Abstract
In females, most genes on the X chromosome are generally assumed to be transcriptionally silenced on the inactive X as a result of X inactivation. However, particularly in humans, an increasing number of genes are known to “escape” X inactivation and are expressed from both the active (Xa) and inactive (Xi) X chromosomes; such genes reflect different molecular and epigenetic responses to X inactivation and are candidates for phenotypes associated with X aneuploidy. To identify genes that escape X inactivation and to generate a first-generation X-inactivation profile of the X, we have evaluated the expression of 224 X-linked genes and expressed sequence tags by reverse-transcription–PCR analysis of a panel of multiple independent mouse/human somatic cell hybrids containing a normal human Xi but no Xa. The resulting survey yields an initial X-inactivation profile that is estimated to represent ≈10% of all X-linked transcripts. Of the 224 transcripts tested here, 34 (three of which are pseudoautosomal) were expressed in as many as nine Xi hybrids and thus appear to escape inactivation. The genes that escape inactivation are distributed nonrandomly along the X; 31 of 34 such transcripts map to Xp, implying that the two arms of the X are epigenetically and/or evolutionarily distinct and suggesting that genetic imbalance of Xp may be more severe clinically than imbalance of Xq. A complete X-inactivation profile will provide information relevant to clinical genetics and genetic counseling and should yield insight into the genomic and epigenetic organization of the X chromosome.
X chromosome inactivation is an extraordinary example of coordinate gene control, resulting in transcriptional silencing of most of the several thousand genes on one X chromosome in mammalian females (1, 2). However, X inactivation is not completely panchromosomal, as evidenced by the increasing number of genes that “escape” X inactivation and are expressed from both active (Xa) and inactive (Xi) chromosomes (3, 4). Such genes reflect different epigenetic responses to X inactivation, determination of the basis for which will be important for dissecting mechanisms of X inactivation. In addition, genes that escape inactivation also have clinical significance as candidates for phenotypes associated with X aneuploidy.
Many approaches have been used to determine whether an X-linked gene is subject to or escapes from inactivation (reviewed in refs. 2, 5). These include analysis of protein polymorphisms to distinguish Xa and Xi expression (6), detection of mosaic expression of a protein in tissues of heterozygous carriers of X-linked disorders (7–9), or comparison of protein or transcript levels among individuals with different numbers of X chromosomes (10–13).
Although these approaches are suitable for studying individual genes, determination of a comprehensive X-inactivation profile of the entire chromosome requires analysis of a large number of genes by using a relatively high-throughput assay. Therefore, we and others have analyzed gene expression from human Xa or Xi chromosomes in rodent/human somatic cell hybrids as a direct measure of a gene’s inactivation status. This method does not require distinction of transcripts on the Xa and Xi and is applicable to all genes that are expressed in these cells (that is, generally all genes that are expressed in fibroblasts). With this approach, each gene is assayed by reverse transcription–PCR (RT-PCR) to determine whether expression is detected only from hybrids retaining Xa chromosomes, in the case of a gene that is subject to X inactivation, or from both Xa and Xi hybrids, indicating a gene that escapes from X inactivation (4, 14–18).
In this study, we have determined the X-inactivation status of 224 X-linked genes and expressed sequence tags (ESTs) by RT-PCR analysis of a panel of multiple Xi hybrids. Together with published information on 23 additional genes, this initial X-inactivation profile is estimated to represent ≈10% of all X-linked transcripts. Of the transcripts analyzed in this study, 34 were expressed in as many as nine Xi hybrids and thus escape inactivation. The genes that escape inactivation are nonrandomly distributed, with the majority of such transcripts mapping to Xp. This distribution suggests that the two arms of the X differ not only in their evolutionary origin (19, 20) but also with respect to epigenetic regulation and implies that genetic imbalance of Xp may be more severe clinically than imbalance of Xq. A complete X-inactivation profile will provide important information for medical genetics and also will provide insight into the role that the genomic organization of the X plays in the regulation of epigenetic silencing on the chromosome.
Materials and Methods
Identification of Genes/ESTs Used in These Studies.
