Abstract
The recognition of mycobacterial cell wall components causes macrophages to secrete tumor necrosis factor α (TNF-α) and other cytokines that are essential for the development of a protective inflammatory response. We show that toll-like receptors are required for the induction of TNF-α in macrophages by Mycobacterium tuberculosis. Expression of a dominant negative form of MyD88 (a signaling component required for toll-like receptor signaling) in a mouse macrophage cell line blocks TNF-α production induced by M. tuberculosis. We identify toll-like receptor-2 (TLR2) as the specific toll-like receptor required for this induction by showing that expression of an inhibitory TLR2 (TLR2-P681H) blocks TNF-α production induced by whole M. tuberculosis. Further, we show that TLR2-dependent signaling mediates responses to mycobacterial cell wall fractions enriched for lipoarrabinomannan, mycolylarabinogalactan–peptidoglycan complex, or M. tuberculosis total lipids. Thus, although many mycobacterial cell wall fractions are identified to be inflammatory, all require TLR2 for induction of TNF-α in macrophages. These data suggest that TLR2 is essential for the induction of a protective immune response to mycobacteria.
Keywords: MyD88, Mycobacterium tuberculosis, tumor necrosis factor α
Mycobacterium tuberculosis is a leading cause of death worldwide, and the incidence of the disease is increasing (1). M. tuberculosis infection induces fever, night sweats, and profound weight loss, and these symptoms are attributed to an increased production of the proinflammatory cytokines, tumor necrosis factor α (TNF-α), and IL-1β by activated mononuclear phagocytes (2, 3). TNF-α production is an early and important event that leads to granuloma formation and a protective host response (4, 5). A wide variety of structurally diverse components derived from mycobacterial cell walls stimulate TNF-α production in macrophages. A key question is whether these structurally diverse mycobacterial components activate macrophages through separate or identical pathways.
The recognition system for the stimulatory mycobacterial cell wall component, liparabinomannan (LAM), has not been defined fully, but it appears to share much in common with the recognition system for lipopolysaccharide (LPS), a stimulatory component of Gram-negative bacteria. Macrophage responses to LPS and LAM are enhanced by LPS-binding protein (LBP), a molecule that binds LPS and transfers it to CD14 (6–9). CD14 is a glycosylphosphatidylinositol-linked cell surface receptor that is required for LPS and LAM signaling (10–12). Together, CD14 and LBP allow cells to respond to extremely low concentrations of LPS and LAM (6, 9). Although CD14 binds directly to LPS, LAM, and other microbial products (11), binding to CD14 is not necessarily coupled to inflammatory signaling. Inflammatory responses are not triggered when CD14 binds to self-phospholipids or apoptotic cells (13, 14).
Despite the common requirement for CD14 in LPS- and LAM-induced signaling, important differences exist. Macrophages from C3H/HeJ mice respond poorly to stimulation by LPS, yet they secrete proinflammatory mediators normally in response to mycobacterial products (15). Conversely, Chinese hamster ovary (CHO) fibroblasts transfected with CD14 respond to LPS (16), but fail to respond to LAM (9). CD14 is an extracellular protein that is unable to interact directly with cytoplasmic signaling components. Recent discoveries suggest that CD14 interacts with toll-like receptors to transmit a signal (17–19). Toll-like receptors comprise a family of innate immune signaling receptors that are related to the Drosophila Toll protein, a molecule that is implicated in defense against fungal infection in the fly (20). Six mammalian homologues of Toll have been identified (21–23). The roles of these receptors in mammalian innate immunity are just beginning to be elucidated. Transfection of either human toll-like receptor-2 (TLR2) or TLR4 into the LPS-unresponsive cell line, HEK 293, permits CD14-dependent NF-κB activation in response to LPS (17–19). The role for TLR4 in LPS-induced activation of macrophages is supported by the demonstration that a point mutation in the gene for TLR4 is associated with LPS hyporesponsiveness in the C3H/HeJ mouse (24, 25), and deletion of the gene results in a similar phenotype (26). The significance of TLR2 in LPS signaling is unclear given the presence of a presumably normal TLR2 gene in the C3H/HeJ mouse. A role for TLR2 in mediating responses from other pathogens is suggested by the observation that lipoteichoic acid and peptidoglycan from Gram-positive bacteria stimulate NF-κB in TLR2-transfected HEK 293 cells (27, 28). Also, specific membrane lipoproteins from Borrelia burgdorferi, M. tuberculosis, and Treponema pallidum signal through TLR2 (29, 30). We recently have demonstrated that toll-like receptors are recruited to phagosomes in macrophages, where they mediate the identification of engulfed pathogens; TLR2 is required for the recognition of Gram-positive bacteria and yeast, whereas TLR4 mediates recognition of Gram-negative bacteria (31).
