Abstract
The high level expression and purification of rat monoamine oxidase B (rMAOB) in the methylotrophic yeast Pichia pastoris is reported. Nearly 100 mg of purified rMAOB is obtained from 130 grams (wet weight) of cells (0.5 liter of culture). The MALDI-TOF mass spectrum of the purified protein shows a single species with a molecular mass of 59.228 ± 0.064 kDa, which agrees with the calculated molecular weight of 59.172 kDa for the rMAOB protein sequence assuming one mole of covalent FAD per mole of the enzyme. Consistent with the MALDI-MS data, purified rMAOB shows a single band near 60 kDa in Coomassie stained SDS/PAGE gel as well as on Western blot analyses performed using antiseras raised against human MAOA and BSA conjugated FAD. A partial amino acid sequence of the purified protein is confirmed to be that of the wild type rMAOB by in-gel trypsin digestion and MALDI-TOF-MS analyses of the liberated peptide fragments. Steady state kinetic data show that purified rMAOB exhibits a Km(amine) of 176 ± 15 µM and a kcat of 497 ± 83 min−1 for benzylamine oxidation, and a Km (O2) of 170 ± 10 µM. Kinetic parameters obtained for purified rMAOB are compared with those reported earlier for recombinant human liver MAOB expressed in Pichia pastoris.
Keywords: Monoamine Oxidase B, Rat Liver MAOB, Pichia pastoris
Monoamine oxidase B (MAOB)1 is an outer mitochondrial membrane bound flavoenzyme responsible for oxidative deamination and subsequent degradation of dopamine and dietary amines in our body [1]. These oxidation processes are important in maintaining physiological levels of dopamine and free dietary amines in our body and proper functioning of the central nervous system. Age related increases in the expression levels of MAOB in neural tissues [2, 3] are implicated in several neurological disorders like, Parkinson’s and Alzheimer’s diseases and selective MAOB inhibitors are clinically proven drug adjuvants for treating these disorders [4, 5].
Owing to their therapeutic applications, developments of MAOB inhibitors with greater clinical potency have gained momentum in recent years. Rat, being one of the most common animal models, is generally used for the screening of potential drug candidates. However, recent quantitative studies have shown differences in inhibitor specificities and properties between human and rat MAOBs despite their ~90% sequence identity [6]. Consistent with these studies, our recent spectroscopic study on human and rat MAOB have shown that the structural properties of their active site cavities differ significantly [7]. These observations therefore raise caution on the applicability of using rat as a model system for screening hMAOB selective inhibitors. Due to the lack of structural information on rMAOB and the lack of enzymological studies on the purified enzyme the reason for the observed differences between human and rat MAOBs are not well understood. Comparative structural and functional studies of purified human and rat MAOB preparations would facilitate interpretation of these observed differences.
One of the major requirements in performing structural and functional studies on any protein is to have a suitable expression system to obtain large quantities of purified protein. Although previous studies have been reported on expression of rMAOB in E. coli [8], and in S. cerevisiae [9, 10], the levels of functional proteins and the lack of details on the properties of the expressed enzyme from these systems provides an incentive to explore other systems for the high level expression and subsequent purification of rMAOB. The methylotrophic yeast Pichia pastoris has been used for heterologous expression of a number of proteins [11, 12] and has been used successfully in our previous work for large scale expression of human monoamine oxidases A and B [13, 14]. The availability of these expression systems provided levels of pure functional enzymes to permit subsequent structural studies [15, 16]. Inspired by this previous success on human MAOs, in this work we have used P. pastoris as the host for the over expression of rMAOB. The results presented here describe a convenient approach for obtaining large amounts of purified rMAOB. The basic functional properties of the purified enzyme with benzylamine as substrate have been determined and compared with the properties of the human enzyme.
Material and Methods
Materials
Expression vector pPIC3.5K, and P. pastoris KM71 strain were purchased from Invitrogen corp. (Carlsbad, CA). A rat liver cDNA library was a kind gift from Dr. Edward Morgan from Department of Pharmacology, Emory University, Atlanta, GA. All media and buffers (YPD, RDB, MGY, MM) for spheroplast transformation and for small scale expression in shake flasks were formulated following the Invitrogen Pichia expression manual using reagents purchased from Difco (Detroit, MI). Fermentation media and protocols are the same as reported earlier for hMAOB and hMAOA expression methods and are described in Invitrogen Pichia fermentation manual. The antibiotic G-418 sulfate (Geneticin®) was purchased from US Biological Inc. Swampscott, MA. Triton X-100, reduced Triton X-100, and Octyl-β-D-glucopyranoside (OGP) was purchased from Sigma. [α, α -2H2]-Benzylamine hydrochloride, used for kinetic isotope effect measurements in benzylamine oxidation by recombinant rMAOB, was synthesized by previous workers in this laboratory as described earlier [17]. Bio-Rad MacroPrep® High-Q resin, used for anion exchange column purification of rMAOB, was purchased from Bio Rad Laboratories (Hercules, CA). All other chemical and reagents used in this work were purchased from commercial sources in the highest purity available.
Cloning of rMAOB gene fron rat cDNA library
The rMAOB gene was cloned from the rat cDNA library by PCR amplification method using a forward primer (5′- GCGG^GATCCATGAGCAACAAAGCGAT-3′) containing a BamH I site (underlined) and a reverse primer (5′-CGCG^AATTCTCAGAAACGTACAAACAGACC-3′) with an EcoR I site (underlined). The amplified rMAOB gene with the incorporated restriction enzyme sites at the two ends was purified and digested with BamH I, EcoR I restriction enzymes for 3 hours at 37 °C. The pPIC3.5K vector was separately isolated and digested with the same restriction enzymes. The digested rMAOB gene was ligated with pPIC3.5K vector into the BamH I and EcoR I sites using T4 DNA ligase. The ligated rMAOB-pPIC3.5K construct was transformed into E. coli DH5α super competent cells and inoculated onto ampicillin containing agar plates. Colonies formed after overnight incubation at 37 °C, were inoculated in 5 ml of ampicillin containing liquid media and grown at 37 °C overnight. DNA was isolated from these cultures and sequenced with an ABI Prism automated DNA sequencer to check for correct insertion.
