Abstract
The end of a telomeric DNA sequence isolated from a polytene chromosome of a hypotrichous ciliate folds back and hybridizes with downstream telomeric sequence to form a t loop that is stable in the absence of protein and DNA cross-linking. The single-stranded, telomeric DNA sequence at the end of a macronuclear molecule does not form a t loop but, instead, is complexed with a heterodimeric, telomere-binding protein. Thus, two mechanisms for capping the ends of DNA molecules are used in the same cell.
The macronuclear genomes of the hypotrichous ciliated protozoans consist of short, gene-size DNA molecules (1) containing telomeric repeats of 36 nt of 3′-G4T4-5′ sequence with the terminal 16 nt forming a 3′ single-stranded overhang (2). The telomeres of the micronuclear chromosomes of these protozoans, on the other hand, contain 5′-C4A4-3′/3′-G4T4-5′ duplex repeats ranging from 3 kbp to 20 kbp (3, 4); whether there is a 3′ single-stranded overhang in the micronuclear telomeres remains unknown. In chromosomes of mammalian cells (HeLa, normal human, and mouse; refs. 5–7) the telomeric (5′-TTAGGG-3′) repeats form long sequences (23–25 kbp mean) with single-stranded, 3′ overhangs. The function of the telomeres is to protect the chromosome ends from recombination, fusion, and from being recognized as broken ends (8). In hypotrich macronuclear DNA, this is accomplished by a heterodimeric, telomere-binding protein, which binds to the 3′ GC-rich telomere overhang and thus protects it from degradation and inappropriate interactions (9, 10). A counterpart of such a protein has not been found in mammalian cells (for review, see ref. 11) but certain telomere-specific duplex DNA-binding proteins have been suggested to play a similar role (12–16). Recently, a radically different model was proposed to account for the protection of mammalian telomeric termini (5). A structure designated as the “t loop” has been observed in the telomeric repeats at the ends of mammalian chromosomes. In the model, derived from electron microscopic observations of mouse and human telomeric DNA cross-linked with psoralen, the 3′ G-rich, single-stranded overhang folds back and invades the double-stranded region of the telomeric DNA, displacing 75–200 nt of the G-rich strand to form a duplex. This folding-back of the 3′ overhang creates a loop of duplex DNA whose size apparently depends on the size of the telomeric sequence.
Materials and Methods
Oxytricha fallax was allowed to conjugate, and the exconjugants in the polytene chromosome stage (45–50 hr postconjugation) were selected with a micropipette. The cells were lysed, the developing nuclei were isolated individually with a micropipette and lysed, and the DNA was prepared for electron microscopy as described in ref. 17. A DNA-cytochrome C film was picked up on parlodian-coated copper grids, shadowed with platinum-palladium alloy, and observed in a Philips 301 Electron Microscope operated at 80 kV. Magnifications were calibrated by using a grating replica, and double-stranded PM2 DNA was used as an internal molecular size standard (3.4 μm or 9.7 kbp). To observe polytene chromosomes, a portion of the lysate used to isolate DNA, but without protease K treatment, was adsorbed to a parlodian-coated electron microscope grid, stained with 1% uranyl acetate, and examined by electron microscopy.
Results
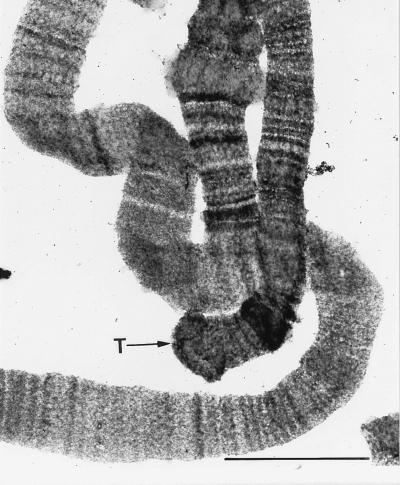
We have observed t loops in the high-molecular-weight duplex DNA isolated from the polytene chromosomes in the developing macronuclei of O. fallax by electron microscopy (Fig. 1). The presence of polytene chromosomes in nuclei that yielded t loops was affirmed as shown in Fig. 2. Telomeres of intact polytene chromosomes appear in the electron micrograph as compact knobs. Because polytene chromosomes represent amplified micronuclear DNA sequences, the t loops observed are presumed to be typical of micronuclear chromosomes.