In this report we demonstrate that upon exposure to M. tuberculosis, macrophages are stimulated to produce the proinflammatory cytokine, TNF-α, in a toll-like receptor-dependent manner. We identify TLR2 as the principle mediator of the proinflammatory signal induced by whole M. tuberculosis. We demonstrate further that three structurally diverse fractions of the mycobacterial cell wall all induce TNF-α production via TLR2. Thus, TLR2 is the principle mediator of macrophage activation in response to mycobacteria, and the signaling mechanism is clearly distinct from LPS-induced signaling that occurs via TLR4.
Materials and Methods
Materials.
Unless stated otherwise, all reagents were from Sigma. Virulent and avirulent, heat-killed M. tuberculosis (H37Rv and H37Ra strains), lipoarabinomannan (AraLAM from rapidly growing mycobacteria species and ManLAM from M. tuberculosis H37Rv), mycolylarabinogalactan–peptidoglycan complex (mAGP), and M. tuberculosis total lipids were obtained from John Belisle through the TB Research Materials and Vaccine Testing Contract (National Institutes of Health, National Institute of Allergy and Infectious Diseases NO1 AI-75320) (32). The mAGP had approximately 5 pg of LPS in 10 μg mAGP as determined by the Chromogenic Limulus Amebocyte Lysate Test (BioWhittaker). In some experiments, polymyxin B (10 μg/ml) was added to the mAGP to inhibit any possible effect from LPS contamination. No differences were detected between polymyxin B-treated and untreated mAGP, indicating that the small amount of LPS detected in the mAGP was not enough to stimulate TNF-α production in RAW cells (data not shown). All other reagents had no detectable LPS.
The mouse macrophage cell line used in this report, RAW-TT10 (31), is a clone of RAW 264.7 (ATCC no. TIB-71) transfected with an expression vector driving synthesis of the Tet-activator protein, a tetracycline-regulatable transcriptional activator (33) that directs expression, in the absence of tetracycline, from the tetracycline-regulated promoter used in the TLR2 and MyD88 expression vectors.
DNA Expression Vectors.
The expression vector pTIGZ2+ (31) uses a tetracycline-regulated promoter (from pTetSplice; GIBCO/BRL) to direct transcription of a bicistronic mRNA encoding the protein of interest followed by a cap-independent translational enhancer region (from pCITE; Novagen) driving translation of enhanced green fluorescent protein (GFP; CLONTECH).
For single-cell TNF-α assays, the coding region of wild-type mouse TLR2 [tagged at its amino terminus with a hemagglutinin (HA) epitope] was cloned into TIGZ2+ (31). The HA-tagged TLR2 dominant negative P681H mutation was generated by PCR, confirmed by sequencing, and cloned into TIGZ2+. The MyD88 dominant negative construct was generated by amplifying the MyD88 C-terminal coding region (amino acids 146–296) by PCR from cDNA of RAW 264.7 cells and cloning the fragment into TIGZ2+ (31).
RAW Cell Transfection.
RAW-TT10 cells were transfected by using a previously described method (34). Briefly, 5 × 106 cells in 0.2 ml of culture medium were mixed together with 10 μg of DNA in 50 μl of PBS in a 0.4-cm gap electroporation cuvette (Bio-Rad). The cells were electroporated in a Gene Pulser apparatus (Bio-Rad) set at 960 μF and 280 V and transferred to 5 ml of fresh medium. The cells were pelleted, resuspended in fresh medium, and transferred to six wells of a 24-well tissue culture plate. After allowing 2 hr for the cells to adhere to the plate, the medium was changed, and experiments were performed 24 hr after transfection. Tetracycline was always absent from the media, resulting in strong activity of the tetracycline-regulated promoter.