Transformation of rMAOB-pPIC3.5K construct in Pichia pastoris
The rMAOB-pPIC3.5K construct was linearized with Sac-I restriction enzyme for integration at the AOX1 locus in the Pichia genome. The linearized DNA was transformed into Pichia (KM71 strain) cells following protocols provided in the Invitrogen Pichia expression kit. Transformants were initially plated on media lacking histidine (RDB) to screen for His+ transformants, and then inoculated onto plates containing increasing amounts (0.25, 0.5, 0.75, and 1 mg/ml) of G-418 sulfate antibiotic to screen for multicopy integrants. The resistance to G-418 sulfate antibiotic is conferred by the kanamycin resistance gene in pPIC3.5K vector and therefore the higher resistance to G-418 sulfate would imply a higher copy number of the inserted rMAOB-pPIC3.5K construct in the Pichia genome.
Screening of expression levels of rMAOB transformants
After 3 to 4 days of incubation at 30 °C on the G-418 sulfate containing plates, single colonies from each plate were inoculated in 100 mL MGY media in 1 liter shake flaks and grown until OD600 reached between 2–6. The cells were harvested by centrifugation at 1500g for 10 minutes and rMAOB expression was induced by re-suspending the cell pellets in 20 ml MM (0.5% methanol) media. The induction continued for four days by supplementing with methanol after every 24 hours. Aliquots (1 ml) of each culture were removed every 24 hours and rMAOB expression levels measured by measuring benzylamine oxidase activity as reported earlier for hMAOB [13]. The best expression level was observed from a single colony obtained from 1.0mg/ml G-418 sulfate plate and was used for large scale fermentation following the protocols as described below.
Fermentation procedure
Fermentation of Pichia pastoris cells containing the gene encoding rMAOB was carried out with a Bioflow 300 fermentor (New Brunswick) equipped with temperature, pH and dissolved oxygen monitoring and regulating system. Autoclaved fermentation basal medium (composition per liter: 27 ml of 85% H3PO4, 0.93 gram of CaSO4, 18.2 gram of K2SO4, 14.9 gram of MgSO4, 7H2O, 4.13 gram of KOH, 40.0 gram of glycerol adjusted to pH 4.5 with 30% ammonium hydroxide) was supplemented with 12 ml/L of trace salts solution (Invitrogen) immediately before inoculation with 5–10% of the initial fermentation volume (~300–400 ml for 6L medium) of a Pichia culture grown in MGY medium (OD600 = 2– 6). Dissolved oxygen concentration in the growth media was maintained at 30 to 100% of air saturation either by adjusting agitation or by supplementing with an air-oxygen mixture through the automated regulator. When the wet cell weight reached 100 g/L, a 50% (v/v) glycerol water solution (supplemented with 12 mL/L trace salt) was fed to the media at an initial flow rate of 18 ml/L/hour. Glycerol feeding stopped when the cell density in the culture doubled (wet cell weight ca. 200 g/L). Induction of rMAOB expression was initiated by 100% methanol (supplemented with 12 mL/L trace salts solution) feeding. The flow rate of methanol feeding was adjusted slowly to a final flow rate of 3 ml/L/hour. At this point, aliquots of 1 ml of cell culture were taken out every 12 hours from the fermentor and assayed for rMAOB expression levels. A maximum expression level was achieved after 80 hours of methanol induction. The cells were harvested by centrifugation (20 minutes) at 1500g and washed once with Buffer A (50 mM sodium phosphate at pH 7.5 containing 5% glycerol, 1 mM PMSF, and 1 mM EDTA). The washed cell pellets were frozen in liquid nitrogen and stored at −80 °C until used for protein purification.
Isolation of intact mitochondria and outer mitochondrial membrane
Intact mitochondria and outer mitochondrial membranes from Pichia pastoris cells expressing recombinant rMAOB were isolated following a method reported previously by Daum et al. [18]. Briefly, the Pichia cell wall was first enzymatically disrupted with Zymolyase (1 mg/g wet cell weight; US Biological, Swampscott, MA) by spheroplasting method. The spheroplasts were then homogenized in a tight fitting Teflon-glass homogenizer and the homogenate was subjected to sequential centrifugations to isolate intact mitochondria [18]. The intact mitochondria were then disrupted by osmotic shock and sonicated to liberate the outer mitochondrial membrane (OMM) from the mitoplasts. OMM vesicles were separated from the mitoplasts on a 30 to 55% (w/v) sucrose density gradient at 20,000 rpm (Beckman SW-28 rotor) for 15 hours. The OMM formed a separate band near 40–45% (w/v) density of sucrose, which contained all of the MAO activity. This layer was diluted with three volumes of 10 mM Tris-HCl buffer, pH 7.4 and pelleted by a one-hour centrifugation at 35,000 rpm using a Beckman Ti-45 rotor. The OMM pellets were suspended in 50 mM potassium phosphate buffer at pH 7.4 containing 10% (v/v) glycerol and kept frozen in small aliquots at −80°C until used.
Purification of rMAOB
Purification of rMAOB was done following the hMAOB protocol [13, 19] with some minor modifications to improve the yield and purity of the enzyme. Approximately 130 gram (wet cell weight) of P. pastoris cells (ca. 0.5 liter of culture) were suspended in Buffer A and broken by mixing with 0.5 mm silica-zirconia beads in a Biospec Biobeater (Bartlesville, OK) at 4 °C. Breakage was carried out with 8 cycles of 2 minutes breaking and 6 minutes cooling on ice. Glass beads were removed by filtration of the homogenate using a layer of Miracloth (Calbiochem, CA). Unbroken cells and large cell debris were removed from the cell lysate by centrifugation at 1000g for 10 minutes at 4 °C. The membrane fractions were pelleted by centrifugation at 100,000g for 30 minutes at 4 °C. The pellet was re-suspended in 0.1 M triethanolamine (TEA), pH 7.2 buffer containing 25 mM CaCl2 (Buffer B). Protein concentrations in the membrane fractions were determined using the biuret assay and the final protein concentration of the suspension was adjusted to 25 mg/ml using Buffer B. The membrane phospholipids were digested by the addition of 1 mg of phopholipase C (Type 1 from C. perfringens, Sigma) and 6700 units of phospholipase A2 (purified from Naja naja venom [20]) per 500 mg of total protein. The mixture was stirred at room temperature for 1 hour in the dark maintaining the pH between 7.0 and 7.2 with 2 N NH4OH. After one hour, the suspension was centrifuged at 100,000g for 15 minutes and the pellet re-suspended in 25 mM potassium phosphate buffer adjusted to pH 7.6 containing 1 mM PMSF (Buffer C) to a final protein concentration of 15 mg/ml. Triton X-100 was added to this mixture to a final concentration of 0.5% (v/v). The mixture was left at room temperature with stirring in the dark for 30 minutes. After 30 minutes of stirring, the mixture was centrifuged at 100,000g for 15 minutes and the bright yellow supernatant (Triton extract) containing rMAOB was collected.