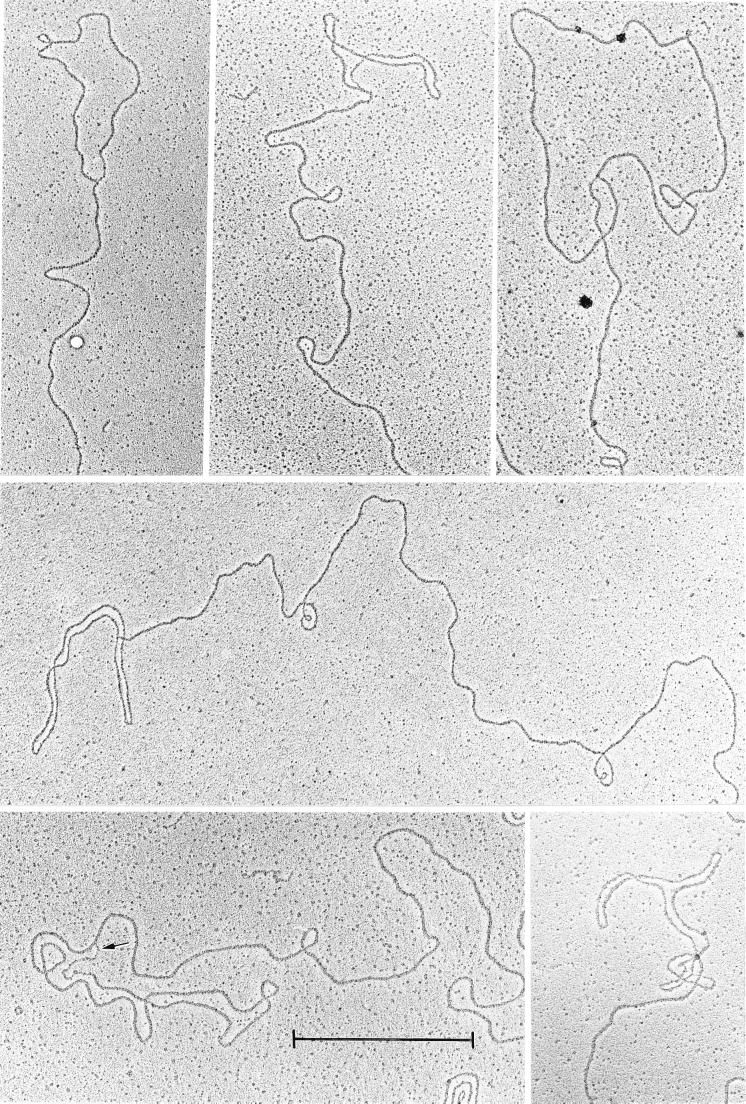
Figure 1.
Electron micrograph of a collection of duplex loops at the molecular ends of polytene chromosome DNA. The arrow points to a single-stranded stretch at the junction of a loop. (Bar = 1 μm.)
Figure 2.
Electron micrograph of polytene chromosomes from a developing macronucleus of O. fallax. T, telomere. (Bar = 0.1 μm.)
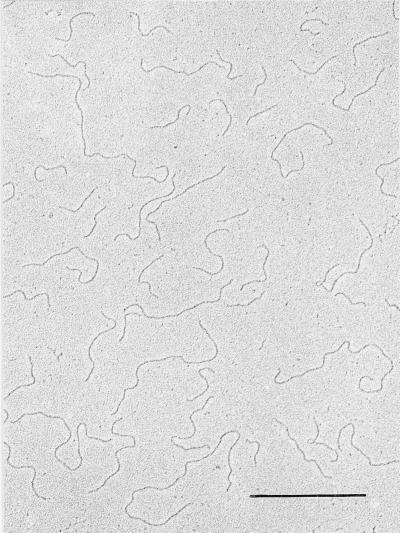
The method used here to extract polytene chromosome DNA is extremely rapid and gentle. The whole procedure from intact cells to DNA on the electron microscope grids consists of three steps and is accomplished within 5 min. The polytene chromosome DNA on the grid revealed many molecules measuring hundreds of micrometers in length with very few ends. A thorough search revealed only 38 free ends among hundreds of duplex strands. Eleven of these strands contained a duplex loop at the end (Fig. 1). Among the 11 molecules observed, the loops ranged in size from 1.6 to 3.5 μm or 4.6 to 10 kbp. These isolated t loops are stable in the absence of protein and of DNA cross-linking. A single-stranded region often is seen in one branch of the loop at its junction; this may represent a partially hybridized, single-stranded overhang (Fig. 1, arrow). Vegetative macronuclear DNA prepared under the same conditions yielded many small molecules with no terminal loops (Fig. 3). This demonstrates a uniqueness of hypotrichous ciliates in possessing two different mechanisms of telomeric protection within the same cell.
Figure 3.
Electron micrograph of DNA prepared from vegetative macronuclei of O. fallax. (Bar = 1 μm.)