The genes and ESTs identified through the Unigene database (http://www.ncbi.nlm.nih.gov/UniGene/Hs.Home.html) each represent unique Unigene clusters and therefore are presumed to represent independent transcripts. Additional genes or ESTs are from the literature. All ESTs were additionally subjected to blast searches (http://www.ncbi.nlm.nih.gov/BLAST/). For cases in which several overlapping ESTs were identified, only one was included in this study. The distribution of the genes assayed in part reflects the current gene distribution in Unigene, including particularly “gene-rich” regions in Xq28 and Xp11. A small number of the genes tested had been previously reported by others to be subject to or to escape X inactivation on the basis of studies of one to four Xi hybrids (e.g., 16, 21, 22). These genes were reanalyzed in our studies to address potential heterogeneity that may not have been detected previously but would be addressed by assaying a larger panel of Xi hybrids. The genes/ESTs analyzed in this study are listed in Table 1, and additional information regarding each gene/EST analyzed, including PCR primers and assay conditions, is available as supplemental material on the PNAS web site (see www.pnas.org). All data are also available from a periodically updated database at our web site (see http://mediswww.meds.cwru.edu/dept/genetics/willard.html).
Table 1.
Inactivation status of 224 X chromosome transcripts assayed in somatic cell hybrids
| Tested | Subject to inactivation | Escape from inactivation | Heterogeneous | |
|---|---|---|---|---|
| Xp | 104 | 70 (7)* | 31 (5)† | 3‡ |
| Xq | 120 | 107 (23)* | 3 (1)† | 10‡ |
| Total | 224 | 177 (30) | 34 (6) | 13 |
Numbers in parentheses reflect number of genes of the total that were concluded to be subject to inactivation despite being expressed in 1 or 2 of the Xi hybrids tested, as discussed in the text.
† Numbers in parentheses reflect number of genes of the total that were concluded to escape inactivation despite absence of expression from 1 or 2 of the Xi hybrids tested, as discussed in the text.
‡ Number of genes that show heterogeneous patterns of X inactivation, defined as being expressed in three to six of the nine Xi hybrids tested.
X-Inactivation Assays in Somatic Cell Hybrids.
Most of the somatic cell hybrids used for these studies have been previously described or were generated by using established protocols (4). Somatic cell hybrid cell lines were maintained, and RNA and DNA were prepared as described (4). For expression assays, cDNA corresponding to 25–250 ng of total RNA was used for amplification reactions. All RNA samples were DNaseI treated before reverse transcription to eliminate any contaminating genomic DNA in RNA preparations. The data reported here subsume and extend data reported previously from our laboratory on a more limited series of hybrids (4, 14, 18, 23).
Integration of X-Inactivation Data and Mapping Information.
Radiation hybrid data were available for most of the Unigene genes/ESTs (see http://www.ncbi.nlm.nih.gov/genemap98). To integrate physical and radiation hybrid mapping information, physical map distances for genes, ESTs, and the markers used to anchor radiation hybrid maps were based on the Integrated 3 Map from the integrated X chromosome database (see http://ixdb.mpimg-berlin-dahlem.mpg.de). Although most markers could be assigned to a ≈5-megabase bin on the chromosome, because radiation hybrid map distances are based on statistical probabilities and carry inherent uncertainty, some genes/ESTs may actually map to an adjacent interval. Pericentromeric ESTs were more precisely mapped within contigs generated in this laboratory (ref. 24 and M. Schuler, personal communication).
X-Inactivation Assays of Transcribed Polymorphisms in Nonrandomly Inactivated Fibroblast Cell Lines.
The panel of 40 primary fibroblast cell lines that are nonrandomly inactivated because of the presence of a structurally abnormal chromosome has been described (25). Assays to differentiate alleles and to determine the relative expression of each allele were developed as described (25). Briefly, a single-cycle primer extension with a 32P-labeled primer was performed before restriction enzyme digestion to prevent heteroduplex formation from complicating analyses. Band intensities were quantitated on a PhosphorImager (Molecular Dynamics). A polymorphism in the EST stSG9723 was identified by comparing EST sequences deposited in GenBank. After amplification with stSG9723-F2: CCACATCTAGATTTCAACCTCC and stSG9723-Bst: GGAAAAAGGAAGGAGCAGGTAAC, alleles were distinguished by digestion with BstEII. To analyze the human homologue of a Drosophila suppressor of position effect variegation, SUVAR39H (WIAF-1863) (26), samples were amplified with SUVAR39-F: GCATAGGGTTGAGGGGTGTA and SUVAR39-R2: TTTGTGCTCACCCTGGTTC and alleles distinguished by digestion with MspI. The polymorphism in the Drosophila fat facets gene homologue, DFFRX (WIAF-572) (26), was tested by amplification with DFFRX-1: ATGAGACCCTGCTTTGAACG and DFFRX-2: TTCCCTTCTGTTGGATCCC, followed by BsaJI digestion to differentiate alleles.