The Triton extract was loaded at flow rate of 1 mL/minute to a Bio-Rad MacroPrep® High-Q anion exchange column equilibrated with Buffer C containing 0.5% (v/v) Triton X-100. The column was then washed with 10 column volumes of Buffer C, followed by 5 column volumes of 25 mM potassium phosphate buffer adjusted to pH 7.6 containing 1 mM PMSF and 0.8% (w/v) OGP. rMAOB was eluted from the column by a linear gradient of 25 mM to 250 mM potassium phosphate buffer adjusted to pH 7.6, containing 20% glycerol, 1 mM PMSF, and 0.8% (w/v) OGP. Elution fractions were tested for benzylamine oxidase activity and optical absorbance of the flavin cofactor. The best fractions with an A280/A450 ratio less than 15 were collected and pooled for immunochemical and activity studies. The buffer in the pooled fractions was exchanged with 50 mM potassium phosphate buffer, pH 7.6, containing 20% (v/v) glycerol, and 0.8% (w/v) OGP and could be stored on ice for nearly up to three months without any significant loss of activity. Protein concentrations in the pooled fractions were determined using Bearden assay [21]. A summary of yields and specific activities of rMAOB in different steps of the purification process is provided in Table 1.
Table 1.
Purification scheme for rat liver MAOB expressed in P. pastoris.
| Step | Total Activity Units (U) a | Specific Activity (U/mg) | %Yield |
|---|---|---|---|
| Cell lysate | 3180 | 0.048 | 100 |
| Membrane fraction | 2520 | 0.049 | 79 |
| After phospholipase digestion | 2360 | 0.038 | 74 |
| Triton extract | 1023 | 0.31 | 32 |
| Pooled High-Q column fractions | 577 | 7.16 | 20 |
1U = 1 µmol product/min at 25 °C in air-saturated 50 mM HEPES, pH 7.5 buffer
Immunochemical analyses
The purified protein was characterized by SDS-polyacrylamide gel electrophoresis followed by either Coomassie blue staining or by Western blot analyses using anti-MAO antisera raised in rabbits against hMAOA, and anti-covalent flavin antisera raised in rabbits against FAD conjugated bovine serum albumin [22].
Protein sequence analyses
The partial amino acid sequence of purified rMAOB was verified by MALDI-TOF-MS analyses of the trypsin-digested peptide fragments on a Bruker Ultraflex- II TOF/TOF MALDI-MS instrument. The protein band was excised from the gel, cut into small cubes and destained by washing 3 times with 10 mM ammonium carbonate in 50% (v/v) acetonitrile/water for 15–20 minutes each. After drying under vacuum, the dry gel pieces were suspended in 20 to 30 µL of 10 mM ammonium carbonate in water solution containing 0.2 – 0.3 µg of procine liver trypsin (Promega) and incubated overnight at 37 °C. The reaction was stopped by adding 1 µL of 10% (v/v) trifluoroacetic acid (TFA). The supernatant from the digestion mixture was removed and saved in a clean microcentrifuge tube. The remaining peptides from the gel pieces were extracted by three incubations with 100 µL of 10 mM ammonium bicarbonate in 60% (v/v) acetonitrile/water for 20 to 30 minutes each. All extracts were collected in the same tube with the supernatant and dried in a speed vac to a final volume of 10 µL. The peptide mixture was desalted using C18 Zip-Tip (Millipore). Peptides were eluted from the Zip-Tip in 4 µL of 70% (v/v) acetonitrile/water mixture containing 0.1% (v/v) TFA. Approximately 1 µL of the eluted peptide mixture was spotted on the MALDI target together with 1 µL of a saturated solution of α-cyano-4-hydroxycinamic acid matrix (Sigma) in 70% (v/v) acetonitrile/water. The MALDI-MS spectrum was recorded in the reflectron mode and the observed peptide mass profile was submitted in the IPI Rat data base using Profound software (http://prowl.rockefeller.edu) to identify the associated protein(s).
Analytical procedures
HPLC analyses of the purified protein were done on a Shimadzu instrument equipped with a SPD-20A prominence UV-Vis detector. Purified protein was loaded on to a Jupiter 5 µm C4 300 Å (150 × 2.0 mm) reversed phase HPLC column (Phenomenex, Inc.) pre-equilibrated with 90% water − 10% acetonitrile mixture (v/v) containing 0.1% (v/v) trifluoroacetic acid (TFA). Elution was carried out by a linear gradient of 10 to 90% acetonitrile in water (v/v) containing 0.1% (v/v) TFA. The elution of the protein from the column was monitored by its absorbance at 280 nm. The MALDI-TOF-MS analyses of the detergent purified protein was performed by first precipitating the protein from its detergent buffer by adding 2 mM trichloroacetic acid (final concentration in the sample) followed by three washings of the precipitate with ultra pure water to remove salts and detergents. The washed protein precipitate was dried under vacuum and re-solubilized in 80% (v/v) formic acid in water. The formic acid solubilized protein was immediately diluted ten-fold with 70% (v/v) acetonitrile (in water) to adjust the final formic acid concentration in the sample to 8% (v/v). One micro liter of the solubilized protein was plated on to a MALDI target together with an equivalent amount of a saturated solution of 3,5-dimethoxy-4- hydroxycinnamic acid (sinapinic acid, Sigma). The MALDI-mass spectrum was recorded on a Bruker Ultrafelx II TOF/TOF instrument.
UV-Visible Spectroscopy
UV-Vis spectra on detergent purified rMAOB samples were recorded using a Varian Cary-50 UV-Visible spectrophotometer.
Activity assays
Activity of purified rMAOB was studied by monitoring the oxidation of benzylamine as substrate. Formation of the oxidation product (benzaldehyde, Δε250 = 12,800 M−1, cm−1) was followed over time using a Perkin Elmer Lambda-2 spectrophotometer. The primary deuterium kinetic isotope effect of benzylamine oxidation was determined using [α, α - 2H2]-benzylamine as substrate. All assays were performed in 50 mM HEPES buffer adjusted to pH 7.5, containing 0.5% (v/v) reduced Triton X-100 (Sigma). One activity unit (U) is defined as the formation of 1 µmol of product/min.
