Abstract
In contrast to naive lymphocytes, memory/effector lymphocytes can access nonlymphoid effector sites and display restricted, often tissue-selective, migration behavior. The cutaneous lymphocyte-associated antigen (CLA) defines a subset of circulating memory T cells that selectively localize in cutaneous sites mediated in part by the interaction of CLA with its vascular ligand E-selectin. Here, we report the identification and characterization of a CC chemokine, cutaneous T cell-attracting chemokine (CTACK). Both human and mouse CTACK are detected only in skin by Southern and Northern blot analyses. Specifically, CTACK message is found in the mouse epidermis and in human keratinocytes, and anti-CTACK mAbs predominantly stain the epithelium. Finally, CTACK selectively attracts CLA+ memory T cells. Taken together, these results suggest an important role for CTACK in recruitment of CLA+ T cells to cutaneous sites. CTACK is predominantly expressed in the skin and selectively attracts a tissue-specific subpopulation of memory lymphocytes.
The exquisite tissue-selective homing of lymphocytes has long been appreciated as central for the control of systemic immune responses. Naive lymphocytes display a relatively homogeneous migration behavior, recirculating through secondary lymphoid tissues, including lymph nodes, Peyer’s patches, tonsils, and spleen, to encounter their cognate antigen. Following antigen encounter and recognition in an appropriate microenvironment, lymphocytes differentiate into effector/memory cells and acquire the ability to access extralymphoid immune effector sites where they are most likely to reencounter their specific antigen (1). Thus, distinct subsets of memory/effector cells exist with tissue-selective patterns of migration (2, 3). A classic example of such tissue-specific homing involves the preferential migration of cutaneous lymphocyte-associated antigen (CLA)+ memory T cells to the skin. The first evidence that a skin-homing subset exists came with the observation that a majority (80–90%) of the T lymphocytes in inflammatory skin lesions express the CLA. In contrast, CLA+ T cells constitute only 10–15% of the pool of circulating T cells and never exceed 5% of lymphocytes within noncutaneous inflamed sites (4). Further implicating the CLA+ subset in cutaneous inflammation are studies showing that specific responses to common skin-associated allergens, including nickel and house dust mite, were restricted to CLA+ T cells (5).
CLA is not simply a marker for this subset of cells but is a ligand for the vascular adhesion molecule E-selectin (6), which is superinduced on vascular endothelium during cutaneous inflammation (7) and is critical for recruiting T cells to sites of cutaneous inflammation (8). It has been suggested that these molecules mediate the selective targeting of memory T cells reactive with skin-associated antigens to cutaneous inflammatory sites. However, it has become increasingly clear that recruitment of leukocytes into tissues involves the sequential engagement of multiple adhesion and signaling receptors (1, 9). Under physiologic flow conditions, leukocytes initially interact with and roll on endothelial cells through constitutively functional (i.e., activation-independent) leukocyte-homing receptors and their ligands; E-selectin is the likely candidate to support recognition and transient adhesion or rolling of CLA+ T cells. This initial adhesion, however, is unstable and must be followed by functional activation of leukocyte integrins, by locally produced activation factors, in order for cells to firmly attach and arrest by means of their vascular ligands. Even this shear-resistant arrest is reversible unless appropriate additional signals and chemotactic factors are present that can drive diapedesis and subsequent localization within the tissues. This multistep process provides a combinatorial mechanism for precise regulation of leukocyte recruitment based on expression of a unique set of adhesion, activation, and chemotactic receptors on the migrating leukocyte subset coupled with selective expression of adhesion, activation, and chemotactic molecules by the tissue (1).
Chemokines are a superfamily of small secreted proteins that attract their targets by interacting with G protein-coupled receptors expressed on the migrating cell (10, 11). The observation that chemokines can trigger activation-dependent adhesion and chemoattract leukocytes led to the hypothesis that this family of molecules provides the key signals that trigger firm arrest and provide chemotactic gradients that direct diapedesis and localization of cells to specific tissue microenvironments. Implicit in the model of differential trafficking is the possibility that there exist tissue-specific chemokines that selectively attract functionally unique subsets of lymphocytes.