Results
Of the total of 224 X-linked transcripts tested, ≈65% represent known protein-coding genes or full-length transcripts with ORFs. The other ≈35% are ESTs. On the basis of their map positions on the X, we estimate that no more than 5% of these may represent nonindependent transcripts. The density of genes and ESTs analyzed in each interval on the chromosome (Fig. 1) largely reflects the distribution of genes on current transcript maps of the X chromosome (27) and accentuates particularly gene-rich regions such as Xp11 and Xq28 (28, 29) and the gene-poor region Xq21 (30, 31).
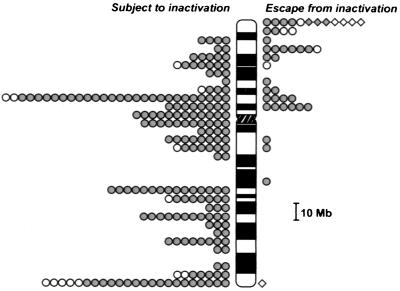
Figure 1.
Gene expression from active and inactive X chromosomes in somatic cell hybrids. Pseudoautosomal genes are indicated with diamonds, and X-specific transcripts are represented as circles. The shaded symbols identify genes/ESTs assayed in this study. Open symbols represent 23 genes that have been published previously (15–17) (see http://mediswww.meds.cwru.edu/dept/genetics/willard.html). Genes showing heterogeneous expression (see text and Table 1) are not included in this figure.
To determine their X-inactivation status, genes were assayed in five to nine rodent/human somatic cell hybrids, each containing a different cytogenetically normal human Xi. Overall, nearly 80% of the genes or ESTs tested (175/224) gave completely concordant results in all hybrids tested; that is, they were either expressed in all Xi hybrids tested (28 transcripts) or silenced in all Xi hybrids tested (147 transcripts) (Table 1). An additional 36 transcripts gave concordant expression patterns in all but one or two hybrids; these one-exception and two-exception categories included genes, for example, that were expressed in eight of nine Xi hybrids tested or in only one of nine hybrids tested. Because the distribution of these genes was nonrandom on the chromosome (Table 1 and see below), we have considered these data as indicating the likely X-inactivation pattern of these particular genes. Thus, overall, of the 224 transcripts tested, 177 appear to be subject to inactivation, whereas 34 escape inactivation (Table 1, Fig. 1). Notably, the status of only 13 genes (6%) was indeterminate, because they were expressed in about half of the hybrids tested. As also described elsewhere (4, 25, 32), such heterogeneous patterns may reflect a naturally occurring heterogeneity in human cells (as demonstrated for the REP1 and TIMP1 genes; refs. 25, 32), occasional reactivation of human X-linked genes in somatic cell hybrids, and/or an innately unstable epigenetic state.
Fig. 1 combines the data reported here with published information on approximately two dozen additional X-linked genes (12, 15, 16, 22, 32–41). Notably, the distribution of genes that escape X inactivation is decidedly nonrandom. For all three categories of genes considered (those that were completely consistent in all hybrids tested as well as the one-exception and two-exception classes), there were many more genes that escape X inactivation on Xp than on Xq. Considering all genes examined, 30% of the genes tested on Xp escaped inactivation, whereas less than 3% of the genes on Xq escaped inactivation (Table 2). In part, this nonrandom distribution reflects a large cluster of genes in and adjacent to the pseudoautosomal region in the distal 15-megabases of Xp, all of which are at least partially expressed from the Xi (42) (Fig. 1). Because all of these genes may have been pseudoautosomal at one point during evolution (43), their expression patterns may be an epigenetic relic of their evolutionary origin. Even excluding these genes, the proportion of genes on Xp that are expressed from the Xi (≈21%) is still significantly higher than the number on Xq (≈3%) (P < 0.001). Other evolutionary studies have concluded that much of human Xp was originally autosomal and was added sometime after marsupials and placental mammals diverged from a common ancestor (19, 20). Because autosomal genes are not normally dosage compensated, our data suggest that many Xp genes may not have acquired the ability to be inactivated and further indicates that the genomic organization of sequences on Xq differs in a characteristic but yet unknown way from genes on Xp or on autosomes (44).