Determination of Km O2
Km of O2 and dKm of O2 were measured using a Clarke-type oxygen electrode (782 Oxygen Meter, Strathkelvin Instruments Limited, www.strathkelvin.com) by measuring the rate of oxygen consumption using protio and deuterio benzylamines as substrates in reaction buffers with varying oxygen concentrations. The dissolved oxygen concentrations in the reaction buffers were adjusted by mixing buffers purged with pure oxygen (1.02 mM in oxygen), pure nitrogen (0.0 mM oxygen) and at air saturation (0.24 mM oxygen) in different proportions. The initial dissolved oxygen concentrations in the reaction buffers were measured polarographically.
Results
Membrane localization and purification of rMAOB
In mammalian tissues MAOB is localized in the outer mitochondrial membrane (OMM) [23]. Recombinant rMAOB, over expressed in Pichia pastoris, is also found to be exclusively localized in the OMM of the Pichia cells. Liberation of the enzyme from its membrane environment requires hydrolysis of the phospholipids by phospholipase treatment followed by extraction with Triton-X 100 detergent. An improvement in enzyme purification over that previously described for recombinant human MAOB [13] is the use of an one step anion exchange column chromatography using Bio-Rad High-Q resin (also now routinely used for the purification of hMAOB), as described in the Methods section. This method allows reproducible fractionation of the Triton-X 100 extract to obtain the enzyme in a pure form and is preferable to the previously published polymer fractionation procedure. The pH conditions of the buffer solutions had to be adjusted for binding of rMAOB to the column. While hMAOB binds to the column at pH 7.2, binding of the rat enzyme requires a higher pH of 7.6.
Characterization of purified rMAOB
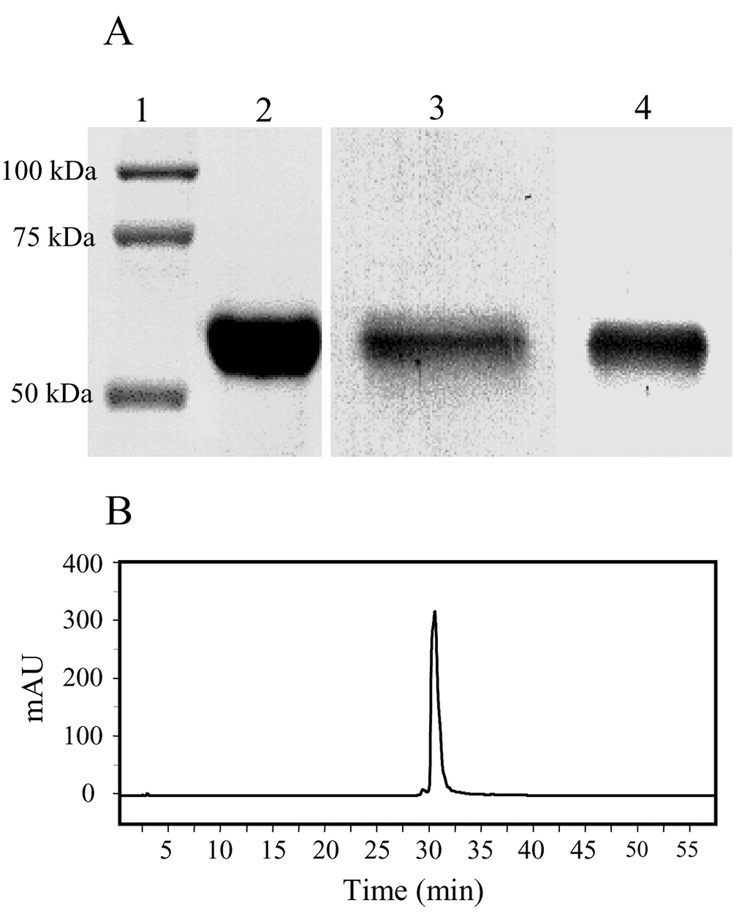
Purified rat MAOB obtained from the P. pastoris expression system shows a single band near 60 kD in the denaturating polyacrylamide gel (Figure 1A, lane 2). Western blot analyses using antiseras raised against human MAOA (hMAOA) or to BSA-conjugated FAD [22] show positive immunochemical reactivities with the purified recombinant rMAOB (Figure 1A, lanes 3 and 4). This data shows the presence of a covalently bound flavin cofactor (Figure 1A, lane 4) in the rMAOB sequence. Precipitation of the purified enzyme with trichloroacetic acid leaves no flavin optical signal in the supernatant, indicating that all flavin in the purified protein is covalently attached, which supports conclusion drawn from the Western blot analysis (Figure 1A, lane 4). Purified enzyme exhibits a stoichiometry of 1:0.91 protein-to-flavin ratio, indicating one mole of covalent FAD per mole of the enzyme. As shown in Table 1, a total of 577 units of rMAOB activity could be obtained in the detergent (OGP) purified form from 130 grams of wet cells (0.5 liter culture), which correlates to 150 –200 mg of purified enzyme isolated per liter of culture.
Figure 1.
(A) SDS-PAGE of purified recombinant rMAOB. (1) Molecular weight markers, (2) Coomassie-stained gel of purified rMAOB. (3) Western blot analyses of purified rMAOB with human MAOA antisera. (4) Western blot analyses with covalent flavin antisera. (B) HPLC chromatogram of purified rMAOB. The elution of the protein was monitored at 280 nm.