Detection of Intracellular TNF-α.
Transiently transfected RAW-TT10 cells were stimulated with the following mycobacteria and mycobacterial products: M. tuberculosis (H37Rv) (100 μg/ml), M. tuberculosis (H37Ra) (100 μg/ml), AraLAM (10 μg/ml), ManLAM (10 μg/ml), mAGPs (100 μg/ml), M. tuberculosis total lipids (100 μg/ml), or LPS (Salmonella minnesota R595, List) (10 ng/ml). The cells were incubated with the indicated stimulant for 4 hr at 37°C in the presence of 5 μg/ml brefeldin A to accumulate intracellular TNF-α. After blocking Fc receptors with 2.4G2 hybridoma supernatant, the cells were fixed with 4% paraformaldehyde in PBS and permeabilized and stained with phycoerythrin-conjugated anti-mouse TNF-α (PharMingen) diluted in 1% FCS and 0.1% saponin in PBS. After two washes, the cells were analyzed by flow cytometry by using a FACScan (Becton Dickinson) and winmdi software (Joseph Trotter, Scripps, La Jolla, CA). Under these conditions, 5–20% of the transiently transfected cells expressed the transgene (as identified by GFP fluorescence). Gating on these cells allowed TNF-α production by transgene-expressing cells to be compared directly with TNF-α production by identically treated cells in the same sample that did not express the transgene. All data presented are representative of at least three independent experiments.
Luciferase Assays.
Cells were transiently transfected, and induction of NF-κB activity was measured as described (31). CHO-K1 cells were transfected with 2 μg of endothelial leukocyte adhesion molecule-1 (ELAM-1) firefly luciferase reporter construct and 0.2 μg of a construct directing expression of Renilla luciferase under control of the constitutively active β-actin promoter, together with 1 μg of murine TLR2 or CD14 expression construct, as indicated in the text. The cells were stimulated with M. tuberculosis as described above, and luciferase activity was measured by using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Background ELAM-luciferase activity was subtracted, and the data are presented as the mean ± SD of triplicate samples.
Results and Discussion
Because macrophages are critical to M. tuberculosis pathogenesis, we used a mouse macrophage cell line to analyze the role of toll-signaling pathways in inflammatory responses to M. tuberculosis. First, we analyzed the role of the toll-like receptor signaling component, MyD88, in the induction of TNF-α by M. tuberculosis. MyD88 is a cytoplasmic adapter protein that links toll-like receptors to IRAK (IL-1 receptor-associated kinase), a serine kinase required for the signal leading to NF-κB activation (35–38). MyD88 associates directly with toll receptors through its C-terminal toll homology domain. When expressed by itself, the C-terminal domain of MyD88 acts as a dominant negative inhibitor of TLR4 and IL-1R signaling presumably by preventing IRAK from associating with the receptors (35–37). By virtue of the conservation of toll homology domains, it is presumed that MyD88 is a common signaling component of all toll-like receptors. Thus, to determine whether M. tuberculosis-induced TNF-α production was mediated by toll-like receptors, we tested the ability of the C terminus of MyD88 (MyD88-DN) to block this stimulation.