Table 2.
X-Inactivation assays in nonrandomly inactivated primary fibroblast cell lines
| Gene | Map position | Distinction of alleles | No. informative female cell lines | Monoallelic expression | Biallelic expression | X-inactivation status |
|---|---|---|---|---|---|---|
| stSG9723 | Xp22.32 | BstEII digestion | 5 | 0 | 5 | Escapes |
| SUVAR39H | Xp11.23 | MspI digestion | 12 | 12 | 0 | Subject |
| DFFRX | Xp11.3 | BsaII digestion | 1 | 0 | 1 | Escapes |
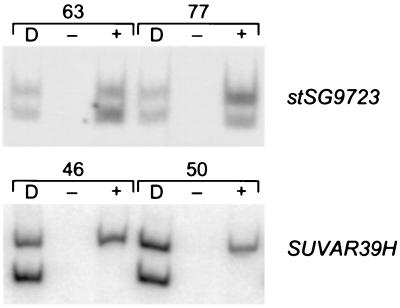
To confirm our findings for some of the genes that are included in the profile, several genes were tested in a complementary system by assaying expressed polymorphisms in a large panel of primary diploid human fibroblast cell lines from individuals with nonrandom X inactivation (25). Monoallelic expression in these cell lines indicates that a gene is subject to inactivation (e.g., SUVAR39H in Fig. 2), whereas biallelic expression demonstrates that a gene is expressed from both the Xa and Xi (e.g., stSG9723 in Fig. 2). Notably, for two Xp genes that escape inactivation in the hybrid system, the human fibroblast data, though limited, were in complete agreement (Table 2, Fig. 2). Overall, for the 10 X-linked genes tested in this system (Table 2, Fig. 2, and ref. 25), expression patterns in fibroblasts were fully consistent with the results from Xi hybrids.
Figure 2.
Gene expression in nonrandomly inactivated primary fibroblast cell lines. For each cell line, DNA and cDNA samples were PCR amplified and then digested with the appropriate restriction enzyme to differentiate alleles. D indicates amplification of DNA and + or − refers to RNA that has been amplified with or without prior reverse transcription. Each cell line tested is identified by a number, as described previously (25).
Discussion
The current study is the most extensive to date (in terms of both the number of genes assayed and the number of hybrids scored) and allows us to evaluate the suitability of the hybrid model system for analyzing X-linked gene expression. Previous reports have suggested that somatic cell hybrids may not recapitulate all features of X inactivation, because gene reactivation after treatment with demethylating agents occurs at a higher frequency in hybrids than in human diploid cells, and as XIST RNA is poorly localized to the Xi in hybrids (45–49). The data presented in this report, however, give credibility to the hybrid system, because ≈150 genes tested are stably inactivated. Additionally, the observation that a disproportionate number of the genes that escape inactivation are located on Xp argues strongly either that X-inactivation status is a stable feature of individual genes, whether in human or hybrid cells, and/or that the propensity of such genes to reactivate at high frequency in hybrids is determined, in large measure, by their map location on the X.
Thirteen additional genes tested showed heterogeneous patterns, being expressed in approximately half of the Xi hybrids tested (Table 1). Such heterogeneity has been demonstrated in human cells for the REP1 and TIMP1 genes (25, 32), but may also reflect occasional reactivation of X-linked genes in somatic cell hybrids. That such patterns are relatively rare further demonstrates that the inactivation status of most X-linked genes is highly stable and is well maintained in somatic cell hybrids and implies, as a corollary, that the mouse/human somatic cell hybrid system is a faithful reporter of X-inactivation status (4). If documented, reactivation in somatic cell hybrids of a specific subset of X-linked genes would argue that Xi hybrids will be an important system to tease apart the epigenetic features required for maintaining inactivation of these specific genes. Nonetheless, because gene reactivation may occur at a higher (albeit still low) level in hybrids (50), it seems advisable to base conclusions about inactivation status on results from multiple independent Xi hybrids.