This study describes the identification of a CC chemokine, cutaneous T cell-attracting chemokine (CTACK), that is expressed in skin and selectively chemoattracts CLA+ memory T cells. CTACK has the characteristics of a molecule that could regulate the specific migration behavior of these skin-homing lymphocytes.
Methods
Sequence Analysis.
tblastn searches of the Human Genome Systems (HGS) and GenBank dbEST databases, with the sequences of known CC chemokines, identified the expressed sequence tags (ESTs) for human and mouse CTACK, respectively. Mouse CTACK cDNA, image consortium clone no. 316475, was obtained from Genome Systems (St. Louis) as an EcoRI–NotI insert in the pT7T3-PacD vector. Human CTACK was obtained from HGS as a SalI–NotI insert in the pSPORT 3.0 vector. The nucleotide sequence of both clones was confirmed by automated sequencing. The signal peptide cleavage sites were predicted by using the SignalP server (http://www.cbs.dtu.dk/signalp/cbssignalp.html). Sequences were aligned by using clustal w.
Mapping.
CTACK was placed on mouse chromosome 4 by interspecific backcross analysis essentially as described (12), except that in this case an ≈400-bp EcoRI/NotI fragment of mouse CTACK cDNA was used for Southern blotting. Fragments of 14.5 and 8.9 kb were detected in BglI-digested C57BL/6J DNA, and fragments of 8.9 and 4.4 kb were detected in BglI-digested Mus spretus DNA. The presence or absence of the 4.4-kb BglI-specific fragment was followed in backcross mice. Recombination distances were calculated with map manager, version 2.6.5.
Expression Studies.
For Southern blotting, 5 μg of each cDNA library were digested with the appropriate restriction enzymes to release the insert, subjected to gel electrophoresis, and transferred to Hybond-N+ membrane. For Northern blotting, all RNAs were isolated by using RNAzol B (Tel-Test, Friendswood, TX) and analyzed by electrophoresis on a 1% formaldehyde/agarose gel and transferred to Hybond-N+ membrane. Northern and Southern blots were hybridized for 16 h at 65°C with 32P-labeled probes obtained by randomly priming (Prime-It; Stratagene) the full-length inserts from mouse or human CTACK clones. After hybridization, blots were washed at high stringency and exposed to film.
Reverse Transcription–PCR (RT-PCR).
RNAs were treated with DNase I and reverse transcribed, and the resulting cDNAs were used as templates for RT-PCR as described (13, 14). Primer pairs used for PCR were as follows: human CTACK (244 bp) sense primer 5′-CTGTACTCAGCTCTACKCGAAAGCC-3′ (position 99–122); antisense primer 5′-GCCCATTTTCCTTAGCATCCC-3′ (position 336–316); human β-actin sense primer 5′-ATCTGGCACCACACCTTCTACKAATGAGCTGCG-3′; and antisense primer 5′-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3′.
Cell and Tissue Samples and Immunohistology.
Adult human primary cells, including keratinocytes, melanocytes, and dermal fibroblasts, were obtained from Clonetics (San Diego) and cultured according to the supplier’s instructions. The epidermal γδ T cell line 7-17 was kindly provided by Richard Boismenu (The Scripps Institute, La Jolla, CA). For cytokine treatment, cells were cultured with 10 ng/ml human tumor necrosis factor-α (TNF-α) plus 3 ng/ml hIL-1β (R & D Systems) in culture medium for the indicated times. Epidermal cell samples were released from the ear skin of BALB/c mice by treating them with 0.5% trypsin in PBS for 30 min at 37°C. Human peripheral blood mononuclear cells (PBMCs) and neutrophils were obtained from healthy donors by erythrocyte sedimentation followed by Ficoll/Histopaque-1077 (Sigma) sedimentation. Freshly drawn blood was used to isolate neutrophils. Human T cells were purified from PBMCs by using T cell- or memory T cell-enrichment columns (R & D Systems), according to the manufacturer’s instructions. A panel of mAbs was produced against mouse CTACK and screened for specificity by ELISA against a panel of chemokines (R & D Systems). These mAbs, along with negative control rat mAbs (PharMingen), were used to stain frozen mouse ear sections with VECTASTAIN ABC Kit (Vector Laboratories), using a goat anti-rat horseradish peroxidase-conjugated secondary Ab and diaminobenzidine as the substrate.