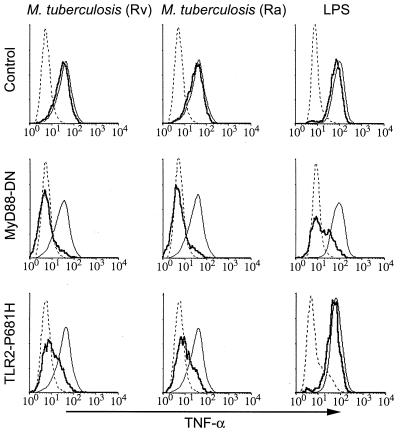
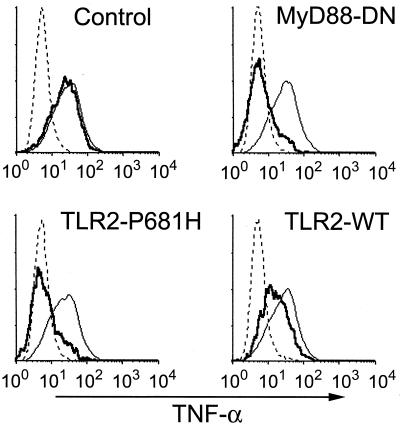
The mouse macrophage cell line RAW-TT10 was transiently transfected with an expression vector encoding a bicistronic mRNA transcript directing expression of both dominant negative MyD88 and GFP. These cells were stimulated with whole, heat-killed, virulent M. tuberculosis (H37Rv), and production of TNF-α was measured by intracellular staining. During analysis of 100,000 cells by flow cytometry, GFP expression correlated precisely with MyD88-DN expression as expected for two products produced from the same mRNA transcript (31). This allowed direct comparison of TNF-α production in MyD88-DN-expressing cells (GFP-positive) with cells in the same sample that did not express the inhibitory protein. Thus, cells that did not express the inhibitory protein (GFP-negative cells) produced TNF-α (Fig. 1). However, MyD88-DN expression (GFP-positive cells) strongly inhibited mycobacteria-induced TNF-α production to background levels (Fig. 1). Expression of MyD88-DN also potently blocked TNF-α induction by avirulent M. tuberculosis (H37Ra) (Fig. 1). The inhibition of toll-like receptor signaling by MyD88-DN was confirmed by the observation that dominant negative protein expression also inhibited TNF-α production in response to LPS, a known stimulator of TLR4 (17, 24–26) (Fig. 1). Transfection of macrophages with a control vector (expressing only GFP) had no effect on TNF-α induction by any of the stimuli (Fig. 1 Top). Thus, mycobacteria stimulate TNF-α production via a MyD88-dependent process, implicating the involvement of a toll-like receptor.
Figure 1.
M. tuberculosis-induced TNF-α production depends on TLR2. RAW-TT10 cells transiently transfected with a control plasmid (Top), MyD88-dominant negative (MyD88-DN, Middle), or TLR2-P681H (Bottom) were analyzed for induction of TNF-α after stimulation with virulent M. tuberculosis (H37Rv), avirulent M. tuberculosis (H37Ra), or LPS. The thin, solid lines indicate TNF-α produced in cells not expressing the transgene, and the bold lines indicate TNF-α produced in cells expressing the indicated transgene. The dotted lines indicate background, unstimulated TNF-α production. In each case, the y axis represents relative cell number.
Because macrophages from mice bearing a dysfunctional TLR4 secrete TNF-α normally in response to mycobacterial products (15), it is unlikely that TLR4 contributes to macrophage activation by mycobacteria. We hypothesized that TLR2, another toll-like receptor implicated in LPS signaling (18, 19), is the toll-like receptor responsible for mycobacterial activation of macrophages. We demonstrated recently that a mutation in mouse TLR2 (TLR2-P681H), a mutation that is analogous to the naturally occurring mutation in the C3H/HeJ mouse TLR4 gene, acts as a dominant negative inhibitor of TLR2 signaling (31). Expression of TLR2-P681H in RAW cells inhibited TNF-α induction in response to both virulent and avirulent M. tuberculosis. Expression of TLR2-P681H had no effect on TNF-α induction by LPS (Fig. 1 Bottom). These data indicate that TLR2 is required for the detection of mycobacteria by macrophages and clearly demonstrate functional specificity of toll-like receptors for particular ligands. Interestingly, whereas dominant negative MyD88 completely blocked M. tuberculosis-induced TNF-α production, inhibition by dominant negative TLR2 was incomplete, suggesting that other toll-like receptors may participate in the detection of mycobacteria by macrophages.
