Abstract
Snf, encoded by sans fille, is the Drosophila homolog of mammalian U1A and U2B′′ and is an integral component of U1 and U2 small nuclear ribonucleoprotein particles (snRNPs). Surprisingly, changes in the level of this housekeeping protein can specifically affect autoregulatory activity of the RNA-binding protein Sex-lethal (Sxl) in an action that we infer must be physically separate from Snf’s functioning within snRNPs. Sxl is a master switch gene that controls its own pre-mRNA splicing as well as splicing for subordinate switch genes that regulate sex determination and dosage compensation. Exploiting an unusual new set of mutant Sxl alleles in an in vivo assay, we show that Snf is rate-limiting for Sxl autoregulation when Sxl levels are low. In such situations, increasing either maternal or zygotic snf+ dose enhances the positive autoregulatory activity of Sxl for Sxl somatic pre-mRNA splicing without affecting Sxl activities toward its other RNA targets. In contrast, increasing the dose of genes encoding either the integral U1 snRNP protein U1-70k, or the integral U2 snRNP protein SF3a60, has no effect. Increased snf+ enhances Sxl autoregulation even when U1-70k and SF3a60 are reduced by mutation to levels that, in the case of SF3a60, demonstrably interfere with Sxl autoregulation. The observation that increased snf+ does not suppress other phenotypes associated with mutations that reduce U1-70k or SF3a60 is additional evidence that snf+ dose effects are not caused by increased snRNP levels. Mammalian U1A protein, like Snf, has a snRNP-independent function.
This study reveals an important molecular aspect of the functioning in vivo of a cog in the basic Drosophila RNA splicing machinery. The conclusions follow from a rather unconventional experimental approach in which dose effects are observed on fly sex determination by wild-type alleles of the gene sans fille (snf) in a genetically sensitized background. These experiments seemed particularly appropriate for an Inaugural Article both because they illustrate a style of analysis characteristic of T.W.C. and because T.W.C. performed much of the work himself. The indirect quality of this kind of genetic analysis and the specialized nature of the tools on which it necessarily relies can make the work challenging to follow. Nevertheless, the genetic approach is worthwhile because it insures in vivo relevance; it facilitates study of molecules that have multiple, often simultaneous functions; it permits manipulation of molecular processes that may be below the level of resolution of more direct biochemical assays; and it minimizes opportunities for experimenter bias.
The two sexes of Drosophila melanogaster are distinguished by a two-fold difference in the dose of a small set of specific X-linked genes—numerator elements—which collectively determine the transcriptional state of the switch gene Sex-lethal (Sxl) through their actions on its “establishment” promoter, SxlPe, during a 45-minute window of time very early in development (1). The double dose of numerator elements in chromosomal females (XX) triggers transcription at SxlPe whereas the single dose in chromosomal males (XY) leaves this promoter off. However, a very different mechanism then operates to maintain the functional state of Sxl that has been triggered by events at SxlPe, and it is in this maintenance process that snf+ gene dose effects are observed.
Maintenance of the sexually determined state for Drosophila is the province of a “maintenance” promoter, SxlPm, which turns on in both sexes just as SxlPe is shutting off in females (1). From this point on, spliced transcripts encoding full-length, feminizing Sxl protein are only generated in XX animals because full-length Sxl protein is required to remove exon 3, whose stop codons would otherwise abort translation of SxlPm-derived mRNAs (Fig. 1). Because this male-specific exon is removed from SxlPe transcripts even in the absence of Sxl, the brief early expression of SxlPe provides a pulse of Sxl protein to XX somatic cells that triggers engagement of a positive autoregulatory feedback loop for the removal of exon 3 from SxlPm transcripts thereafter. Female cells are thereby locked into the exon 2-exon 4 splicing mode that ensures continued production of Sxl. In contrast, male somatic cells lack the SxlPe-derived protein trigger and by default lock into the alternative exon 2-3-4 splicing mode that does not generate full-length Sxl protein. Sxl in females imposes the female rate of X chromosome dosage compensation and induces female differentiation by controlling pre-mRNA splicing and/or translation for more functionally specialized switch-gene targets such as male-specific-lethal-2 (msl-2) and transformer (tra) (1).
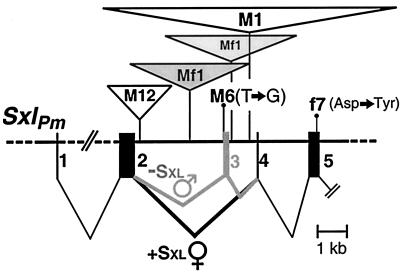
Figure 1.
Molecular lesions associated with four gain-of-function (M-type, male-lethal) and one loss-of-function (f-type, female-lethal) Sxl alleles used in this study. Inverted triangles represent insertions of DNA drawn to scale. The part of Sxl shown includes the region of sex-specific alternative splicing. Sxl protein imposes the exon 2–4 (female) splicing mode, which generates more full-length Sxl protein. Without Sxl, the exon 2–3-4 (male, “nonproductive”) splicing mode ensues, with exon 3 aborting translation. Because these four incompletely constitutive SxlM alleles relax but do not eliminate this autoregulatory requirement for Sxl, the level of female splicing reached in males and thus the degree of developmental disruption caused by the mutants is influenced by factors like Snf that affect positive autoregulation.
Discovery of the involvement of snf in Sxl regulation began with the observation that females heterozygous for both Sxl− and X chromosome deficiencies removing snf were partially sex transformed and/or inviable (2). Attention was drawn to snf1621, a female sterile mutation that interacted with Sxl− just like the deficiencies and was suppressed by gain-of-function (g.o.f.) Sxl alleles (3, 4). However, the inference that snf+ interacts with Sxl+ was confounded by the subsequent discovery that snf1621 is not a straightforward loss-of-function (l.o.f.) allele and that true null snf alleles fail to exhibit dominant synergism with Sxl− (5). Although studies with a partial l.o.f. allele ultimately reestablished that snf is involved in Sxl regulation in the germline and probably also in the soma (6), it could no longer be assumed that the regulatory relationship between snf and Sxl was as specific or as strong as it first appeared or that inferences from snf1621 were straightforward.
Work we present here exclusively with wild-type snf alleles reestablishes that the snf-Sxl regulatory relationship is both strong and specific. Particularly notable is the fact that, although Sxl interacts with a variety of RNAs to control a diversity of functions, only the autoregulatory aspect of Sxl is affected by increased Snf. This observation adds to evidence that the functional relationship between these two genes is very different from that between Sxl and other genes that affect Sxl pre-mRNA splicing (see 7).
Snf is the fly homolog of mammalian U1A and U2B′′ and hence is an integral component of U1 and U2 small nuclear ribonucleoprotein particles (snRNPs) that participates in all pre-mRNA splicing, not just that for Sxl (5). Although RNase-sensitive complexes between Snf and Sxl free of other U1 and U2 snRNP components had been observed, chemical crosslinking studies suggested that these complexes had dissociated from snRNPs, supporting the prevailing idea that Snf participates in Sxl splicing autoregulation only as an integrated component of U1 or U2 snRNPs (6, 8).