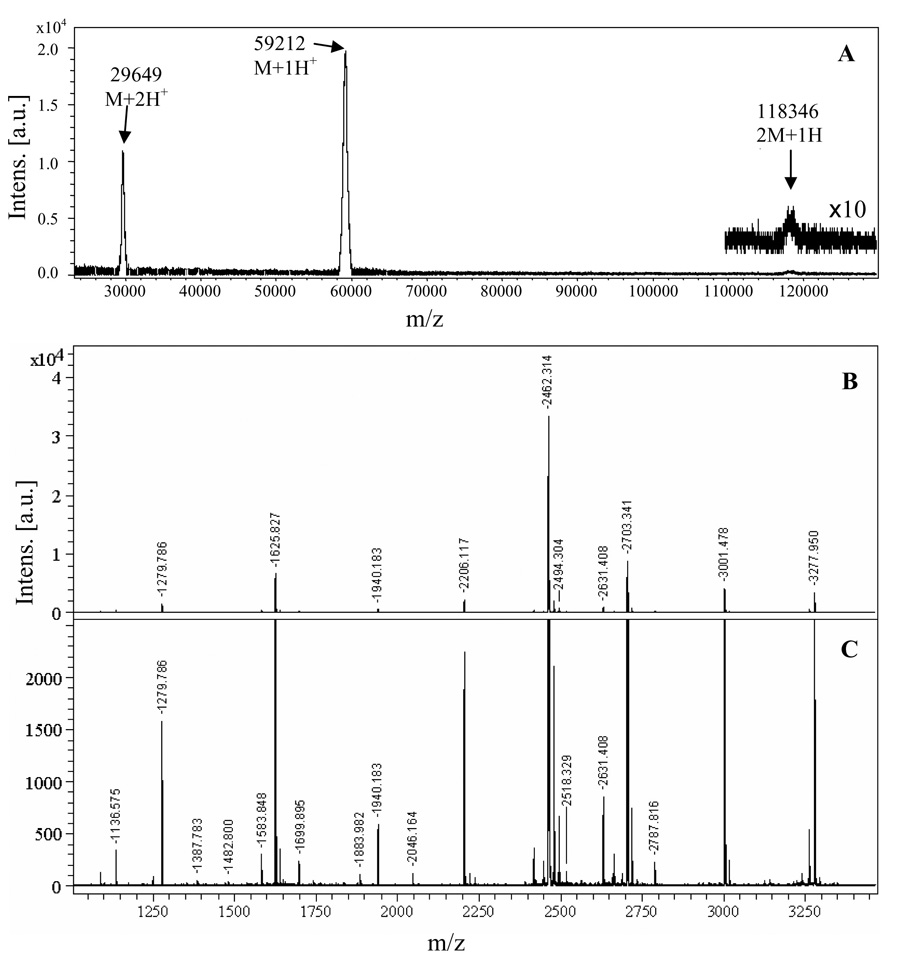
Homogeneity of the purified protein is also demonstrated by HPLC analyses, as shown in Figure 1B. Consistent with the highly pure HPLC elution profile, MALDI-TOF-mass spectrum (Figure 2A) of the purified rMAOB shows three peaks near 29.649 kDa (M +2H+), 59.212 kDa (M+1H+), and 118.346 (2M+1H+) kDa, which correspond to a single species with a molecular mass of ca. 59.228 ± 0.064 kD. This value is within the experimental uncertainty of the calculated molecular weight of 59.172 kDa based on the rMAOB amino acid sequence and the presence of one mole of covalent flavin cofactor per mole of the purified protein. The observed molecular weight of the recombinant rMAOB (Figure 2A) thus suggests no additional post-translational modifications, such as covalent attachment of lipids or glycosylations, other than covalent flavinylation.
Figure 2.
(A) Baseline subtracted MALDI-TOF mass spectrum of purified rMAOB. Mass peaks corresponding to a doubly charged monomer (M+2H+) at 29.649 kDa, a singly charged monomer (M+1H+) at 59.212 kDa, and a singly charged dimer (2M+1H+) at 118.346 kDa are observable. These three peaks correspond to a single species with an average molecular mass of 59.228 ± 0.064 kDa. (B) Peptide mass profile of trypsin digested recombinant rMAOB shown in original intensity scale, Mono-isotopic peptide mass peaks (M+1H+) are labeled (C) The same peptide mass spectrum as shown in panel B in an expanded intensity scale to show the peaks with weaker intensities.
Analysis of protein sequence
The amino acid sequence of the purified enzyme was verified by in-gel trypsin digestion and MALDI-TOF-MS analyses of tryptic peptide fragments. Figure 2B and Figure 2C show the peptide mass profile of the trypsin digested rMAOB in two different intensity (Y) scales. Peptide mass peaks with relatively weaker intensities are shown with an expanded Y-scale in the panel C of Figure 2. A search in the IPI Rat data base (allowing up to a maximum of one missed cuts and a mass tolerance of ±1 Da) using Profound software (http://prowl.rockefeller.edu) with the observed mono-isotopic mass peaks shows a 44% sequence coverage of the rMAOB protein sequence. The matched observed and theoretical mass peaks are listed in Table 2. The missing peptides in the mass spectrum of the tryptic digest could be due to incomplete digestions (missed cleavage) and/or due to hydrophobic nature of these peptides, which make them hard to elute from the de-salting C18 Zip-Tip column prior to mass analysis.
Table 2.
Observed and computed monoisotopic masses of the tryptic peptides of recombinant rMAOB expressed in P. pastoris.
| Mono isotopic Mass (Da) |
Error (Da) | Residues |
Missed Cut | Peptide sequence | ||
|---|---|---|---|---|---|---|
| Observed | Computed | Start | End | |||
| 1135.567 | 1135.562 | 0.005 | 210 | 220 | 0 | IGGSGQVSER |
| 1174.632 | 1174.594 | 0.039 | 198 | 208 | 0 | IISTTNGGQER |
| 1278.778 | 1278.755 | 0.023 | 272 | 282 | 0 | IHHSPPLPILR |
| 1386.775 | 1386.794 | −.019 | 259 | 271 | 0 | YVISAIPPVLGMK |
| 1481.792 | 1481.835 | −0.043 | 82 | 93 | 1 | VNEVERLIHFVK |
| 1582.840 | 1583.699 | −0.859 | 137 | 149 | 0 | APLAEEWDYMTMK |
| 1624.819 | 1624.784 | 0.035 | 53 | 67 | 0 | YVDLGGSYVGPTQNR |
| 1833.812 | 1833.790 | 0.022 | 121 | 136 | 0 | TMDEMGQEIPSDAPWK |
| 1882.974 | 1882.942 | 0.033 | 371 | 386 | 0 | VLNSQEALQPVHYEEK |
| 1939.175 | 1939.150 | 0.026 | 495 | 513 | 0 | LLGLTTILSATALGFLAHK |
| 2045.156 | 2045.126 | 0.030 | 231 | 248 | 0 | LERPVIHIDQTGENVVVK |
| 2461.306 | 2461.221 | 0.085 | 101 | 120 | 0 | GPFPPVWNPITYLDYNNLWR |
| 2702.333 | 2702.206 | 0.127 | 421 | 445 | 0 | IFFAGTETASHWSGYMEGAVEAGER |
| 3000.470 | 3000.381 | 0.089 | 421 | 448 | 1 | IFFAGTETASHWSGYMEGAVEAGERAAR |
| 3276.942 | 3276.629 | 0.313 | 457 | 484 | 0 | IPEDEIWQPEPESVDVPARPITNTFLER |
Based on the sequence homology between human and rat MAOBs, the flavin cofactor in rMAOB should bind to the protein backbone by a thioether linkage through Cys-397. The corresponding flavinylated tryptic peptide (formed by residues 387 to 412) should exhibit a peak at ca. 3787.7 Da. We did not observe a peptide peak of this size in the MALDI-mass spectrum of the trypsin digested rMAOB. If the thioether linkage between the flavin cofactor and Cys397 of the peptide is cleaved during sample preparations and/or mass analyses, the resulting de-flavinated peptide should exhibit a peak near 3004.3 Da. In the MALDI-mass spectrum shown in Figure 2B (and 2C), a peak with monoisotopic mass of 3000.47 Da is observed, which corresponds to the peptide fragment formed by residues 421 to 448 (considering a missed cleavage at residue 445 due to the presence of a negatively charged Glu444 residue immediately before the Arg445 residue). It is possible that the peaks associated with the higher isotopic masses of this peptide (421–448) mask the peak originating from the de-flavinated peptide fragment (387–412), thereby obscuring its detection. A similar situation is also observed for MALDI-TOF-MS analyses of trypsin digested human MAOB (not shown), where the peptide fragment formed by the residues 421–448 (considering a missed cut after residue Arg445) has a computed molecular weight of 3030.392 Da, which is close to the computed mass (3032.309 Da) of the de-flavinated form of the peptide fragment formed by residues 387 to 412. The failure to observe the intact flavin peptide in the MALDI-MS analysis of tryptic digests of either human or rat MAOB may be a consequence of photolytic cleavage of the thio-ether (or its oxidation product) linkage between Cys397 and FAD on laser excitation (ND : YAG laser. λ = 355 nm).