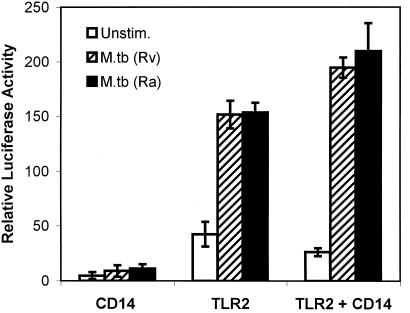
To confirm a role for TLR2 in mycobacteria-induced signaling, we measured mycobacteria-induced activation of NF-κB in transiently transfected CHO cells. CHO cells recently have been shown to lack functional endogenous TLR2 (39) and do not demonstrate measurable NF-κB activation when stimulated with mycobacteria (not shown). Expression of murine CD14, a molecule known to be required for some TLR2 responses (18, 19, 27, 28, 30), was not sufficient to confer mycobacteria sensitivity on the CHO cells (Fig. 2). Expression of murine TLR2 conferred mycobacteria-induced NF-κB activation, and this stimulation was facilitated marginally by coexpression of CD14 (Fig. 2).
Figure 2.
Expression of TLR2 is sufficient to mediate M. tuberculosis-induced NF-κB activation in CHO cells. The relative activity of a NF-κB-luciferase reporter was assessed in CHO cells transiently expressing the indicated constructs before stimulation (open bars) or after stimulation with heat-killed virulent (hatched bars, H37Rv) or avirulent (filled bars, H37Ra) M. tuberculosis.
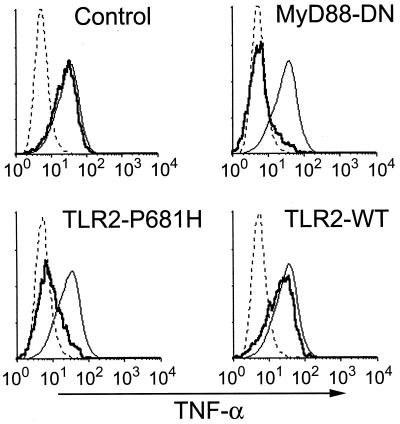
The mycobacterial cell wall can be separated into these major fractions: LAM, mAGP, and total lipids. The most well-characterized and potent mycobacterial cell wall inflammatory component is LAM. LAM consists of a mannose-rich core carbohydrate, with highly branched arabinofuranosyl side chains, linked to acyl groups consisting of palmitic and tuberculostearic acids. Interestingly, LAM isolated from virulent M. tuberculosis has mannose-capped arabinofuranosyl ends (ManLAM) and is nonstimulatory (ref. 40 and data not shown). LAM derived from avirulent M. tuberculosis and nonpathogenic, rapidly growing mycobacterial species lacks extensive mannose capping (AraLAM) and potently stimulates TNF-α production in macrophages. AraLAM strongly induced TNF-α in RAW cells, and this induction was inhibited strongly by MyD88-DN or TLR2-P681H expression (Fig. 3). AraLAM-induced TNF-α production was unaffected by transfection with a control plasmid (Fig. 3). These data demonstrate that TLR2 is required for LAM-induced activation of macrophages.
Figure 3.
TNF-α production induced by LAM depends on TLR2. RAW-TT10 cells transiently transfected with a control plasmid, MyD88-dominant negative (MyD88-DN), wild-type TLR2, or dominant negative TLR2 (TLR2-P681H) were analyzed for induction of TNF-α after stimulation with AraLAM. Histograms are formatted as described in Fig. 1.
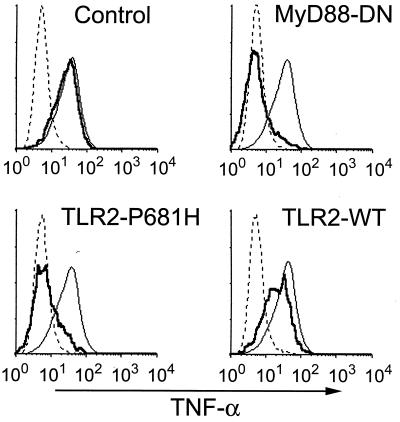
Because virulent M. tuberculosis lacks stimulatory LAM, other cell wall components derived from virulent M. tuberculosis must be responsible for MyD88- and TLR2-dependent TNF-α production. We examined the proinflammatory activity of M. tuberculosis total lipids. The M. tuberculosis cell wall contains a complex mixture of biologically active lipids including trehalose dimycolate, sulfolipids, and phosphatidyl inositol mannoside (32). The total lipid fraction from M. tuberculosis strongly induced TNF-α production by RAW cells, and this stimulation was blocked by expression of either MyD88-DN or TLR2-P681H (Fig. 4). Expression of wild-type TLR2 had little or no effect (Fig. 4). Thus, TLR2 plays an important role in the detection of mycobacterial-derived lipids in macrophages.