Below, we document dose effects of snf+ that are incompatible with a role for Snf in Sxl regulation only as part of U1 and/or U2 snRNPs. The inference of a snRNP-independent role for Snf is bolstered by the striking contrast observed between the behavior of Snf and behavior of two other integral U1 and U2 snRNP proteins.
Materials and Methods
Flies were raised at 25°C in uncrowded conditions on a standard cornmeal, yeast, sucrose, and molasses medium. The criterion for viability was eclosion. All mutations and chromosomes are described in FlyBase (9) except as indicated. U1-70K62 is a P-mobilization, ry−, partial revertant of U1-70K1 that leaves the protein coding region intact (10, 11). U1-70K1 is a ry+-marked P-element insertion 115 bp upstream of the translation start site (10).
Transgenes.
The snf+ transgene also carries a wild-type copy of deadhead, which encodes a thioredoxin homolog (12). The vir+ transgene carries the 6-kb vir transcription unit within a 10-kb genomic fragment and fully rescues vir− (13). The 3.6-kb genomic fragment in the noi+ transgene fully rescues the most extreme noi mutant alleles (14). The U1-70K+ transgene (11) includes 6 kb of Drosophila sequence upstream of the translation start site, then codons for residues 1–352. Codons for the 96-residue Drosophila C terminus have been replaced by those for residues 394–437 of the human U1-70K C terminus followed by a FLAG tag. This transgene complements the recessive lethal U1-70K1.
Molecular Characterization of New SxlM Alleles.
DNA isolated from SxlMf1/Y, SxlM12/Y, and SxlM6/SxlfP7bo animals was scanned for gross DNA changes by Southern blots and PCR amplification (Ampli Taq DNA polymerase from Perkin–Elmer) using a set of 14 primer pairs that provide full coverage of the Sxl transcription unit (−2, 260 to +22, 430 with 0 as the SxlPm transcription start site). Regions including gross changes were amplified by long-range PCR (Elongase Amplification System of GIBCO/BRL), were gel isolated, and then were partially sequenced (Applied Biosystems Prism 377 DNA sequencer with the Big Dye Terminator Cycle Sequencing Ready Reaction kit).
Like all previously reported g.o.f. alleles (15), SxlMf1 and SxlM12 were associated with gross DNA changes in the vicinity of the male-specific exon 3, which is at 9,299–9,489 (Fig. 1). SxlMf1 carried two insertions: a 4.1-kb doc transposon between 8,241 and 8,299 and a 4.3-kb insertion between 9,575 and 9,594 unlike any known transposon but present in more than one copy in wild-type animals. SxlM12 is associated with a hobo insertion between 6,572 and 6,581.
SxlM6 appears to be a T9312G point mutation disrupting the most 3′ U in the polypyrimidine tract for the 3′ splice site of exon 3. SxlM6 has no gross DNA changes. The mutation at 9,312 was found in the course of sequencing most of the 4-kb region (6,395–10,409) between exons 2 and 4 (gaps remain between 8,169 and 8,339 and between 8,929 and 9,049). In connection with the characterization of an unusual SxlM6 male-viable derivative, all SxlM6 Sxl protein coding regions were also sequenced and found to be wild type. The SxlM6 T to G change may promote the exon-2-exon-4 (female) splice by decreasing the strength of either the competing exon-2-exon-3 (male) splice, which relies on this polypyrimidine tract, or the competing exon-3-exon-4 (male) splice, which may conceivably depend on an exon-bridging interaction across exon 3 to help define the 5′ splice site of exon 3.
Phenotypic Characterization of New SxlM Alleles.
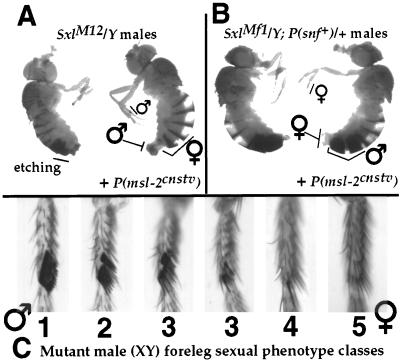
The three new g.o.f. alleles used here were recovered as partial suppressors of sisA1 female-specific lethality (16). SxlM6, like SxlM1, is a dominant, male-specific lethal, suppressible by snf1621, whereas SxlM12 and SxlMf1 are not lethal to males by themselves. SxlM12 is the first case of an insertion between exons 2 and 3 causing (partially) constitutive female-specific expression. SxlM12 disrupts sex-specific regulation much more in abdominal histoblasts than in imaginal discs. This fact is most evident (Fig. 2A, right fly) in the presence of H83M2 (17), a msl-2cnstv transgene partially suppressing SxlM12/Y dosage compensation upsets that otherwise lead to frequent abdominal etching (Fig. 2A, left fly) that obscures sexual phenotype (see below). For SxlM12/Y males with H83M2, 72% (n = 60) of their fifth and sixth hemitergites were completely feminized. In contrast, none of their forelegs (imaginal disc derivatives) were fully female (n = 30), and 77% were entirely male.
Figure 2.
Males showing the reciprocal, tissue-specific feminizing effects of SxlM12 and SxlMf1. For each pair, the male on the right carries a msl-2cnstv transgene to suppress dosage compensation upsets and hence more fully reveal the extent of feminization (see text). SxlM12 predominately affects abdominal histoblast derivatives (tergites and sternites) whereas SxlMf1 predominately affects imaginal disc derivatives (e.g., forelegs and genitalia). Sexual phenotype was quantified by using the scale shown in C for the foreleg sexcomb region. The sexcomb is a row of distinctive male-specific bristles, with each comb-tooth bristle being the product of a single differentiating cell. In these cases, intersexuality was of the mosaic type (see text). Forelegs of SxlMf1 males from the various crosses in Fig. 3 illustrate the full range of sexual transformation observed: (1) none, fully male with at least eight teeth and no breaks; (2) slight, mostly male but with one break and no fewer than seven comb teeth; (3) intermediate, more than three comb teeth and either multiple breaks or fewer than seven teeth; (4) severe, mostly female but with one to three comb teeth; (5) complete, entirely female.
The SxlM12 tissue bias cannot be attributed to the weakness of this g.o.f. allele because the other new male-viable g.o.f. allele, SxlMf1, is even weaker, yet it is biased in the opposite direction: little disruption of abdominal histoblasts, but clear effects on imaginal discs. Because defects of any kind are rare for SxlMf1 males, this bias is more apparent when the phenotype of SxlMf1 is enhanced by increased snf+ dose and H83M2. All such males (Fig. 2C, right fly) have completely female genitalia and forelegs (n = 30 animals) yet nearly normal male abdomens (43% of hemitergites fully masculine, and the rest only slightly feminized). Without H83M2, most of these males lacked part or all of their terminalia (Fig. 2C, left fly). The “M(ale)f(emale)” designation for SxlMf1 reflects the fact that this allele has l.o.f. as well as g.o.f. character, reflected in the low viability and fertility of SxlMf1/Sxl− females (data not shown). Although SxlMf1 carries two insertions, the tissue bias of this allele must be attributable to the doc transposon between exons 2 and 3 because the phenotype of SxlMf2, an allele with the identical doc insertion but no insertion between exons 3 and 4, has somewhat less g.o.f. character but otherwise closely resembles that of SxlMf1 (data not shown).