Catalytic activity
The catalytic properties of purified recombinant rMAOB is determined using benzylamine as substrate and compared (Table 3) with those reported previously for recombinant human MAOB expressed in P. pastoris [13, 24]. The observed Km(benzylamine) of the P. pastoris expressed rMAOB (176 ± 15 µM) is similar to the value reported earlier for rMAOB (140 ± 10 µM) expressed in S. cerevisiae [9]. Like human MAOB [13], rMAOB exhibits an observable primary kinetic isotope effect with [α, α -2H2] benzylamine as substrate. As evident from the kinetic data (Table 3), the Dkcat and D[kcat/Km(amine)] of human and rat MAOBs are similar in magnitude, indicating that Cα-H bond cleavage constitutes the rate limiting step in catalysis. Since D[kcat/Km(amine)] values reflect only those catalytic steps that are involved up to and including the first irreversible step in catalysis; this result indicates that the aggregate of rate constants of the catalytic steps prior to Cα-H bond cleavage in benzylamine oxidation by rat MAOB are similar to those reported for human MAOB. The Km(O2) of rMAOB is ca. two-fold lower than observed for the human enzyme. This is consistent with the higher kcat value exhibited by rMAOB for benzylamine oxidation at an oxygen concentration equal to that of air saturation (0.24 mM). Both human and rat MAOBs show considerable effects of deuterio substrate on Km(O2). The D[kcat/Km(O2)] values (Table 3) of rMAOB is found to be slightly lower (0.62 ± 0.14) compared to that of the human enzyme (1.0 ± 0.34).
Table 3.
Comparison of steady state kinetic parameters for recombinant human and rat liver MAOBs expressed in P. pastoris. The kcat and Km (Benzylamine) values are determined at an [O2] of 0.24 mM.
| Recombinant human livera | Recombinant rat liver | |
|---|---|---|
| Benzylamine | ||
| Km (amine) mM | 0.15 ± 0.01 | 0.176 ± 0.015 |
| kcat min−1 | 300 ± 10 | 497 ± 83 |
| Km (O2) mM | 0.33 ± 0.07 | 0.17 ± 0.01 |
| [α, α -2H2]Benzylamine | ||
| Km (amine) mM | 0.146 ± 0.01 | 0.26 ± 0.02 |
| kcat min−1 | 64 ± 5 | 149 ± 24 |
| Km (O2) mM | 0.074 ± 0.023 | 0.03 ± 0.006 |
| Deuterium isotope effect | ||
| Dkcat | 4.7 ± 0.5 | 3.4 ± 0.7 |
| D[kcat/Km(amine)] | 4.6 ± 0.46 | 4.9 ± 0.4 |
| D[kcat/Km(O2)] | 1.0 ± 0.34 | 0.62 ± 0.14 |
| Apparent Kd (µM) | 147 ± 25 | 304 ± 84 |
The values for the human enzyme are calculated from the values listed in Ref. [13].
The steady state isotope effect data presented in Table 3 could be used to calculate the apparent binding constants (Kd) of benzylamine to the human and rat MAOB active sites using the equation reported earlier by Klinman and Matthews (Eq. 1, [25]), and shown in our previous work to be equal to the Ks values determined from reductive half reaction data on bovine MAOB [17].
| (1) |
The parameter Kd in Eq. 1 represents the dissociation constant of the substrate from the enzyme-substrate complex prior to the isotope sensitive Cα-H bond cleavage step. The Kd values calculated for the two enzymes show (Table 3) that the binding of substrate (benzylamine) in the human MAOB active site is ca. 2 fold tighter compared to that in the rat MAOB active site.
Optical Spectroscopic Study
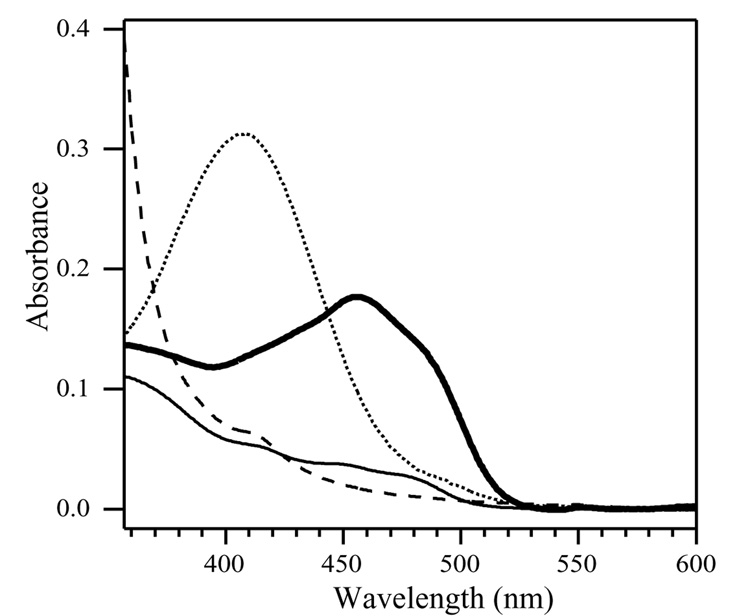
Detergent purified rMAOB shows an optical absorbance spectrum typical of oxidized flavin cofactor (bold solid line, Figure 3). Addition of benzylamine to an anaerobic sample in a sealed cuvette results in reduction of the flavin cofactor to the two electron reduced hydroquinone form (solid line in Figure 3). No intermediate formation of a semiquinone species was observed. The small level (5–10%) of flavin 450 nm absorption observed in the benzylamine reduced sample (solid line) relative to that observed after reduction with sodium dithionite (dashed line in Figure 3) suggests the presence of a small level (<10%) of non-functional enzyme in the sample.