Discussion
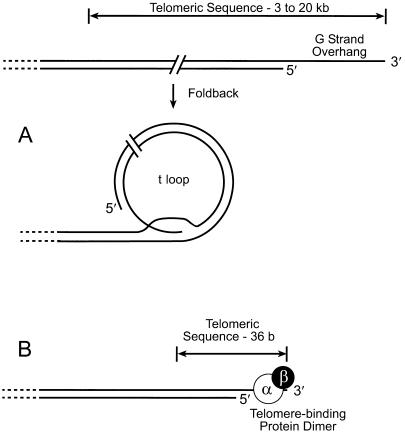
The duplex loops observed in the polytene chromosome DNA appear identical to t loops as defined by Griffith et al. (5). Their frequency of occurrence (∼29%) is too high to be explained by a random yet precise overlapping of ends with duplex regions. That these structures were not seen with other high-molecular-weight DNAs prepared by procedures (e.g., phenol extraction) that inevitably cause considerable DNA breakage, or with macronuclear DNA in the present study, further suggests that these loops are specific structures. Therefore, we assert that these t loops are typical of natural ends of micronuclear DNA. Micronuclear chromosomes are known to contain telomeric repeats (5′-C4A4-3′/3′-G4T4-5′) ranging from 3 to 20 kb. The size of the t loops observed is consistent with the size of the micronuclear telomeres. It is not known whether a 3′ single-stranded overhang exists in micronuclear telomeres, but the formation of t loops is consistent with their presence, and single-stranded regions are evident at the junction of most loops. Because polytene chromosomes are formed by multiple replications of micronuclear chromosomal DNA, it is highly likely that polytene chromosomes possess the same telomeric structure as micronuclear chromosomes, although the telomeric DNA could have become longer or shorter during the multiple replications of polytenization. The t loops observed here are stable in the absence of a cross-linking agent (5) or any protein, because proteinase K was used in preparing the DNA. However, the telomeric repeat-binding factor TRF2 greatly enhances the in vitro formation of t loops in mammalian DNA (5). The ends of the short, macronuclear DNA molecules of hypotrichs terminate with very short telomeric sequence, i.e., 20 bp of 5′-C4A4-3′/3′-G4T4-5′ and a 3′ tail of 16 nt of 3′-G4T4-5′. As pointed out by Griffith et al. (5), macronuclear telomeric DNA is too short to form t loops; the single-stranded tail conceivably could fold back into the duplex region of the telomere, but this is likely precluded by telomere-binding protein. We conclude that the two fundamentally different mechanisms of telomere-capping modeled in Fig. 4 occur in the same cell. In a micronucleus, chromosomal telomeres are capped with a t loop (Fig. 4A); in a macronucleus, the telomeric DNA of the millions of short DNA molecules is capped by the heterodimeric telomere-binding protein, forming a complex with the single-stranded, G-rich 3′ overhang (19).
Figure 4.
Models of two different mechanisms for capping telomeric repeats in the micronucleus and macronucleus. (A) Formation of a t loop by a fold-back of the telomeric DNA in a micronuclear chromosome. The single-stranded, G-rich 3′ overhang hybridizes with the C-rich chain in a proximal segment of telomere, displacing the original G-rich chain. Redrawn from Griffith et al. (5). (B) Formation of a terminal complex between heterodimeric, telomere-binding protein and the single-stranded, G-rich 3′ overhang of the telomere in a macronuclear DNA molecule. Redrawn from Froelich-Ammon et al. (18).
Acknowledgments
We thank Gayle Prescott and A. Murti for manuscript preparation and A. Murti for photographic help. This work was supported by the Cancer Center Support (CORE) Grant CA-21765 from the National Institutes of Health and American Lebanese Syrian-associated charities to K.G.M. and National Institute of General Medical Sciences Grant GM56161 to D.M.P.
References
- 1.Prescott D M. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klobutcher L A, Swanton M T, Donini P, Prescott D M. Proc Natl. Acad Sci USA. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jahn C L. Exp Cell Res. 1988;177:162–175. doi: 10.1016/0014-4827(88)90034-1. [DOI] [PubMed] [Google Scholar]
- 4.Dawson D, Herrick G. Cell. 1984;36:171–177. doi: 10.1016/0092-8674(84)90086-2. [DOI] [PubMed] [Google Scholar]
- 5.Griffith J D, Comeau L, Rosenfield S, Stansel R M, Bianchi A, Moss H, de Lange T. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 6.Kipling D, Cooke H J. Nature (London) 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 7.Zijlmans J M, Martens U M, Poon S S, Rapp A K, Tanke H J, Ward R K, Landsdor P M. Proc Natl Acad Sci USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greider C W. Cell. 1999;97:419–422. doi: 10.1016/s0092-8674(00)80750-3. [DOI] [PubMed] [Google Scholar]
- 9.Fang G, Cech T R. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 69–105. [Google Scholar]
- 10.Price C. Nature (London) 1999;397:213–214. doi: 10.1038/16598. [DOI] [PubMed] [Google Scholar]
- 11.deLange T. Semin Cell Biol. 1996;7:23–29. [Google Scholar]
- 12.Chong L, van Steensel B, Broccoli D, Erdjunment-Bromage H, Hamish J, Tempst P, de Lange T. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 13.Bilaud T, Brun C, Ancelin K, Koering C E, Laroche T, Gilson E. Nat Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 14.Broccoli D, Smorgorzewska A, Chong L, de Lange T. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 15.van Steensel B, Smogorzewska A, de Lange T. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 16.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 17.Murti K G, Prescott D M. Mol Cell Biol. 1983;3:1562–1566. doi: 10.1128/mcb.3.9.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Froelich-Ammon S J, Dickinson B A, Bevilacqua J M, Schultz S C, Cech T R. Genes Dev. 1998;12:1504–1514. doi: 10.1101/gad.12.10.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicke B J, Willis M D, Koch T H, Cech T R. Biochemistry. 1994;33:3364–3373. doi: 10.1021/bi00177a030. [DOI] [PubMed] [Google Scholar]