Chemotaxis.
Recombinant mouse CTACK was produced in Escherichia coli and purified by R & D Systems, as previously described for other chemokines (15). For T cell chemotaxis assays, column-purified human T cells (90% pure) or CD4+ memory T cells (92% pure) in DMEM (pH 6.9)/1% BSA were added to the top chamber of 3-μm pore polycarbonate Transwell culture insert (Costar) and incubated with the indicated concentrations of purified chemokine in the bottom chamber for 3 hr. For B cell and monocyte assays, PBMCs were added to the top chamber and incubated for 3 hr (B cells) or 1 hr (monocytes). For neutrophil assays, purified neutrophils were added to a chamber with a 1-μm pore polyethylene terephthalate membrane and incubated for 90 min. The number of migrating cells of each subtype was determined by multiparameter flow cytometry with fluorochrome-conjugated Abs (all from PharMingen) against CLA (HECA-452), CD4, CD8, CD45RA, and CD45RO for T cells, CD19 for B cells, CD14 for monocytes, and CD66b for neutrophils. A known number of 15-μm microsphere beads (Bangs Laboratories, Fishers, IN) was added to each sample before analysis to determine the absolute number of migrating cells.
Results
Identification of CTACK.
Searching the HGS database, we identified an EST encoding a human CC chemokine. This EST contains an ORF encoding a putative protein of 112 amino acids with a predicted 24-amino acid signal peptide at the N terminus (Fig. 1a). A mouse clone was identified in the public database of ESTs encoding a protein with 78% amino acid similarity to the human clone (Fig. 1a). The putative mature proteins are 84% similar. We designated this chemokine cutaneous T cell-attracting chemokine (CTACK). The CC chemokines with the greatest similarity to CTACK are thymus-expressed chemokine (TECK), 6Ckine, and macrophage inflammatory protein (MIP)-3β (Fig. 1b). The human and mouse CTACK clones map to syntenic chromosomal regions, human segment 9p13 and the proximal region of mouse chromosome 4, respectively (Fig. 1c). 6Ckine and MIP-3β also map to these chromosomal regions. A search of GenBank with the sequence of human CTACK revealed that the gene for human CTACK overlaps with the extreme 3′ end [after the poly(A) signal] of the IL-11 receptor α-chain gene, but on the opposite strand.
Figure 1.
(a) Amino acid sequence alignment of mouse (m) and human (h) CTACK. (b) Amino acid sequences of CTACK aligned with other CC chemokines, hTECK, hMIP-3β, h6Ckine, and human regulated on activation normal T expressed and secreted (hRANTES). Dark-shaded boxes indicate identical amino acids. Conserved amino acids are shaded light gray. Arrow denotes the predicted signal peptide cleavage site. Asterisks are above conserved cysteines. (c) Mouse CTACK maps in the proximal region of mouse chromosome 4. CTACK was placed on mouse chromosome 4 by interspecific backcross analysis. The segregation patterns of CTACK and flanking genes in 98 backcross animals that were typed for all loci are shown at the top of the figure. Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J × M. spretus) F1 parent. The shaded boxes represent the presence of a C57BL/6J allele, and white boxes represent the presence of an M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 4 linkage map showing the location of CTACK in relation to linked genes is shown at the bottom of the figure with recombination distances between loci in centimorgans shown to the left and the positions of loci in human chromosomes shown to the right.
The Expression of CTACK Is Highly Tissue-Specific.