The X-inactivation profile presented here has implications for understanding both chromosomal mechanisms of X inactivation and the role that X inactivation plays in individuals with X aneuploidy and in female carriers of X-linked disorders. The X-linked genes analyzed here and elsewhere (12, 15, 16, 22, 32–41) (Fig. 1) represent an estimated 10% of the genes expected to be on the chromosome (27), and accordingly several considerations must be taken into account to extrapolate these data to the entire X chromosome. The overall estimate that nearly 35% of Xp genes escape inactivation (considering all genes shown in Fig. 1) may be inflated by the unique characteristics of Xp22.3 discussed above and of a multigene domain in Xp11.2 (18). To counter this potential bias, we considered 90 genes or ESTs (selected randomly and distributed along the chromosome) for which there was no prior X-inactivation information available. In this unbiased gene set, 19% of genes on Xp escaped inactivation, whereas only a single Xq gene escaped inactivation (P < 0.01). This comparison confirms the significant distinction between Xp and Xq, but indicates that the overall proportion of Xp genes that escape inactivation may be somewhat lower than apparent in Fig. 1.
Another feature of X inactivation that may influence estimates are mechanism(s) of escape from inactivation. In addition to Xp22.3, genes that escape inactivation are clustered in at least two other regions of the chromosome, suggesting that at least for these genes, X-linked gene expression is controlled at the level of chromosomal domains (3, 18, 23). Regional control has not been addressed for other genes that escape inactivation and may increase estimates, as more closely linked genes are analyzed in the future. On the basis of these considerations, it is possible that there are hundreds of genes that are expressed from the Xi chromosome. Because many of these genes do not have Y-linked homologues (3, 51, 52), this finding suggests that strict dosage compensation may not be necessary for all genes on the chromosome. Nevertheless, the large number of genes that escape inactivation and their nonrandom distribution on the chromosome has implications for counseling individuals with structurally abnormal X chromosomes and predicts that aneuploidy for Xp would be more severe than aneuploidy for Xq. Indeed, relatively mild or normal phenotypes have been reported for several cases of 47,XX, i(Xq) (with four copies of Xq arm material and only two copies of Xp), in contrast to the severe mental retardation and dysmorphism seen in females with a 48,XXXX karyotype (with four copies of both Xp and Xq material) (53, 54).
In contrast to the human X chromosome, it has been suggested that fewer genes are expressed from the mouse Xi (55). However, the number of genes that have been tested in mouse is quite low (3, 55, 56) and has been restricted mainly to homologues of the human genes that escape inactivation. This present study of human X-linked genes suggests that location on the X may be a primary determinant as to whether a gene will escape inactivation. Because there have been multiple evolutionary rearrangements between mouse and human X chromosomes (57), comparisons between species should be made with caution. Ultimately, the question of how many mouse X-linked genes escape inactivation will be addressed only by an extensive profile of a large number of mouse genes, selected without reference to their human counterparts.
Finally, with the increasing availability of genomic resources (26, 27, 58), a comprehensive X-inactivation profile of the entire X chromosome is a timely and logical extension of the human X genome project. Such a profile will integrate the nucleotide sequence of the chromosome with a functional understanding of how the chromosome behaves as part of the X-inactivation process. Given the range of patterns of X-linked gene expression demonstrated here and elsewhere (4, 25), a complete profile of the X chromosome will benefit from the two complementary approaches illustrated here, screening both a large number of Xis isolated in somatic cell hybrids and a large panel of nonrandomly inactivated human fibroblast lines. The complete X-inactivation profile should provide relevant clinical information as well as give insight into the genomic and epigenetic organization of the X chromosome.
Supplementary Material
Acknowledgments
We thank M. Schueler and J. Amos-Landgraf for initial screening and mapping of several primer pairs used in these studies, G. Compitello for technical assistance, and K. Boycott and T. Bech-Hansen for providing several unpublished ESTs. This work was supported by National Institutes of Health grant GM45441 to H.F.W.