Results
Increased Zygotic snf+ Dose Enhances SXL Autoregulatory Activity but not SXL Sex Determination or Dosage Compensation Activities.
Positive autoregulation of Sxl was first deduced from phenotypic assays of adults (18). In these assays, products from the mutant allele Sxlf7,M1 induced female-specific expression of an Sxl+ allele in trans in a situation in which that Sxl+ allele would not otherwise have been active because the X chromosome signal normally required for female expression was too low. The M1 mutation in Sxlf7,M1 causes this allele’s pre-mRNA to be spliced in the female mode even in males, but the missense mutation f7 eliminates somatic sex-determination activity and greatly reduces dosage compensation and autoregulation activities of the female proteins produced (Fig. 1) (15). Sxlf7,M1/Y males are phenotypically wild-type even if they also carry an Sxl+ allele in trans because the constitutively expressed autoregulatory activity of a single copy of Sxlf7,M1 is not normally sufficient to induce female-specific expression of Sxl+. However, duplicating region 3E-4F, the region of X in which snf+ resides, was seen to boost autoregulation so that even a single copy of Sxlf7,M1 would activate Sxl+ in males, feminizing them and lowering their viability (19).
Table 1 shows that this 3E-4F dose effect can be attributed to snf. Males with one copy each of Sxlf7,M1 and Sxl+ were fully viable if they carried only the endogenous copy of snf+ (class 3), but with one additional copy of snf+ carried on a transgene (class 4), 75% of the animals died, and two extra copies were invariably lethal (class 5). Two controls established that this male-specific lethality is caused by stimulation by snf+ of an interaction between the mutant and wild-type Sxl alleles (autoregulation) rather than enhancement of dosage compensation activities from either Sxl allele alone. First, Sxlf7,M1 males tolerate even four extra copies of snf+ without ill effects so long as there is no Sxl+ allele present (class 2). Second, males with even an extra copy of Sxl+ are fully viable with four extra copies of snf+ so long as Sxlf7,M1 is absent (class 6). Indeed, Sxl+/Y males are fully viable and somatically wild-type with 10 extra copies of snf+ (data not shown).
Table 1.
Increased zygotic snf+ dose enhances Sxl autoregulation assayed by Sxlf7,M1-induced activation of Sxl+ in trans
| Progeny class (cross*) | Sxl alleles present in males | P(snf+) dose | Relative viability† |
|---|---|---|---|
| 1 (A) | Sxlf7,M1 | 2 | 114% |
| 2 (B) | Sxlf7,M1 | 4 | 70% |
| 3 (A) | Sxlf7,M1 and Sxl+ | 0 | 100% |
| 4 (A) | Sxlf7,M1 and Sxl+ | 1 | 25% |
| 5 (A) | Sxlf7,M1 and Sxl+ | 2 | 0% |
| 6 (C) | Sxl+ and Sxl+ | 4 | 113% |
A, w cm Sxlf7,M1 ct6v; P{snf+w+mC}108/+; Dp(1; 3)sn13a1, cm+Sxl+ct+/+ ☿☿ × ♂♂ w cm Sxlf7,M1ct6v/Y; P{snf+w+mC}108/+. B, w cm Sxlf7,M1ct6v/w, P{snf+w+mC}108 and P{snf+w+mC}19/CyO ☿☿ × ♂♂ w/Y; P{snf+w+mC}108 and P{snf+w+mC}19. C, Binsinscy/Dp(1;1)jnR1-A, y wjt cm Sxl+Sxl+v, P{snf+w+mC}108 and P{snf+w+mC}19/CyO ☿☿ × ♂♂ Dp(1;1)jnR1-A, y wjt cm Sxl+Sxl+v/Y; P{snf+w+mC}108 and P{w+mC snf+}19/+.
† Males relative to these female sibs (n ♀): A, Sxlf7,M1/Sxlf7,M1; P{snf+}/P{snf+}; Dp(Sxl+)/+ (117 = 100%). B, Sxlf7,M1 or +/+; P{snf+}P{snf+}/P{snf+}P{snf+} (97 = 200%). C, Sxl+Sxl+/Sxl+Sxl+; P{snf+}P{snf+}/P{snf+}P{snf+} (31 = 100%).
Because assessment of effects on sexual phenotype that arise from Sxl+ activation in males is complicated by accompanying dosage compensation upsets that reduce cell and organism viability, only data on viability are presented in Table 1 to simplify the presentation. As expected, feminization by the activated Sxl+ allele was observed (data not shown) and was more evident in the presence of H83M2, hereafter designated P(msl-2cnstv), a transgene (17) that reduces dosage compensation upsets caused by inappropriate expression of Sxl in males. P(msl-2cnstv) constitutively expresses the male-specific product of msl-2, thereby counteracting repression of the endogenous msl-2 by full-length Sxl that is a normal part of dosage compensation for XX animals but is inappropriate for XY individuals.
Data in Table 2 address the question of whether the snf+ dose effect that stimulates autoregulation—Sxl’s action on Sxl transcripts—might also stimulate Sxl’s action on transcripts from tra and msl-2, its two other known targets, which control somatic sex determination and dosage compensation respectively (1). In this experiment, effects on Sxl mRNA splicing were not a consideration because the only source of Sxl female-specific protein was a cDNA expression construct without introns that was driven by the hsp70 promoter (20). For males with no extra snf+, (class A), the Sxl transgene reduced viability to 30%, indicating a moderate upset in dosage compensation. Because these transgenic males were heterozygous for tra−, they also displayed an intermediate sexual phenotype, reflecting an incomplete shift in tra transcript splicing toward the female mode (tra+ transgenic males were too strongly feminized to be useful). Such intermediate phenotypes should be maximally sensitive to changes in the effectiveness of Sxl protein; nevertheless, there was no significant enhancement of either feminization or lethality when three extra copies of snf+ were added (class B). Thus, although extra snf+ enhances Sxl autoregulatory activity, it does not enhance Sxl’s sex determination or dosage compensation activities.
Table 2.