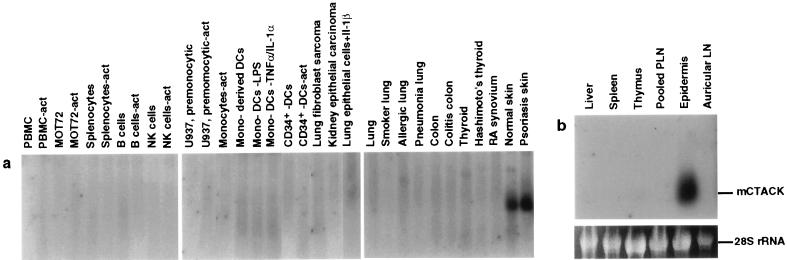
The vast majority of the CTACK ESTs were found in libraries derived from skin. Although a few CTACK ESTs were found in non-skin-associated tissues, such as liver, our expression studies suggest that CTACK is predominantly expressed in skin. More than 70 human cDNA libraries were probed by Southern blotting, including 46 libraries from hematopoetic and nonhematopoetic cells (including activated and resting cells) and 25 libraries from normal tissue and diseased tissue. CTACK hybridized exclusively with libraries derived from normal or psoriatic skin (Fig. 2a). Additionally, CTACK did not hybridize with a CLONTECH multiple tissue Northern blot containing RNA from 12 human tissues (including heart, brain, skeletal muscle, colon, thymus, spleen, kidney, liver, small intestine, placenta, lung, and peripheral blood leukocytes; data not shown). Southern blotting with mouse CTACK revealed no significant hybridization with over 45 mouse libraries derived from a wide variety of cells and tissues (including libraries derived from all the organs listed above for the CLONTECH blot; data not shown); however, this panel did not include a skin-derived library. To determine whether mouse CTACK is also expressed in skin, we performed Northern blot analysis with RNA generated from the epidermis of mouse ear skin or from several other organs. An RNA species of 0.8 kb is readily detected in the epidermis and not in the other organs tested (Fig. 2b).
Figure 2.
Expression of CTACK in skin. (a) Southern blot analysis of CTACK expression in cDNA libraries generated from human samples including: PBMC and PBMC-act, resting or activated (act), with anti-CD3 and phorbol 12-myristate 13-acetate (PMA); T cells (MOT72 and MOT72-act) resting or activated with anti-CD28 and anti-CD3; total splenocytes resting or activated with anti-CD40 and IL-4; pooled B cell lines, pooled natural killer (NK) cell clones, and U937 (premonocytic cell line), all resting or activated with PMA and ionomycin; elutriated monocytes activated with lipopolysaccharide (LPS), IFN-γ, and anti-IL-10; monocyte-derived dendritic cells (mono-derived DCs) resting or activated with either LPS or TNF-α plus IL-1α; CD34+ stem cell-derived dendritic cells (CD34+-DCs) resting or activated with PMA and ionomycin; MRC5 cells (lung fibroblast sarcoma); CHA cells (kidney epithelial carcinoma); A549 cells (lung epithelial) treated with IL-1β, adult organs and tissues, diseased and normal as indicated above each lane. (b) Northern blot analysis of CTACK expression in mouse tissues and organs, including liver, spleen, thymus, peripheral lymph nodes (PLN), epidermis of ear skin, and ear draining lymph nodes (Auricular LN). Ethidium bromide-stained 28S rRNA is shown as a loading control.
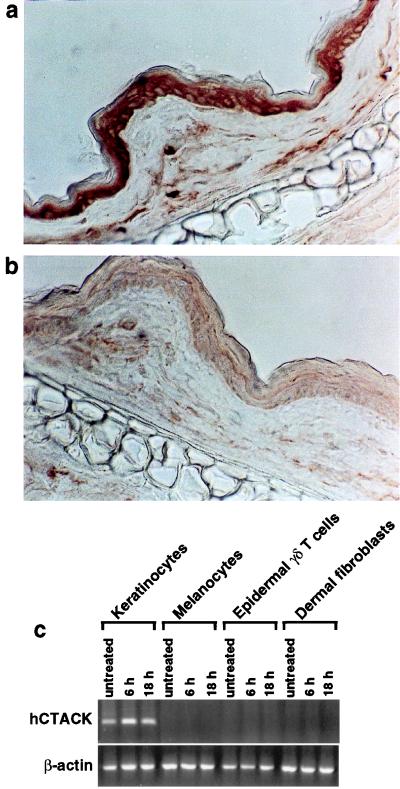