Abbreviations
- EST
expressed sequence tag
- Xa
active X chromosomes
- Xi
inactive X chromosomes
Footnotes
See commentary on page 14180.
References
- 1.Lyon M F. Nature (London) 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Willard H F. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 719–735. [Google Scholar]
- 3.Disteche C. Trends Genet. 1995;11:17–22. doi: 10.1016/s0168-9525(00)88981-7. [DOI] [PubMed] [Google Scholar]
- 4.Brown C J, Carrel L, Willard H F. Am J Hum Genet. 1997;60:1333–1343. doi: 10.1086/515488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown C J, Willard H F. Adv Dev Biol. 1993;2:37–72. [Google Scholar]
- 6.Davidson R G, Nitowsky H M, Childs B. Proc Natl Acad Sci USA. 1963;50:481–485. doi: 10.1073/pnas.50.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurko O, Hoffman E P, McKee L, Johns D R, Kunkel L M. Am J Hum Genet. 1989;44:820–826. [PMC free article] [PubMed] [Google Scholar]
- 8.Brown R M, Dahl H H M, Brown G K. Genomics. 1989;4:174–181. doi: 10.1016/0888-7543(89)90297-8. [DOI] [PubMed] [Google Scholar]
- 9.Arahata K, Ishihara T, Kamakura K, Tsukahara T, Ishiura S, Baba C, Matsumoto T, Nonaka I, Sugita H. N Engl J Med. 1989;320:138–142. doi: 10.1056/NEJM198901193200302. [DOI] [PubMed] [Google Scholar]
- 10.Beutler E, Yeh M, Fairbanks V F. Proc Natl Acad Sci USA. 1962;48:9–16. doi: 10.1073/pnas.48.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyon M F. Biol Rev Camb Philos Soc. 1972;47:1–35. doi: 10.1111/j.1469-185x.1972.tb00969.x. [DOI] [PubMed] [Google Scholar]
- 12.Schneider-Gadicke A, Beer-Romero P, Brown L G, Nussbaum R, Page D C. Cell. 1989;57:1247–1258. doi: 10.1016/0092-8674(89)90061-5. [DOI] [PubMed] [Google Scholar]
- 13.Fisher E M C, Beer-Romero P, Brown L G, Ridley A, McNeil J A, Lawrence J B, Willard H F, Bieber F R, Page D C. Cell. 1990;63:1205–1218. doi: 10.1016/0092-8674(90)90416-c. [DOI] [PubMed] [Google Scholar]
- 14.Brown C J, Flenniken A M, Williams B R G, Willard H F. Nucleic Acids Res. 1990;18:4191–4195. doi: 10.1093/nar/18.14.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tribioli C, Mancini M, Plassart E, Bione S, Rivella S, Sala C, Torri G, Toniolo D. Hum Mol Genet. 1994;3:1061–1067. doi: 10.1093/hmg/3.7.1061. [DOI] [PubMed] [Google Scholar]
- 16.Esposito T, Gianfrancesco F, Ciccodicola A, D’Esposito M, Nagaraja R, Mazzarella R, D’Urso M, Forabosco A. Genomics. 1997;43:183–190. doi: 10.1006/geno.1997.4797. [DOI] [PubMed] [Google Scholar]
- 17.D’Esposito M, Matarazzo M R, Ciccodicola A, Strazzullo M, Mazzarella R, Quaderi N A, Fujiwara H, Ko M S, Rowe L B, Ricco A, et al. Hum Mol Genet. 1997;6:1917–1923. doi: 10.1093/hmg/6.11.1917. [DOI] [PubMed] [Google Scholar]
- 18.Miller A P, Willard H F. Proc Natl Acad Sci USA. 1998;95:8709–8714. doi: 10.1073/pnas.95.15.8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spencer J A, Watson J M, Graves J A M. Genomics. 1991;9:598–604. doi: 10.1016/0888-7543(91)90352-f. [DOI] [PubMed] [Google Scholar]
- 20.Graves J A, Disteche C M, Toder R. Cytogenet Cell Genet. 1998;80:94–103. doi: 10.1159/000014963. [DOI] [PubMed] [Google Scholar]