Increased snf+ dose affects neither sex determination nor dosage compensation activities of a Sxl cDNA expression construct
| P(snf+) dose | Phenotype of Sxl−/Y; P(hsp70:SxlcF1) tra+/tra− partially feminized males*
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Viability relative to XY siblings with no P(hsp70:Sxl), (n siblings) | Sexual phenotype, n = 25; ±SEM
|
|||||||
| Foreleg, percent intersex (n male sexcomb teeth) | Terminalia
|
Abdominal bristle number, male = 0
|
Pigmentation of hemitergites 5 and 6, percent all female (percent intersex) | |||||
| Anus, percent all female (percent intersex) | Genitalia, percent all female (percent intersex) | (percent relative to pseudofemale control†)
|
||||||
| 7th hemitergite | 6th hemisternite | 7th hemisternite | ||||||
| 0 (class A) | 30% | 100% | 0% | 0% | 10.6 ± 0.4 | 7.9 ± 0.1 | 3.0 ± 0.1 | 86% |
| (90) | (1.6 ± 0.2) | (100%) | (100%) | (45%) | (92%) | (58%) | (14%) | |
| 3 (class B) | 36% | 100% | 12% | 8% | 9.6 ± 0.5 | 8.3 ± 0.2 | 3.1 ± 0.2 | 91% |
| (105) | (1.5 ± 0.2) | (88%) | (92%) | (41%) | (96%) | (61%) | (9%) | |
From the cross: y w cm Sxlf1ct6sn3/Binsinscy, y w sn B; TM3,Ser P{hsp70:SxlcF1 w+mC}19/+ ☿☿ × ♂♂ w/Y; P{snf+w+mC}P{snf+w+mC} tra1 P{snf+w+mC}/tra1. The reduced dose of tra+ moderated the otherwise overly strong feminizing effect of hsp70:SxlcF1.
† Pseudofemales were tra1/tra1 males (XY) feminized by a transgene constitutively expressing female-specific tra+ product.
Materials and Methods describes three new g.o.f. (M) alleles that are used for the autoregulation assays that follow: SxlM12 and SxlMf1 are male viable whereas SxlM6 is male viable only if snf is also mutated. Although Sxlf7,M1 provided a graphic introduction to Sxl autoregulation, these newer alleles are more convenient for such assays because only a single Sxl allele need be present: the Sxl allele stimulated by Sxl protein is also the allele initially generating that Sxl protein. As was true for Sxlf7,M1, the female Sxl protein initially generated by these new g.o.f. alleles undoubtedly arises from a partial relaxation of the normal autoregulatory rules for SxlPm-derived pre-mRNA sex-specific splicing. Ambiguities that would otherwise arise in interpreting the results of such single allele assays are eliminated by the demonstration in Table 2 that snf+ stimulates only the autoregulatory activity of wild-type Sxl. Although females can be used for autoregulation studies, males are used below because the silence of SxlPe in this sex simplifies the analysis.
Increased zygotic dose of snf+ but not vir+ Enhances the Gain-of-Function Character of SxlM12 and SxlMf1.
Table 3 shows that SxlM12 males are extremely sensitive to increased zygotic snf+ dose: Those with the normal single copy of snf+ were fully viable (class 1), but those with one extra copy invariably died (class 2). SxlM12 male viability was also reduced simply by the introduction of a Sxl+ allele in trans (class 3), showing that SxlM12 has some autoregulatory activity even when the dose of snf+ is wild-type; however, the deleterious effect of the added Sxl+ allele was much less than that of the extra copy of snf+.
Table 3.
Increased zygotic snf+ dose kills SxlM12/Y and SxlMf1/Y males
| Progeny class (cross†) | Males
|
Siblings for viability reference*
|
||||
|---|---|---|---|---|---|---|
| Sxl allele(s) | P(snf+) dose | Other key mutations | Relative viability | Zygotic genotype | n | |
| 1 (A) | SxlM12 | 0 | 93% | SxlM12/+ ♀ | 1,044 | |
| 2 (A) | SxlM12 | 1 | <0.1% | SxlM12/+; P(snf+)/+ ♀ | 1,086 | |
| 3 (B) | SxlM12 and Sxl+ | 0 | 33% | X∧X/Y ♀ | 1,101 | |
| 4 (C) | SxlM12 | 2 | vir2f/vir2f | 93% | SxlM12/+; P(snf+) vir2f/+ ♀ | 208 |
| 5 (D) | SxlM12 | 0 | P(vir+)/+ | 118% | +/Y; P(vir+)/+ ♂ | 61 |
| 6 (E) | SxlMf1 | 0 | 97% | X∧X/Y ♀ | 196 | |
| 7 (E) | SxlMf1 | 1 | 57% | X∧X/Y; P(snf+)/+ ♀ | 396 | |
| 8 (E) | SxlMf1 | 2 | 3% | X∧X/Y; P(snf+)P(snf+)/+ ♀ | 234 | |
For crosses A, C, and D, the 100% value for male viability equals this sibling class. For crosses B and E, expected sex-chromosome segregation ratios are not 1:1; hence, a multiplier for estimating the 100% value from the sibling class was determined from control crosses of experimental females to w/Y males [cross B: 0.87 (617♂/709♀); cross E: 1.44 (141♂/98♀)].
† A, w SxlM12 ct6 ☿☿ × ♂♂ w/Y; P{snf+w+mC}108/+. B, y w f: =/y+ct+Y, Sxl+ ☿☿ × ♂♂ w SxlM12ct6/Y. C, w SxlM12ct6/Binsinscy; P{snf+w+mC}108 vir2fbw/CyO ☿☿ × ♂♂ w SxlM12ct6/Y; P{snf+w+mC}108 vir2fbw. D, y w f: =/Y ☿☿ × ♂♂ w SxlM12ct6/Y; P{vir+w+mC}6.2/+. E, y w f: =/Y; P{snf+w+mC}108 and P{snf+w+mC}19/+ ☿☿ × ♂♂ w SxlMf1ct6/Y.
SxlMf1 males also respond to increased snf+ dose. More than half survived with one extra copy of snf+ (Table 3, class 7) and 3% tolerated even two extra doses (class 8). Nevertheless, survivors exhibited a wide range of imaginal disc defects, including thin or missing bristles, as well as small and rough eyes. The feminizing effect of increased snf+ dose was apparent for the foreleg region even without efforts to suppress dosage compensation upsets. This effect is quantified in Fig. 3A. With no extra copy of snf+, 60% of the forelegs of SxlMf1 males had entirely normal sexcombs (class 1), and no foreleg was completely feminized (class 5). The average sex score was 1.4. When one extra copy of snf+ was added, only 1% of the legs were entirely male, whereas 11% were entirely female, and most were intersexual (classes 2–4), for an average sex score of 3.8. Intersexuality was predominately of the “mosaic” type in which individual cells are either male or female rather than intermediate. Mosaic intersexuality signals ambiguity among cells with respect to engagement of the Sxl positive feedback loop, but within individual cells the expression state of Sxl is unambiguously either male or female (1). Because SxlMf1/Y foreleg cells engaging the female-specific Sxl splicing feedback loop do differentiate, such engagement is likely to occur only late in imaginal disc growth when the resulting upsets in dosage compensation would be less deleterious (18, 21). Because snf+ dose effects on viability are less severe and probably occur later for SxlMf1 than for SxlM12, the adult male phenotype of SxlMf1 might reveal subtle changes in the level of Sxl autoregulation that would be obscured by lethality or maternal effects in the case of SxlM12.
