Biol. 1995;15:2625–2634. doi: 10.1128/mcb.15.5.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMahon A P. Trends Genet. 1992;8:236–242. [Google Scholar]
- 42.Slusarski D C, Yang-Snyder J, Busa W B, Moon R T. Dev Biol. 1997;182:114–120. doi: 10.1006/dbio.1996.8463. [DOI] [PubMed] [Google Scholar]
- 43.Slusarski D C, Corces V G, Moon R T. Nature (London) 1997;390:410–413. doi: 10.1038/37138. [DOI] [PubMed] [Google Scholar]
- 44.Gao P, Malbon C C. J Biol Chem. 1996;271:30692–30698. doi: 10.1074/jbc.271.48.30692. [DOI] [PubMed] [Google Scholar]
- 45.Gao P, Malbon C C. J Biol Chem. 1996;271:9002–9008. doi: 10.1074/jbc.271.15.9002. [DOI] [PubMed] [Google Scholar]
- 46.Wang H Y, Watkins D C, Malbon C C. Nature (London) 1992;358:334–337. doi: 10.1038/358334a0. [DOI] [PubMed] [Google Scholar]
- 47.Galvin-Parton P A, Watkins D C, Malbon C C. J Biol Chem. 1990;265:17771–17774. [PubMed] [Google Scholar]
- 48.Wang H, Lipfert L, Malbon C C, Bahouth S. J Biol Chem. 1989;264:14424–14431. [PubMed] [Google Scholar]
- 49.Andrade-Gordon P, Strickland S. Biochemistry. 1986;25:4033–4040. doi: 10.1021/bi00362a007. [DOI] [PubMed] [Google Scholar]
- 50.Bahouth S W, Park E A, Beauchamp M, Cui X, Malbon C C. Receptors Signal Transduction. 1996;6:141–149. [PubMed] [Google Scholar]
- 51.Watkins D C, Johnson G L, Malbon C C. Science. 1992;258:1373–1375. doi: 10.1126/science.1455234. [DOI] [PubMed] [Google Scholar]
- 52.Watkins D C, Moxham C M, Morris A J, Malbon C C. Biochem J. 1994;299:593–596. doi: 10.1042/bj2990593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Macke J P, Abella B S, Andreasson K, Worley P, Gilbert D J, Copeland N G, Jenkins N A, Nathans J. J Biol Chem. 1996;271:4468–4476. doi: 10.1074/jbc.271.8.4468. [DOI] [PubMed] [Google Scholar]
- 54.Vinson C R, Conover S, Adler P N. Nature (London) 1989;338:263–264. doi: 10.1038/338263a0. [DOI] [PubMed] [Google Scholar]
- 55.Wong S K, Parker E M, Ross E M. J Biol Chem. 1990;265:6219–6224. [PubMed] [Google Scholar]
- 56.Wong S K, Ross E M. J Biol Chem. 1994;269:18968–18976. [PubMed] [Google Scholar]
- 57.Kobilka B K, Kobilka T S, Daniel K, Regan J W, Caron M G, Lefkowitz R J. Science. 1988;240:1310–1316. doi: 10.1126/science.2836950. [DOI] [PubMed] [Google Scholar]
- 58.Olah M E. J Biol Chem. 1997;272:337–344. doi: 10.1074/jbc.272.1.337. [DOI] [PubMed] [Google Scholar]
- 59.Olah M E, Stiles G L. Methods Mol Biol. 1997;83:25–43. doi: 10.1385/0-89603-495-X:25. [DOI] [PubMed] [Google Scholar]
- 60.Verrall S, Ishii M, Chen M, Wang L, Tram T, Coughlin S R. J Biol Chem. 1997;272:6898–6902. doi: 10.1074/jbc.272.11.6898. [DOI] [PubMed] [Google Scholar]
- 61.Kozell L B, Machida C A, Neve R L, Neve K A. J Biol Chem. 1994;269:30299–30306. [PubMed] [Google Scholar]
- 62.Stroop S D, Kuestner R E, Serwold T F, Chen L, Moore E E. Biochemistry. 1995;34:1050–1057. doi: 10.1021/bi00003a040. [DOI] [PubMed] [Google Scholar]
- 63.Kleuss C, Hescheler J, Ewel C, Rosenthal W, Schultz G, Wittig B. Nature (London) 1991;353:43–48. doi: 10.1038/353043a0. [DOI] [PubMed] [Google Scholar]
- 64.Kleuss C, Schultz G, Wittig B. Methods Enzymol. 1994;237:345–355. doi: 10.1016/s0076-6879(94)37074-5. [DOI] [PubMed] [Google Scholar]