To look more precisely at the distribution of CTACK in skin, we performed immunohistochemistry using a panel of mAbs generated against mouse CTACK. While the staining intensity was variable between the mAbs, all gave a similar staining pattern and showed predominant staining in the outermost cell layers of the skin corresponding to the epidermis and rare positive cells in the dermis (Fig. 3). To determine the cellular origin of human CTACK, we looked for CTACK mRNA in cultured human skin-derived cells that were either left untreated or treated with TNF-α plus IL-1β for 6 h or 18 h. These proinflammatory cytokines are important primary regulators of immune responses in the skin and can induce the expression of other inflammatory mediators, including chemokines (16–18). CTACK message is detected by RT-PCR in untreated human keratinocytes, the predominant cell type in the epidermis (Fig. 3c) and is up-regulated after 8 hr of cytokine treatment. Competitive, semiquantitative RT-PCR analysis confirmed this observation and demonstrated an 8- to 30-fold induction in CTACK message (data not shown). In contrast, other skin-derived cell types, such as melanocytes, epidermal γδ T cells, and dermal fibroblasts, do not express CTACK under any of the conditions tested (Fig. 3c). Additionally, CTACK is not detected in libraries derived from resting or activated dendritic cells (Fig. 2a). Together, these results indicate that both human and mouse CTACK are predominantly expressed in skin by cells of the epidermis, as they were not detected in any of the other tissues or cells examined. This is in contrast to other chemokines, such as IP-10 and IL-8, that are expressed in skin (16, 18) but are also broadly expressed outside this tissue (19).
Figure 3.
Expression of CTACK. (a) Frozen sections of mouse skin (ears) were immunostained with a panel of anti-CTACK mAbs. A representative mAb (isotype IgG2b) is shown and revealed predominant staining in the epidermis and scattered positive cells in the dermis. (b) A control rat mAb (IgG2b) is shown for comparison. (c) RT-PCR analysis of CTACK expression in primary keratinocytes, primary melanocytes, 7-17 cells (epidermal γδ T cell line), and primary dermal fibroblasts either untreated or treated with TNF-α and IL-1β for the indicated times.
CTACK Selectively Chemoattracts Skin-Homing T Cells.
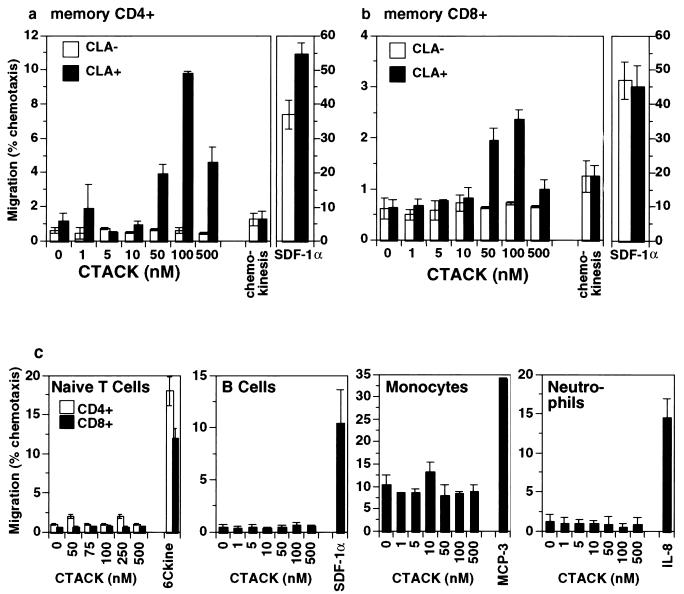
The abundant expression of CTACK in the skin makes it a potential candidate for selective recruitment of CLA+ skin-homing memory T cells. We therefore investigated the ability of CTACK to chemoattract leukocyte subsets from peripheral blood from healthy donors. CTACK failed to attract CD4+ or CD8+ naive (CD45RA+) T cells, B cells, monocytes, or neutrophils (Fig. 4c). CTACK attracts CLA+ but not CLA−CD4+ and CD8+ memory (CD45RO+) T cells (Fig. 4 a and b). A range of 2–24% of the input CD4+CLA+ memory T cell population migrated at this concentration (observed over seven independent experiments), perhaps suggesting that CTACK attracts a subset within the CLA+ cell population whose size may vary from donor to donor. These results are in striking contrast with those using the known T cell chemoattractant 6Ckine (20–22), which lacks specificity for CLA+ cells, as it preferentially attracts CLA−CD45RO+ memory T cells; and stromal cell-derived factor (SDF)-1α (23), which attracts CLA− and CLA+ CD45RO+ memory T cells equally well (Fig. 4 a and b). CLA+ cells migrate only in the presence of a gradient of CTACK, indicating that the response is chemotactic and not chemokinetic (Fig. 4 a and b). (Lack of an anti-mouse CLA Ab precludes similar analysis in the mouse.) Thus, CTACK attracts only CLA+ memory T cells out of all the leukocytes tested, demonstrating remarkable target cell specificity.