Figure 3.
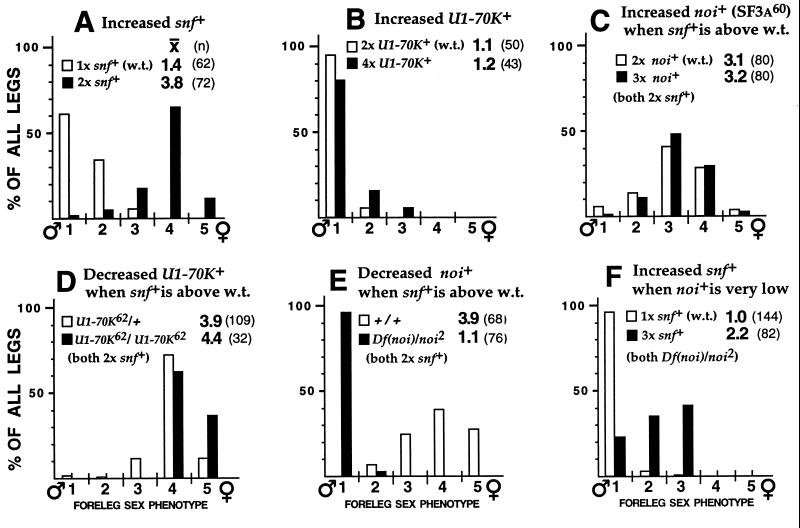
Effects of changes in the dose of genes encoding the U1 snRNP protein U1-70k or the U2 snRNP protein SF3a60 (noi) on the sexual differentiation of SxlMf1/Y male forelegs. Phenotype classes are defined in Fig. 2. Crosses: (A) w SxlMf1ct6/FM7c, B ☿☿ × ♂♂ w/Y; P{snf+w+mC}108/+. (B) w SxlMf1 ct6/w; P{U1-70K+w+mC}Hcter/+ ☿☿ × ♂♂ w/Y; P{U1-70K+w+mC}Hcter/+. (C) w SxlMf1ct6/w; P{noi+w+mC}A71/+ ☿☿ × ♂♂ w/Y; P{snf+w+mC}108. (D) w SxlMf1ct6/w; U1-70K62/SM6β Cy, Roi ☿☿ × ♂♂ w/Y; U1-70K62 P{snf+w+mC}108/+. (E) w SxlMf1 ct6/w; noi2-P{w+mC}/TM2,Ubx ☿☿ × ♂♂ w/Y; P{snf+w+mC}108/+; Ki/Df(3R)noi-D. (F) y w/w SxlMf1ct6; TM3, Sb/noi2-P{w+mC} ☿☿ × ♂♂ w/Y; P{snf+w+mC}108 and P{snf+w+mC}19/+; Df(3R)noi-D./TM3, Sb.
Differences between equivalent genotypes across Fig. 3—such as between A and B controls (open bars)—show how sensitive this assay can be to uncontrolled differences in genetic background. Hence, experiments testing the effects of particular variables should always be designed so that key comparisons can be made among siblings, preferably ones who do not differ with respect to balancer chromosomes.
Studies with virilizer (vir) show that enhancement of SxlM phenotypes by increased gene dose is not a feature of all genes that facilitate Sxl autoregulation. Like snf, vir is a pleiotropic gene essential for both sexes that functions with Sxl in Sxl somatic autoregulation, but, unlike snf, vir also functions with Sxl in somatic sex determination and dosage compensation (22). Evidence that vir is less functionally specific than snf is also provided by the observation that mutations in vir but not in snf affect alternative splicing for Ultrabithorax (7). vir2f is a rare female-specific lethal allele that is like the rare snf1621 allele in suppressing SxlM1 male-specific lethality. Relatedness of snf and vir function is indicated further in Table 3 by the fact that loss of female-specific vir gene function in males suppresses snf+ dose effects: Although a single extra copy of snf+ killed all SxlM12;vir+ males, the same males homozygous for vir2f tolerated even two extra copies of snf+ without ill effects (class 4). Notwithstanding these similarities, increased vir+ dose had no deleterious effect on SxlM12 males (class 5).
Transgenes Used To Increase snf+ Dose Are Half as Active in Males as the Endogenous (X-Linked) snf+ Allele.
To better understand the magnitude of the snf+ dose effect, we used the SxlMf1 foreleg feminization assay to calibrate the activity of the small genomic fragment on the snf+ transgenes relative to that of the endogenous snf+ allele. If the transgenes were functionally equivalent to the endogenous allele, the sexual phenotype of Df(1)snf-J210 SxlMf1/Y; P(snf+)/+ males should be identical to that of snf+ SxlMf1/Y males. Df(1)snf-J210 is a 3.2-kb deletion of the entire snf transcription unit. P(msl-2cnstv) was included in the genotypes not only to minimize distortions caused by dosage compensation upsets but also to make the snf+ SxlMf1/Y phenotype more intersexual and thereby increase the resolution of the assay.
Df(1)snf-J210 SxlMf1/Y; P(snf+)108/+ males were considerably more masculine than snf+ SxlMf1/Y males (sexcomb scores of 1.0, n = 60, vs. 2.1, n = 60, respectively), but Df(1)snf-J210 SxlMf1/Y; P(snf+)108/P(snf+)108 males (2.3, n = 44) were nearly identical to the snf+ control. Because two copies of the transgene match the feminizing activity of the single endogenous allele, the transgene must be half as active as the endogenous locus. This result is expected if the transgene is not dosage compensated. The specific autosomal site of insertion of the transgene did not appear to have a large effect on its somatic activity as assayed by effects on Sxl autoregulation (data not shown).
Increased Maternal as well as Zygotic snf+ Dose Enhance SxlM6.
Experiments with SxlM12 (not shown) suggested that there might also be a maternal effect of increased snf+ dose. A sensitized genotype involving SxlM6 proved to be most effective at establishing this point. Although SxlM6 is lethal to males, its lethality is completely suppressed by snf1621 (Table 4, class 1)—but only if the mothers carry no more than a wild-type dose of snf+. From mothers with a single extra maternal copy of the snf+ transgene, only 15% of snf1621SxlM6/Y sons survived (class 2) even though those sons carried no snf+ allele themselves. No such sons survived from mothers with two extra snf+ copies (class 3). snf1621SxlM6/Y males also carrying P(snf+) died regardless of maternal snf genotype (not shown).
Table 4.
Increased maternal snf+ dose kills sensitized sons
| Progeny class (cross†) | P{snf+} dose in mothers | snf1621SxlM6/Y sons, relative viability* | X∧X/Y siblings as viability reference, n |
|---|---|---|---|
| 1 (A) | 0 | 104% | 270 |
| 2 (B) | 1 | 15% | 296 |
| 3 (C) | 2 | 0% | 137 |
These sons did not carry P{snf+}. The 100% viability values for males were estimated by multiplying the number of female sibs with the same snf+ genotype by the ♂/♀ ratio for the progeny from y w f:=/Y; P{snf+w+mC}108 P{snf+w+mC}19/+ ☿☿ × ♂♂ w1118/Y.