Figure 4.
Induction of TNF-α in response to M. tuberculosis total lipids depends on TLR2. RAW-TT10 cells transiently transfected with a control plasmid, MyD88-dominant negative (MyD88-DN), wild-type TLR2, or dominant negative TLR2 (TLR2-P681H) were analyzed for induction of TNF-α after stimulation with M. tuberculosis lipids. Histograms are formatted as described in Fig. 1.
Next, we examined the stimulatory activity of the cell wall core mAGP. This complex contains major structural components of the cell wall including mycolic acids, arabinogalactan, and peptidoglycan (32). Like AraLAM and M. tuberculosis total lipids, mAGP induced TNF-α production in RAW cells, and this stimulation was blocked by MyD88-DN and TLR2-P681H expression (Fig. 5). Wild-type TLR2 expression mildly inhibited mAGP-induced TNF-α, consistent with our previous observations that, in some instances, overexpression of the wild-type receptor can be mildly inhibitory (31). Peptidoglycan is likely to be one stimulatory component of mAGP. It recently has been shown that peptidoglycan purified from Staphylococcus aureus, a Gram-positive bacterium, can stimulate a NF-κB reporter in fibroblasts transfected with TLR2 (refs. 27 and 28 and data not shown).
Figure 5.
TNF-α produced in response to mAGP depend on TLR2. RAW-TT10 cells transiently transfected with a control plasmid, MyD88-dominant negative (MyD88-DN), wild-type TLR2, or dominant negative TLR2 (TLR2-P681H) were analyzed for induction of TNF-α after stimulation with mAGP. Histograms are formatted as described in Fig. 1. Similar results were obtained in the presence or absence of polymyxin B.
Our murine macrophage system indicates that macrophages detect whole M. tuberculosis via a TLR2-dependent pathway. In contrast, TLR2 is not required for LPS signaling, a process that is blocked by expression of dominant negative murine TLR4 (P712H) (31). These observations are consistent with reports showing that a mutation in the TLR4 gene is responsible for the LPS hyporesponsiveness of the C3H/HeJ mouse (24, 25) and that these mice are responsive to mycobacterial products (15). Recently, it has been shown that a dominant negative form of human TLR2 (lacking the C-terminal 13 aa) blocked stimulation of IL-12 and inducible nitric oxide synthase (iNOS) in macrophages by a lipoprotein derived from M. tuberculosis. Interestingly, this mutant form of human TLR2 also blocked LPS signaling (29), an observation consistent with several other studies suggesting a role for TLR2 in LPS detection (18, 19, 27, 28). The cause of the greater TLR2 ligand specificity observed in our murine macrophage system is not known. It is possible that the mouse TLR2 used in the data presented here exhibits greater selectivity than the human TLR2 used in other studies (18, 19, 27–29). It also is possible that the discrepancy is caused by different methods of measuring TLR2 signaling; we measured TNF-α production, whereas others have examined IL-12 and iNOS induction in macrophages (29) or NF-κB reporter activity in fibroblasts (18, 19, 27, 28).
Our data demonstrate that macrophages use TLR2 to detect a variety of mycobacterial cell wall fractions that are complex mixtures of sugars, lipids, and peptides. Each cell wall fraction used in this study is enriched with a structurally distinct set of stimulatory compounds. The observation that TLR2 is required for proinflammatory responses to all of these mycobacterial cell wall fractions, as well as responses to Gram-positive bacteria, lipoteichoic acid, peptidoglycan, and yeast cell walls, suggests a relatively broad ligand specificity for this receptor (27, 28, 31). This contrasts with TLR4, for which the only know ligand is LPS. This implies a selective advantage for an innate immune recognition system that can distinguish LPS from other microbial products. Independent regulation of toll-like receptor gene expression may confer selective pathogen responsiveness to different cells. This may be important for modulating responses to commensal and pathogenic organisms. Similarly, independent toll-like receptors that are specific for different microbial products may permit inflammatory responses to be tailored to particular organisms. However, to date, no difference has been demonstrated between TLR2 and TLR4 signal transduction.