Figure 3.
Optical absorption spectra of oxidized (bold solid line), benzylamine reduced (solid line), dithionite reduced (dashed line) and pargyline inhibited (dotted line) recombinant rMAOB. All proteins samples were prepared in 50 mM potassium phosphate buffer pH 7.0, containing 0.8% (w/v)of OGP. Protein concentrations in all samples were adjusted to 1 mg/mL prior to spectral measurements.
Like human MAOB, recombinant rMAOB is irreversibly inhibited with pargyline with the formation of a covalent N5 flavocyanine adduct. The optical spectrum of pargyline inhibited rMAOB is shown in Figure 3 (dotted line) shows a characteristic absorption band near 410 nm with relative extinction coefficient (ε410 ~ 21.8 mM−1 cm−1) similar to those reported for human MAOA and for MAOB.
Discussion
Pichia pastoris has been successfully used as a system of choice for heterologous expressions of several different proteins [11, 12]. The major factors, which make the P. pastoris expression system better than the other yeast systems (e.g. Saccharomyces cerevisiae), are: (1) its preference for respiratory growth, which allows it grow to a higher cell density in the culture, (2) the function of the strong AOX1 promoter, which allows high level (> 30% of the total expressed proteins in the cells) expression of the foreign genes when induced with methanol [26, 27], (3) simplicity in genetic manipulation and (4) the ability of P. pastoris to carry out several eukaryotic post-translations modifications like glycosylation (both O- and N-linked), disulfide bond formations etc. [11]. In the context of MAO expression, a major advantage of using the P. pastoris expression system is its preference for respiratory growth, which requires it to contain higher number of mitochondria per cell (compared to S. cerevisiae), thereby increasing the yield of MAO expression. In the present work we have described a convenient method for the expression and purification of rMAOB in high yield using this (P. pastoris) expression system. Although methods involving bacterial (E.coli) and yeast (S. cerevisiae) expression systems have been reported previously [8–10], the levels of expression and the yields of the purified active enzyme obtained from those systems were too low to provide sufficient amounts of purified enzyme for direct structural and functional studies.
The results presented above describe an efficient approach to obtain purified rMAOB in homogeneous form with >90% functionality. The recombinant rMAOB obtained from P. pastoris expression system exhibits similar Km value for benzylamine (Table 3) as reported earlier for the enzyme purified from S. cerevisiae expression system [9]. The purification scheme discussed in Methods section is modified for pH and buffer conditions relative to that previously published for human MAOA and MAOB [13, 14, 19]. As mentioned above, unlike human MAOA and MAOB, purification of rat MAOB requires using a higher pH (7.5–7.6) for efficient binding and subsequent washings in the anion exchange high-Q column. This modification is critical. Triton extracts made at lower pH values (7.0 and 25 mM KPi, like that used for human enzymes) show no binding of rMAOB to the resin. Lowering the salt concentrations to 10 mM partially solved this problem by allowing partial binding to the resin, but the final purified protein obtained by this method was less pure as it eluted from the anion exchange column very early in the elution fractions. This differential behavior of human and rat MAOBs could be due to differences in their respective pI values. The amino acid sequence of hMAOB predicts a pI of 7.2, whereas the sequence for rMAOB predict a pI of 8.3. This suggests that rMAOB is slightly more basic than hMAOB, requiring an elevated pH to have enough surface residues in anionic forms to enable efficient binding to the resin.
Both human and rat MAOBs show large steady state primary kinetic isotope effect with [α, α -2H2]-benzylamine as substrate, indicating that the Cα-H bond cleavage is the rate limiting step in catalysis for the two enzymes. These results suggest that the overall mechanism of benzylamine oxidation catalyzed by rMAOB follows a pathway similar to that observed for hMAOB [13]. Based on the steady state kinetic parameters listed in Table 3, binding of benzylamine into the hMAOB active site is slightly tighter (ca. 2 fold) compared to that in the rMAOB active site, suggesting a subtle difference in their active site structures. This is consistent with the differences in inhibitors specificities reported earlier for the two enzymes, as well as with the differences reported earlier between human and rat MAOB active site cavities by EPR spectroscopic studies using a spin labeled inhibitor.
As shown in Table 3, the Km(O2) of rMAOB is observed to be two-fold lower than human MAOB, which is consistent with its higher kcat value observed for benzylamine oxidation (Table 3) at [O2] of air saturation. This difference in Km(O2) values between the two enzymes is also consistent with higher specific activity (~ 7.1 U/mg) observed for the purified rMAOB (Table 1) compared to that reported earlier (3.0 U/mg) for human MAOB [13]. A further understanding of the kinetic properties of rMAOB requires further studies with other possible substrates and will be addressed in future work.
Acknowledgements
We thank Dr. Edward Morgan for providing us the rat cDNA library. This work was supported by NIH grant GM29433 (DEE).
Footnotes
This work was supported by National Institute of Health Grant GM-29433 (DEE).