† A, y w f:=/Y ☿☿ × ♂♂ y w snf1621SxlM6/Y. B, y w f:=/Y; P{snf+w+mC}108 or P{snf+w+mC}19/+ ☿☿ × ♂♂ same as A. C, y w f:=/Y; P{snf+w+mC}108 P{snf+w+mC}19/+ ☿☿ × ♂♂ same as A. Females for these three crosses were siblings.
Increased Dose of Genes Encoding Other Integral U1 and U2 snRNP Proteins Has no Effect on SxlM/Y Males.
Data in Table 5 and Fig. 3 show that the dramatic dose effects displayed by snf+ are not a general feature of genes encoding U1 or U2 snRNP proteins. Although SxlM12 males do not tolerate a single extra transgenic copy of snf+, Table 5 shows that they are essentially fully viable even with two extra copies of a transgene encoding U1-70k (class 3). Similarly, an extra copy of a noisette+ (noi) transgene encoding the U2 snRNP integral protein SF3a60 had no significant effect on SxlM12 male viability (class 5). Data in Fig. 3 show that SxlMf1 males are likewise unaffected by either transgene: Neither two extra copies of U1-70K+ (B) nor one extra copy of noi+ (C) had a significant effect on sexual phenotype.
Table 5.
Increased dose of genes ecoding two other U1 and U2 snRNP proteins does not kill SxlM12 males
| Progeny class (cross*) | Transgene(s) present (noi = SF3a60) | SxlM12/Y males relative viability | Sxl+/Y siblings viability reference, n |
|---|---|---|---|
| 1 (A) | none | 108% | 74 |
| 2 (A) | 1 × (U1-70K+) | 101% | 135 |
| 3 (A) | 2 × (U1-70K+) | 73% | 89 |
| 4 (B) | none | 84% | 195 |
| 5 (B) | 1 × (noi+) | 102% | 189 |
A, w SxlM12ct6/w; P{U1-70K+w+mC}Hcter/+ ☿☿ × ♂♂ w/Y; P{U1-70K+w+mC}Hcter/+. B, w SxlM12ct6/w; P{noi+w+mC}A71/+ ☿☿ × ♂♂ w/Y.
A Genetic Test for snRNP-Independence of the Snf-Sxl Functional Interaction.
Is the effect of snf+ dose on Sxl autoregulation caused by an effect on the concentration of functional U1 and/or U2 snRNPs, or, instead, do these dose effects reflect the participation of Snf in Sxl autoregulation as a separate protein independent of snRNPs? Although it seemed unlikely a priori that an increase in the level of just the one snRNP protein would induce the cell to increase the level of these multiprotein complexes above wild-type, a test of this point seemed important.
If snf+ dose effects on Sxl autoregulation were a consequence of increases in the level of U1 or U2 snRNPs, then those dose effects should be eliminated if the concentration of some other component of U1 or U2 snRNPs were reduced to a level that made that component, rather than Snf+, limiting for the formation and/or stability of functional snRNPs. In such a mutant situation, the one copy of snf+ present in the wild-type male should provide more than enough product to accommodate the reduced level of functional snRNPs assembled—a level now determined by some other gene. If, on the other hand, increased snf+ dose does not change the level of available snRNPs, snf+ dose effects should persist even when other mutations limit the level of snRNPs.
This genetic test is complicated by the fact that the sex-specific regulation of Sxl involves a delicate balance between competing reactions in which U1 and U2 snRNPs participate to define alternative splice sites. Hence, by either model, one might expect to see some effect on Sxl autoregulation by a reduction in either U1 or U2 snRNPs sufficiently severe to impact general splicing. Nevertheless, so long as the effect on autoregulation caused by disruption of general splicing is not too severe, the snRNP-independent model for the Snf-Sxl interaction predicts that snf+ dose effects will still be seen, though perhaps with a shift in the phenotypic curve, whereas the snRNP-dependent model predicts they will not be seen.
The design of such a suppression test must necessarily rely on mutations in snRNP protein encoding genes that are only partial l.o.f. (hypomorphic) because null mutations are lethal. The U1-70K62 allele used below (10, 11) is hypomorphic based on the fact that it fails to complement the recessive lethal U1-70K1 and is not fully viable when homozygous (Table 6, class A). A strongly hypomorphic condition for SF3a60 is generated by the noi2/Df(noi) genotype, which causes a developmental delay of several days and aborts spermatogenesis (14). It is important to note that the mutant phenotypes of these hypomorphic alleles must reflect changes in the quantity rather than the quality of the snRNP proteins because the lesions do not disrupt protein coding sequences.
Table 6.
Mutations lowering levels of other integral U1 or U2 snRNP proteins do not suppress snf+ dose effects on SxlM12/Y male viability
| Progeny class (Cross†) | Experimental males
|
Control Sxl+ male sibs*, n | |||
|---|---|---|---|---|---|
| Sxl allele | snRNP locus genotype | P{snf+} copies | Relative viability | ||
| 1 (A) | SxlM12 | U1-70K62/U1-70K62 | 1 | 1% | 71 |
| 2 (A) | SxlM12 | +/+ | 0 | 84% | 117 |
| 3 (A) | Sxl+ | U1-70K62/U1-70K62 | 1 | 44% | 161 |
| 4 (B) | SxlM12 | noi2/Df(noi) | 1 | 0% | 240 |
| 5 (B) | SxlM12 | noi2/Df(noi) | 0 | 80% | 225 |
| 6 (B) | Sxl+ | noi2/Df(noi) | 1 | 90% | 267 |
For classes 3 and 6, these are U1-70K62/+ and noi2/+, respectively. For all others, autosomal genotypes match experimentals. Sxl genotype was inferred from the closely linked (0.9 cM) ct marker, except that rare Sxl+-ct recombinants were recognized by their lack of the abdominal etching diagnostic for SxlM12.
† A, w SxlM12ct6/w; U1-70K62/SM6b, Cy Roi ☿☿ × ♂♂ w/Y; U1-70K62 P{snf+w+mC}108/+. B, w SxlM12ct6/w; noi2{PlacW,w+mC}/TM2, Ubx ☿☿ × ♂♂ w/Y; P{snf+w+mC}108/+; Df(3R)noi-D/st Ki pp ry.
Because little is known about the regulation of snRNP levels in vivo, can one ignore complex alternative scenarios in which a mutation lowering the level of the other snRNP protein would not eliminate a dose effect of Snf on snRNPs but instead would just make both proteins rate-limiting for active snRNP formation? Fortunately, the fact that the two hypomorphic alleles described above have measurable phenotypes allows a test of this possibility. If the U1-70K and noi mutant phenotypes reflect lowered levels of U1 and U2 snRNPs respectively, and if the dose of snf+ still influences U1 or U2 snRNP levels when one or the other of these snRNP proteins is clearly limiting, then increasing snf+ dose should partially suppress either the U1-70K or the noi hypomorphic phenotype.