It is not clear how TLR2 is involved in proinflammatory signaling by multiple structurally diverse ligands. It is possible that TRL2 directly binds these diverse ligands. Alternately, TLR2 may bind other endogenous macrophage proteins that are responsible for the specific detection of the ligands. This second model is especially attractive because an endogenous protein, spaetzle, is the ligand for Drosophila Toll, the prototypic toll family receptor (41). TLR2 may oligomerize with other toll-like receptors, thus accounting for its apparently broad ligand specificity. However, the observation that TLR2 does not block LPS signaling (viaTLR4) suggests that this is not a general property of toll-like receptors.
In summary, we have shown that macrophages use TLR2 to recognize mycobacteria and induce TNF-α, an important event in the elaboration of a protective inflammatory response to mycobacterial infection. Thus, TLR2-dependent signals are likely to participate in innate inflammatory responses to mycobacteria in a manner that is analogous to the role of TLR4 in host response to Gram-negative bacterial infections (42).
Acknowledgments
Lynn Hajjar and Anne Stevens provided murine TLR2 and MyD88 cDNAs. This work was supported in part by grants from the National Institutes of Health (AI25032 and AI32972). D.M.U. is an Irvington Institute postdoctoral fellow.
Abbreviations
- TNF-α
tumor necrosis factor α
- LAM
liparabinomannan
- LPS
lipopolysaccharide
- CHO
Chinese hamster ovary
- TLR2
toll-like receptor-2
- mAGP
mycolylarabinogalactan–peptidoglycan complex
- GFP
green fluorescent protein
References
- 1.Hirsch C S, Johnson J L, Ellner J J. Curr Opin Pulmonary Med. 1999;5:143–150. doi: 10.1097/00063198-199905000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Ellner J J, Wallis R S. Rev Infect Dis. 1989;11, Suppl. 2:S455–S459. doi: 10.1093/clinids/11.supplement_2.s455. [DOI] [PubMed] [Google Scholar]
- 3.Rook G A, Taverne J, Leveton C, Steele J. Immunology. 1987;62:229–234. [PMC free article] [PubMed] [Google Scholar]
- 4.Flynn J L, Goldstein M M, Chan J, Triebold K J, Pfeffer K, Lowenstein C J, Schreiber R, Mak T W, Bloom B R. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 5.Kindler V, Sappino A P, Grau G E, Piguet P F, Vassalli P. Cell. 1989;56:731–740. doi: 10.1016/0092-8674(89)90676-4. [DOI] [PubMed] [Google Scholar]
- 6.Mathison J, Wolfson E, Steinemann S, Tobias P, Ulevitch R. J Clin Invest. 1993;92:2053–2059. doi: 10.1172/JCI116801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu B, Hailman E, Wright S D. J Clin Invest. 1997;99:315–324. doi: 10.1172/JCI119160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu W, Soprana E, Cosentino G, Volta M, Lichenstein H S, Viale G, Vercelli D. J Immunol. 1998;161:4244–4251. [PubMed] [Google Scholar]
- 9.Savedra R, Jr, Delude R L, Ingalls R R, Fenton M J, Golenbock D T. J Immunol. 1996;157:2549–2554. [PubMed] [Google Scholar]
- 10.Ulevitch R J, Tobias P S. Curr Opin Immunol. 1999;11:19–22. doi: 10.1016/s0952-7915(99)80004-1. [DOI] [PubMed] [Google Scholar]
- 11.Pugin J, Heumann I D, Tomasz A, Kravchenko V V, Akamatsu Y, Nishijima M, Glauser M P, Tobias P S, Ulevitch R J. Immunity. 1994;1:509–516. doi: 10.1016/1074-7613(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Doerfler M, Lee T C, Guillemin B, Rom W N. J Clin Invest. 1993;91:2076–2083. doi: 10.1172/JCI116430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P Y, Kitchens R L, Munford R S. J Biol Chem. 1998;273:24309–24313. doi: 10.1074/jbc.273.38.24309. [DOI] [PubMed] [Google Scholar]