- 21.de Conciliis L, Marchitiello A, Wapenaar M C, Borsani G, Giglio S, Mariani M, Consalez G G, Zuffardi O, Franco B, Ballabio A, et al. Genomics. 1998;51:243–250. doi: 10.1006/geno.1998.5348. [DOI] [PubMed] [Google Scholar]
- 22.Franco B, Meroni G, Parenti G, Levilliers J, Bernard L, Gebbia M, Cox L, Maroteaux P, Sheffield L, Rappold G A, et al. Cell. 1995;81:15–25. doi: 10.1016/0092-8674(95)90367-4. [DOI] [PubMed] [Google Scholar]
- 23.Carrel L, Clemson C M, Dunn J M, Miller A P, Hunt P A, Lawrence J B, Willard H F. Hum Mol Genet. 1996;5:391–401. doi: 10.1093/hmg/5.3.391. [DOI] [PubMed] [Google Scholar]
- 24.Miller A P, Gustashaw K, Wolff D J, Rider S H, Monaco A P, Eble B, Schlessinger D, Gorski J L, van Ommen G J, Weissenbach J, et al. Hum Mol Genet. 1995;4:731–739. doi: 10.1093/hmg/4.4.731. [DOI] [PubMed] [Google Scholar]
- 25.Carrel L, Willard H F. Proc Natl Acad Sci USA. 1999;96:7364–7369. doi: 10.1073/pnas.96.13.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D G, Fan J B, Siao C J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 27.Deloukas P, Schuler G D, Gyapay G, Beasley E M, Soderlund C, Rodriguez-Tome P, Hui L, Matise T C, McKusick K B, Beckmann J S, et al. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- 28.Schindelhauer D, Hellebrand H, Grimm L, Bader I, Meitinger T, Wehnert M, Ross M, Meindl A. Genome Res. 1996;6:1056–1069. doi: 10.1101/gr.6.11.1056. [DOI] [PubMed] [Google Scholar]
- 29.Maestrini E, Tamanini F, Kioschis P, Gimbo E, Marinelli P, Tribioli C, D’Urso M, Palmieri G, Poustka A, Toniolo D. Hum Mol Genet. 1992;1:275–280. doi: 10.1093/hmg/1.4.275. [DOI] [PubMed] [Google Scholar]
- 30.Mumm S, Molini B, Terrell J, Srivastava A, Schlessinger D. Genome Res. 1997;7:307–314. doi: 10.1101/gr.7.4.307. [DOI] [PubMed] [Google Scholar]
- 31.Philippe C, Arnould C, Sloan F, van Bokhoven H, van der Velde-Visser S D, Chery M, Ropers H H, Gilgenkrantz S, Monaco A P, Cremers F P. Genomics. 1995;27:539–543. doi: 10.1006/geno.1995.1089. [DOI] [PubMed] [Google Scholar]
- 32.Anderson C L, Brown C J. Am J Hum Genet. 1999;65:699–708. doi: 10.1086/302556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, Maestrini E, Pieretti M, Taillon-Miller P, et al. Nature (London) 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 34.Gianfrancesco F, Esposito T, Montanini L, Ciccodicola A, Mumm S, Mazzarella R, Rao E, Giglio S, Rappold G, Forabosco A. Hum Mol Genet. 1998;7:407–414. doi: 10.1093/hmg/7.3.407. [DOI] [PubMed] [Google Scholar]
- 35.Rao E, Weiss B, Fukami M, Rump A, Niesler B, Mertz A, Muroya K, Binder G, Kirsch S, Winkelmann M, et al. Nat Genet. 1997;16:54–63. doi: 10.1038/ng0597-54. [DOI] [PubMed] [Google Scholar]
- 36.Scheibel K, Weiss B, Wohrle D, Rappold G. Nat Genet. 1993;3:82–87. doi: 10.1038/ng0193-82. [DOI] [PubMed] [Google Scholar]
- 37.Esposito T, Gianfrancesco F, Ciccodicola A, Montanini L, Mumm S, D’Urso M, Forabosco A. Hum Mol Genet. 1999;8:61–67. doi: 10.1093/hmg/8.1.61. [DOI] [PubMed] [Google Scholar]