Lowered Levels of the U1 snRNP Protein U1-70k Do Not Block snf+ Dose Effects on Sxl Autoregulation.
Data in Table 6 show that snf+ dose effects persist even when the level of U1-70k is reduced, thus favoring the snRNP-independent model for the Snf-Sxl interaction. Increased snf+ dose kills SxlM12/Y males even when they are homozygous for l.o.f. mutations predicted to lower the level of U1 snRNPs (class 1). One SxlM12 male mutant for U1-70K did manage to escape the lethal effect of increased snf+ dose in this experiment, but the fact that he was much more malformed and feminized than any of his brothers makes it unlikely that his survival signals any significant suppression of the snf+ dose effect. Extra copies of snf+ do not appear to suppress the U1-70K62 phenotype because the viability of Sxl+ males homozygous for U1-70K62 and also carrying an extra copy of snf+ (44%, class 3) was no higher than that observed for males with no extra copy of snf+ generated in a cross between U1-70K62/CyO males and females (data not shown).
The extent of feminization of SxlMf1 males provides a convenient assay for effects of snRNP mutations that might be too weak to rescue SxlM12 males. Even by this sensitive assay (Fig. 3D), impairment of U1-70K function failed to suppress the snf+ dose effect: SxlMf1/Y males homozygous for U1-70K62 were at least as feminized by an extra copy of snf+ as males that were heterozygous for this recessive mutant allele (sexcomb scores of 4.4 vs. 3.9 respectively).
Lowered Levels of the U2 snRNP Protein SF3a60 Affect Sxl Autoregulation but Do Not Eliminate snf+ Dose Effects.
Table 6 shows that SxlM12/Y males mutant for noi and hence deficient for SF3a60 are as sensitive to the killing effects of increased snf+ dose (compare classes 4 and 5) as those that are noi+ (Table 3, class 2). Moreover, increased Snf does not suppress noi: The sterility and developmental delay observed for Sxl+/Y mutant noi males with an extra copy of snf+ (class 6) were as severe as for their noi mutant sibs wild-type for snf (data not shown).
The sensitive phenotypic assay with SxlMf1 gave a strikingly different result: Mutations in noi prevented feminization that would otherwise have accompanied the addition of an extra copy of snf+ (Fig. 3E). However, data in Fig. 3F show that this block stems from a shift in the snf+ dose–response curve rather than elimination of snf+ dose effects: Two extra copies of snf+ were sufficient to feminize SxlMf1 males even in a noi mutant background. These effects of noi mutations were strictly recessive (data not shown). The same two extra copies of snf+ did not reduce the developmental delay or sterility caused by noi.
Lowering SF3a60 activity reduced but did not eliminate the dose effect of snf+ on viability as well as sex: The partially feminized SxlMf1/Y; Df(noi)/noi2 males with two extra copies of snf+ shown in Fig. 3F (black bars) were 31% as viable as their Df(noi)/noi2 brothers with no extra snf+ (white bars). Recall that SxlMf1 males with two extra copies of snf+ but not mutant for noi were only 3% as viable as controls (Table 3, class 8). In summary, reducing SF3a60 does impair Sxl alternative splicing, but increasing the level of Snf enhances autoregulation even in this splicing-impaired background, consistent with predictions for a snRNP-independent role for Snf.
Discussion
From the effects of raising the dose of the wild-type snf gene above normal levels, we infer that the integral snRNP protein encoded by snf acts outside of the snRNP in controlling pre-mRNA splicing for Sxl, the master regulator of fruit fly sex determination. One would not pick snf as a gene likely to display phenotypic effects of increased dose because snf encodes only one of many proteins that make up U1 and U2 snRNPs. In the genetically sensitized system used here to reveal snf+ dose effects, these complex multimeric assemblies are at levels that suffice for all of the needs of the organism. We show that such dose effects are not typical of integral snRNP proteins because increasing the dose of the gene encoding the U1 protein U1-70k or that encoding the U2 protein SF3a60 had no effect on Sxl autoregulation. This negative result is particularly meaningful in light of our demonstration that lowering the level of SF3a60 does interfere with Sxl autoregulation but does not eliminate the effects of increased snf+ dose.
Could the influence of increased snf+ dose reflect a quirk of fruit fly regulatory circuitry in which snRNP levels are tied to U1A/U2B" levels? A priori, this would seem a disadvantageous strategy for the fly to use. Because most RNA splicing involves a sensitive balance between competing potential splice sites that one might expect to be affected by changes in the levels of these two snRNPs, one would expect regulatory circuitry to insulate the general splicing system from perturbation, not tie it to a single gene product in this way. Moreover, because we observe a maternal effect of increased snf+ dose that is nearly as striking as the zygotic dose effect, such a sensitive regulatory connection would have to operate both maternally during oogenesis to govern subsequent snRNP levels in the embryo and zygotically to govern snRNP levels at later stages. Two experimental observations argue against such a tie to snf. First, although we see striking effects on Sxl by even a single extra copy of snf+ in various sensitized situations, males and females wild-type for Sxl can carry as many as 10 extra copies of the same snf+ construct and be fully viable. Second, and most damaging for this unlikely hypothesis, increasing snf+ dose does not suppress the mutant phenotypes caused by decreasing the level of U1-70k or SF3a60.
If, on the other hand, Snf functions specifically in Sxl autoregulation not as an integral component of U1 or U2 snRNPs but as an individual protein, the snf+ dose effects would not be reflecting changes in functional snRNP levels, but simply the established tendency of metazoan gene product levels to be roughly proportional to structural gene dose. Dose effects in this case would be indicating Snf’s key participation in the process by which Sxl protein inhibits the male Sxl pre-mRNA splice by binding to RNA, a process likely to directly involve relatively few proteins.
The fly’s use of U1A/U2B′′ as an alternative splicing factor in sex determination would not be the first case of an integral spliceosomal protein acting outside of the snRNPs. Non-snRNP mammalian U1A negatively regulates its level by binding to sites in U1A pre-mRNA to block polyadenylation (23). U1A may also function more generally to couple splicing and 3′ end formation (24, 25). Such pleiotropy raises the possibility of an undiscovered world of biological functions for integral snRNP proteins operating as free agents. Because these proteins also have essential housekeeping functions, their other roles might not be easily revealed in vivo. Positive autoregulation gives the Sxl assay used here an extremely nonlinear character that surely facilitated study of biochemical effects that might otherwise have been too small to detect.
How might Snf be involved in Sxl autoregulation? There is evidence that a small fraction of Snf is in proximity to Sxl on RNA (8). Previous models (6, 8) assumed that any interaction between Snf and Sxl occurred with Snf acting as part of U1 or U2 snRNPs and that this interaction was preceded by Sxl binding to pre-mRNA between exons 3 and 4 to block the male splice. Through an interaction between Snf within the snRNPs and Sxl bound to RNA surrounding the male exon, an abortive presplicing complex for exon-3 was proposed to form, allowing the alternative exon 2–4 female-specific splice to proceed by default.