- 14.Devitt A, Moffatt O D, Raykundalia C, Capra J D, Simmons D L, Gregory C D. Nature (London) 1998;392:505–509. doi: 10.1038/33169. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda-Fujita T, Kotani S, Tsujimoto M, Ogawa T, Takahashi I, Takada H, Shimauchi H, Nagao S, Kokeguchi S, Kato K, et al. Microbiol Immunol. 1987;31:289–311. doi: 10.1111/j.1348-0421.1987.tb03091.x. [DOI] [PubMed] [Google Scholar]
- 16.Golenbock D T, Liu Y, Millham F H, Freeman M W, Zoeller R A. J Biol Chem. 1993;268:22055–22059. [PubMed] [Google Scholar]
- 17.Chow J C, Young D W, Golenbock D T, Christ W J, Gusovsky F. J Biol Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 18.Kirschning C J, Wesche H, Merrill Ayres T, Rothe M. J Exp Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang R B, Mark M R, Gray A, Huang A, Xie M H, Zhang M, Goddard A, Wood W I, Gurney A L, Godowski P J. Nature (London) 1998;395:284–288. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 20.Lemaitre B, Reichhart J M, Hoffmann J A. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhary P M, Ferguson C, Nguyen V, Nguyen O, Massa H F, Eby M, Jasmin A, Trask B J, Hood L, Nelson P S. Blood. 1998;91:4020–4027. [PubMed] [Google Scholar]
- 22.Rock F L, Hardiman G, Timans J C, Kastelein R A, Bazan J F. Proc Natl Acad Sci USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi O, Kawai T, Sanjo H, Copeland N G, Gilbert D J, Jenkins N A, Takeda K, Akira S. Gene. 1999;231:59–65. doi: 10.1016/s0378-1119(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 24.Qureshi S T, Lariviere L, Leveque G, Clermont S, Moore K J, Gros P, Malo D. J Exp Med. 1999;189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poltorak A, He X, Smirnova I, Liu M Y, Huffel C V, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 26.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 27.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning C J. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimura A, Lien E, Ingalls R R, Tuomanen E, Dziarski R, Golenbock D. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 29.Brightbill H D, Libraty D H, Krutzik S R, Yang R B, Belisle J T, Bleharski J R, Maitland M, Norgard M V, Plevy S E, Smale S T, et al. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 30.Hirschfeld M, Kirschning C J, Schwandner R, Wesche H, Weis J H, Wooten R M, Weis J J. J Immunol. 1999;163:2382–2386. [PubMed] [Google Scholar]
- 31.Underhill D M, Ozinsky A, Hajjar A M, Stevens A, Wilson C B, Bassetti M, Aderem A. Nature (London) 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 32.Besra G S. Methods Mol Biol. 1998;101:91–107. doi: 10.1385/0-89603-471-2:91. [DOI] [PubMed] [Google Scholar]
- 33.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Underhill D M, Chen J, Allen L A, Aderem A. J Biol Chem. 1998;273:33619–33623. doi: 10.1074/jbc.273.50.33619. [DOI] [PubMed] [Google Scholar]
- 35.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway C A., Jr Mol Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 36.Muzio M, Ni J, Feng P, Dixit V M. Science. 1997;278:1612–1615. [Google Scholar]
- 37.Wesche H, Henzel W J, Shillinglaw W, Li S, Cao Z. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 38.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 39.Heine H, Kirschning C J, Lien E, Monks B G, Rothe M, Golenbock D T. J Immunol. 1999;162:6971–6975. [PubMed] [Google Scholar]
- 40.Brown M C, Taffet S M. Infect Immunol. 1995;63:1960–1968. doi: 10.1128/iai.63.5.1960-1968.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morisato D, Anderson K V. Cell. 1994;76:677–688. doi: 10.1016/0092-8674(94)90507-x. [DOI] [PubMed] [Google Scholar]
- 42.Cross A, Asher L, Seguin M, Yuan L, Kelly N, Hammack C, Sadoff J, Gemski P., Jr J Clin Invest. 1995;96:676–686. doi: 10.1172/JCI118110. [DOI] [PMC free article] [PubMed] [Google Scholar]