- 38.Ellison J, Passage M, Yu L-C, Yen P, Mohandas T K, Shapiro L. Somat Cell Mol Genet. 1992;18:259–268. doi: 10.1007/BF01233862. [DOI] [PubMed] [Google Scholar]
- 39.Yen P H, Ellison J, Salido E C, Mohandas T, Shapiro L. Hum Mol Genet. 1992;1:47–52. doi: 10.1093/hmg/1.1.47. [DOI] [PubMed] [Google Scholar]
- 40.D’Esposito M, Quaderi N A, Ciccodicola A, Bruni P, Esposito T, D’Urso M, Brown S D. Mamm Genome. 1996;7:533–535. doi: 10.1007/s003359900157. [DOI] [PubMed] [Google Scholar]
- 41.Vermeesch J R, Petit P, Kermouni A, Renauld J-C, Van Den Berghe H, Marynen P. Hum Mol Genet. 1997;6:1–8. doi: 10.1093/hmg/6.1.1. [DOI] [PubMed] [Google Scholar]
- 42.Migeon B R, Shapiro L J, Norum R A, Mohandas T, Axelman J, Dabora R L. Nature (London) 1982;299:838–840. doi: 10.1038/299838a0. [DOI] [PubMed] [Google Scholar]
- 43.Ellis N, Yen P, Neiswanger K, Shapiro L J, Goodfellow P N. Cell. 1990;63:977–986. doi: 10.1016/0092-8674(90)90501-5. [DOI] [PubMed] [Google Scholar]
- 44.White W M, Willard H F, Van Dyke D L, Wolff D J. Am J Hum Genet. 1998;63:20–28. doi: 10.1086/301922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gartler S M, Goldman M A. Dev Genet. 1994;15:504–514. doi: 10.1002/dvg.1020150609. [DOI] [PubMed] [Google Scholar]
- 46.Migeon B R. Nature (London) 1972;239:87–89. doi: 10.1038/239087a0. [DOI] [PubMed] [Google Scholar]
- 47.Dyer K A, Canfield T K, Gartler S M. Cytogenet Cell Genet. 1989;50:116–120. doi: 10.1159/000132736. [DOI] [PubMed] [Google Scholar]
- 48.Clemson C M, Chow J C, Brown C J, Lawrence J B. J Cell Biol. 1998;142:13–23. doi: 10.1083/jcb.142.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen R S, Canfield T K, Stanek A M, Keitges E A, Gartler S M. Proc Natl Acad Sci USA. 1998;95:5133–5138. doi: 10.1073/pnas.95.9.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kahan B, DeMars R. Proc Natl Acad Sci USA. 1975;72:1510–1514. doi: 10.1073/pnas.72.4.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitchell M J, Wilcox S A, Watson J M, Lerner J L, Woods D R, Scheffler J, Hearn J P, Bishop C E, Graves J A. Hum Mol Genet. 1998;7:429–434. doi: 10.1093/hmg/7.3.429. [DOI] [PubMed] [Google Scholar]
- 52.Jegalian K, Page D C. Nature (London) 1998;394:776–780. doi: 10.1038/29522. [DOI] [PubMed] [Google Scholar]
- 53.Ocrant I, Bangs C D, Johnston K M, Wilson D M, Hintz R L, Rosenfeld R G, Donlon T A. Am J Med Genet. 1989;32:536–539. doi: 10.1002/ajmg.1320320422. [DOI] [PubMed] [Google Scholar]
- 54.King C R, Schimke R N. J Med Genet. 1982;19:467–469. doi: 10.1136/jmg.19.6.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashworth A, Rastan S, Lovell-Badge R, Kay G. Nature (London) 1991;351:406–408. doi: 10.1038/351406a0. [DOI] [PubMed] [Google Scholar]
- 56.Adler D A, Bressler S L, Chapman V M, Page D C, Disteche C M. Proc Natl Acad Sci USA. 1991;88:4592–4595. doi: 10.1073/pnas.88.11.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amar L C, Dandolo L, Hanauer A, Cook A R, Arnaud D, Mandel J L, Avner P. Genomics. 1988;2:220–230. doi: 10.1016/0888-7543(88)90006-7. [DOI] [PubMed] [Google Scholar]
- 58.Collins F S, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.