In light of the data reported here, it now appears that Snf may bind with Sxl to pre-mRNA flanking the male exon, perhaps each facilitating or stabilizing the other’s binding. By this model, it would not be surprising if the consequences of such an association were most significant at low concentrations of Sxl, such as those which surely prevail in the sensitized situations describe here. In addition to stabilizing Sxl binding, or even as an alternative to it, non-snRNP Snf associating with Sxl may be necessary to inhibit further spliceosomal complex assembly around the male-specific exon 3. Perhaps independent Snf protein interacting with Sxl bound to the pre-mRNA interferes with an essential association that Snf in the snRNPs themselves would need to have with other splicing factors to define exon 3 splice sites.
The dose-sensitive involvement of snf in somatic Sxl autoregulation described here is one of the strongest similarities between the regulation of sex-specific gene expression in the soma and in the germ line. It was shown earlier that simply increasing the dose of snf+ in an otherwise wild-type fly can trigger female-specific splicing of Sxl transcripts in male germ cells (26). For the soma, increasing snf+ alone will not suffice to engage the autoregulatory splicing loop; however, somatic Sxl regulation can be made nearly as sensitive to increased snf+ dose as germline Sxl regulation by alleles such as SxlMf1 that are so weak that they do not lower male viability or fertility by themselves. The ease with which Sxl splicing control in the soma can be made to respond to the dose of RNA splicing factors favors the idea that the ancestral system controlling the sex-specific expression of Sxl in both the germline and the soma might have been based entirely on dose effects of RNA splicing factors.
In view of the central and remarkably specific role snf plays in controlling sex-specific expression of Sxl, it is a curious coincidence that the only genus known to use Sxl as a master sex switch is also the only genus with a species known to use a single protein, Snf, for tasks that two proteins, U1A and U2B′′, handle in species as diverse as potatoes and humans (27). Learning how closely the evolution of Sxl as the master sex-determination gene for Drosophila was paralleled by the evolution of this difference in integral U1 and U2 snRNP proteins might suggest what the driving forces were that led to both changes.
Acknowledgments
Authors are listed in order of contribution. We are extremely grateful to Drs. S. Mount, B. Suter, H. Salz, D. Pauli, D. Bopp, and R. Nöthiger for generously providing mutants and transgenes, often before publication. D.Z.R. thanks K. Breger for assistance with crosses. We appreciate the many helpful comments on the manuscript given by Drs. B. Meyer and D. Rio and by current members of Clinelab: J. Dines, L. Megna, L. Wrischnik, and L. Sefton. This work was supported by National Institutes of Health Grant GM23468 to T.W.C.
Abbreviations
- l.o.f.
loss-of-function
- g.o.f.
gain-of-function
- snRNP
small nuclear ribonucleoprotein particle
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected April 30, 1996
References
- 1.Cline T W, Meyer B J. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 2.Steinmann-Zwicky M, Nöthiger R. Cell. 1985;42:877–887. doi: 10.1016/0092-8674(85)90284-3. [DOI] [PubMed] [Google Scholar]
- 3.Steinmann-Zwicky M. EMBO J. 1988;7:3889–3898. doi: 10.1002/j.1460-2075.1988.tb03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliver B, Perrimon N, Mahowald A. Genetics. 1988;120:159–171. doi: 10.1093/genetics/120.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flickinger T W, Salz H K. Genes Dev. 1994;8:914–925. doi: 10.1101/gad.8.8.914. [DOI] [PubMed] [Google Scholar]
- 6.Salz H K, Flickinger T W. Genetics. 1996;144:95–108. doi: 10.1093/genetics/144.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnette J M, Hatton A R, Lopez A J. Genetics. 1999;151:1517–1529. doi: 10.1093/genetics/151.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshpande G, Samuels M E, Schedl P D. Mol Cell Biol. 1996;16:5036–4047. doi: 10.1128/mcb.16.9.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.FlyBase. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tseng C, Nikiforova O, Mancebo R, Mount S. Annual Drosophila Research Conference. Vol. 39. Washington, DC: Genetics Soc. Am.; 1998. p. a159. [Google Scholar]
- 11.Mancebo R. Ph.D. thesis. New York: Colombia Univ.; 1995. [Google Scholar]
- 12.Larochelle S, Pandur J, Fisher R P, Salz H K, Suter B. Genes Dev. 1998;12:370–381. doi: 10.1101/gad.12.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schütt C, Hilfiker A, Nöthiger R. Development (Cambridge, UK) 1998;125:1501–1507. doi: 10.1242/dev.125.8.1501. [DOI] [PubMed] [Google Scholar]
- 14.Meyer V, Oliver B, Pauli D. Mol Cell Biol. 1998;18:1835–1843. doi: 10.1128/mcb.18.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein M, Lersch R A, Subrahmanyan L, Cline T W. Genetics. 1995;139:631–648. doi: 10.1093/genetics/139.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbash D A, Cline T W. Genetics. 1995;141:1451–1471. doi: 10.1093/genetics/141.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley R L, Solovyeva I, Lyman L M, Richman R, Solovyev V, Kuroda M I. Cell. 1995;81:867–877. doi: 10.1016/0092-8674(95)90007-1. [DOI] [PubMed] [Google Scholar]
- 18.Cline T W. Genetics. 1984;107:231–277. doi: 10.1093/genetics/107.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cline T W. Genetics. 1988;119:829–862. doi: 10.1093/genetics/119.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell L R, Horabin J I, Schedl P, Cline T W. Cell. 1991;65:229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- 21.Belote J M. Genetics. 1983;105:881–896. doi: 10.1093/genetics/105.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilfiker A, Amrein H, Dübendorfer A, Schneiter R, Nöthiger R. Development (Cambridge, UK) 1995;121:4017–4026. doi: 10.1242/dev.121.12.4017. [DOI] [PubMed] [Google Scholar]
- 23.Boelens W C, Jansen E J, van Venrooij W J, Stripecke R, Mattaj I W, Gunderson S I. Cell. 1993;72:881–892. doi: 10.1016/0092-8674(93)90577-d. [DOI] [PubMed] [Google Scholar]
- 24.Gunderson S I, Vagner S, Polycarpou-Schwarz M, Mattaj I W. Genes Dev. 1997;11:761–773. doi: 10.1101/gad.11.6.761. [DOI] [PubMed] [Google Scholar]
- 25.Lutz C S, Cooke C, O’Connor J P, Kobayashi R, Alwine J C. RNA. 1998;4:1493–1499. doi: 10.1017/s1355838298981183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hager J H, Cline T W. Development (Cambridge, UK) 1997;124:5033–5048. doi: 10.1242/dev.124.24.5033. [DOI] [PubMed] [Google Scholar]
- 27.Polycarpou-Schwarz M, Gunderson S I, Kandels-Lewis S, Séraphin B, Mattaj I W. RNA. 1996;2:11–23. [PMC free article] [PubMed] [Google Scholar]