Abbreviations: MAO – Monoamine oxidase, MALDI-MS – Matrix Assisted Laser Desorption/Ionization Mass Spectrometry, OGP - Octyl-β-D-glucopyranoside, BSA – Bovine Serum Albumin, HPLC – High Performance Liquid Chromatography, OMM – Outer Mitochondrial Membrane, Da – Dalton.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Silverman RB. Radical Ideas about Monoamine Oxidase. Acc. Chem. Res. 1995;28:335–342. [Google Scholar]
- 2.Kumar MJ, Nicholls DG, Andersen JK. Oxidative a-Ketoglutarate Dehydrogenase Inhibition via Subtle Elevations in Monoamine Oxidase B Levels Results in Loss of Spare Respiratory Capacity: Implications for Parkinson'ls Disease. J. Biol. Chem. 2003;278:46432–46439. doi: 10.1074/jbc.M306378200. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy BP, Ziegler MG, Alford M, Hansen LA, Thal LJ, Masliah E. Early and persistent alterations in prefrontal cortex MAO A and B in Alzheimer's disease. J. Neural Transmission. 2003;110:789–801. doi: 10.1007/s00702-003-0828-6. [DOI] [PubMed] [Google Scholar]
- 4.Youdim MBH, Edmondson D, Tipton KF. The therapeutic potential of monoamine oxidase inhibitors. Nat. Rev. Neurosci. 2006;7:295–309. doi: 10.1038/nrn1883. [DOI] [PubMed] [Google Scholar]
- 5.Thomas T. Monoamine oxidase-B inhibitors in the treatment of Alzheimers disease. Neurobiol. Aging. 2000;21:343–348. doi: 10.1016/s0197-4580(00)00100-7. [DOI] [PubMed] [Google Scholar]
- 6.Novaroli L, Daina A, Favre E, Bravo J, Carotti A, Leonetti F, Catto M, Carrupt P-A, Reist M. Impact of Species-Dependent Differences on Screening, Design, and Development of MAO B Inhibitors. J. Med. Chem. 2006;49:6264–6272. doi: 10.1021/jm060441e. [DOI] [PubMed] [Google Scholar]
- 7.Upadhyay AK, Wang J, Edmondson DE. Comparison of the structural properties of the active site cavities of human and rat monoamine oxidase A and B in their soluble and membrane-bound forms. Biochemistry. 2008;47:526–536. doi: 10.1021/bi7019707. [DOI] [PubMed] [Google Scholar]
- 8.Hirashiki I, Ogata F, Ito A. Rat monoamine oxidase B expressed in Escherichia coli has a covalently-bound FAD. Biochem. Mol. Biol. Intl. 1995;37:39–44. [PubMed] [Google Scholar]
- 9.Tsugeno Y, Hirashiki I, Ogata F, Ito A. Regions of the molecule responsible for substrate specificity of monoamine oxidase A and B: a chimeric enzyme analysis. J. Biochem. 1995;118:974–980. doi: 10.1093/jb/118.5.974. [DOI] [PubMed] [Google Scholar]
- 10.Tsugeno Y, Ito A. A key amino acid responsible for substrate selectivity of monoamine oxidase A and B. J. Biol. Chem. 1997;272:14033–14036. doi: 10.1074/jbc.272.22.14033. [DOI] [PubMed] [Google Scholar]
- 11.Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 12.Boeer E, Steinborn G, Kunze G, Gellissen G. Yeast expression platforms. Appl. Microbiol. Biotechnol. 2007;77:513–523. doi: 10.1007/s00253-007-1209-0. [DOI] [PubMed] [Google Scholar]
- 13.Newton-Vinson P, Hubalek F, Edmondson DE. High-level expression of human liver monoamine oxidase B in Pichia pastoris. Prot. Expr. Purif. 2000;20:334–345. doi: 10.1006/prep.2000.1309. [DOI] [PubMed] [Google Scholar]
- 14.Li M, Hubalek F, Newton-Vinson P, Edmondson DE. High-level expression of human liver monoamine oxidase A in Pichia pastoris: comparison with the enzyme expressed in Saccharomyces cerevisiae. Prot. Expr. Purif. 2002;24:152–162. doi: 10.1006/prep.2001.1546. [DOI] [PubMed] [Google Scholar]
- 15.Binda C, Newton-Vinson P, Hubalek F, Edmondson DE, Mattevi A. Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nat. Str. Biol. 2002;9:22–26. doi: 10.1038/nsb732. [DOI] [PubMed] [Google Scholar]
- 16.De Colibus L, Li M, Binda C, Lustig A, Edmondson DE, Mattevi A. Three-dimensional structure of human monoamine oxidase A (MAO A): Relation to the structures of rat MAO A and human MAO B. Proc. Natl. Acad. Sci. USA. 2005;102:12684–12689. doi: 10.1073/pnas.0505975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker MC, Edmondson DE. Structure-activity relationships in the oxidation of benzylamine analogues by bovine liver mitochondrial monoamine oxidase B. Biochemistry. 1994;33:7088–7098. doi: 10.1021/bi00189a011. [DOI] [PubMed] [Google Scholar]
- 18.Daum G, Boehni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 19.Hubalek F, Binda C, Khalil A, Li M, Mattevi A, Castagnoli N, Edmondson DE. Demonstration of Isoleucine 199 as a Structural Determinant for the Selective Inhibition of Human Monoamine Oxidase B by Specific Reversible Inhibitors. J. Biol. Chem. 2005;280:15761–15766. doi: 10.1074/jbc.M500949200. [DOI] [PubMed] [Google Scholar]
- 20.Cremona T, Kearney EB. Studies on the Respiratory Chain-Linked Reduced Nicotinamide Adenine Dinucleotide Dehydrogenase. Vi. Further Purification and Properties of the Enzyme from Beef Heart. J. Biol. Chem. 1964;239:2328–2334. [PubMed] [Google Scholar]
- 21.Bearden JC., Jr Quantitation of submicrogram quantities of protein by an improved protein-dye binding assay. Biochim. Biophys. Acta. 1978;533:525–529. doi: 10.1016/0005-2795(78)90398-7. [DOI] [PubMed] [Google Scholar]
- 22.Barber MJ, Eichler DC, Solomonson LP, Ackrell BA. Anti-flavin antibodies. Biochem. J. 1987;242:89–95. doi: 10.1042/bj2420089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry MD, Juorio AV, Paterson IA. The functional role of monoamine oxidases A and B in the mammalian central nervous system. Prog. Neurobiol. 1994;42:375–391. doi: 10.1016/0301-0082(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 24.Li M, Binda C, Mattevi A, Edmondson DE. Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins. Biochemistry. 2006;45:4775–4784. doi: 10.1021/bi051847g. [DOI] [PubMed] [Google Scholar]
- 25.Klinman J, Matthews RG. Calculation of substrate dissociation constants from steady-state isotope effects in enzyme-catalyzed reactions. J. Am. Chem. Soc. 1985;107:1058–1060. [Google Scholar]
- 26.Roggenkamp R, Janowicz Z, Stanikowski B, Hollenberg CP. Biosynthesis and regulation of the peroxisomal methanol oxidase from the methylotrophic yeast Hansenula polymorpha. Mol. Gen. Genet. 1984;194:489–493. doi: 10.1007/BF00425563. [DOI] [PubMed] [Google Scholar]
- 27.Egli T, Van Dijken JP, Veenhuis M, Harder W, Fiechter A. Methanol metabolism in yeasts: regulation of the synthesis of catabolic enzymes. Arch. Microbiol. 1980;124:115–121. [Google Scholar]