Figure 4.
CTACK preferentially attracts CLA+ memory T cells. (a) Migration (% of each phenotype added to the upper chamber) of CD4+ memory T cell populations in response to CTACK. (b) Migration of CD8+ memory T cell populations in response to CTACK. Chemotaxis to the optimal concentration of SDF-1α (5 × 10−8 M) is shown for comparison. Chemokinesis was measured by using the optimal chemotactic concentration of CTACK on both sides of the filter. Data from a representative experiment are shown. The composite mean and SD of the migration indices of seven independent experiments at the maximally effective concentration of CTACK for CD4+ memory cells: CLA+ = 10.14 ± 1.13 vs. CLA− = 1.02 ± 0.19 (P < 2 × 10−5), and for CD8+ cells: CLA+ = 4.26 ± 1.93 vs. CLA− = 1.07 ± 0.09 (P < 0.005). (c) CTACK does not attract CD4+ or CD8+ naive T cells, B cells, monocytes, or neutrophils. As positive controls, the following chemokines were used: 6Ckine (5 × 10−7) for naive T cells, SDF-1α (5 × 10−8 M) for B cells, monocyte chemoattractant protein-3 (1 × 10−9 M) for monocytes, and IL-8 (1 × 10−9 M) for neutrophils.
Discussion
The CLA+ memory T cell subset constitutes a skin-associated population of memory cells that preferentially extravasate at normal (24) and chronically inflamed cutaneous sites (4) and has been shown to be involved in local immunity and inflammatory cutaneous reactions (5). The localization specificity of CLA+ lymphocytes may be determined by regulating one or more steps along the recruitment pathway, including adhesion to the endothelium, transendothelial migration, chemotaxis within the skin to sites of immune or inflammatory activity, and interaction with tissue components including cells or matrix, which would serve to retain CLA+ lymphocytes in the skin. It has been proposed that CLA targets memory T cells to cutaneous sites by interaction with its vascular ligand E-selectin (6, 25); however, CLA/E-selectin binding cannot fully explain the skin-specific homing of CLA+ memory T cells for several reasons. First, neutrophils express CLA (26), yet they do not preferentially migrate to skin, and, second, E-selectin is induced on inflamed endothelium in both cutaneous and noncutaneous sites (27).
Here, we report the identification of a chemokine, CTACK, that may provide a skin-specific cue involved in localization of CLA+ memory T cells to the epidermis. This inference is supported by several observations. We show that CTACK is readily detected in the skin of both humans and mice and that expression was not detected in any other tissue examined. On closer examination of the cell types that make up the skin, we observed that CTACK is expressed in the epidermis by keratinocytes; an Ab to CTACK stains predominantly the epidermis of the skin, and RT-PCR detects CTACK in keratinocytes, but not melanocytes, dermal fibroblasts, or epidermal γδ T cells. Additionally, in vitro functional studies show that CTACK attracts preferentially the skin-homing subset of memory T cells, whereas it does not attract naive T cells, CLA− T cells, B cells, monocytes, or neutrophils. Together, these characteristics suggest that this molecule may play a key role in the trafficking of CLA+ T cells to skin.
The constitutive expression of CTACK by epidermal cells suggests that CTACK may be involved in basal trafficking of T cells through the skin during normal immune surveillance. However, CTACK can be superinduced by the proinflammatory cytokines TNF-α and IL-1β, which, together with the dramatic up-regulation of vascular E-selectin, may recruit memory T cells to the skin during cutaneous inflammation.
As discussed above, recruitment of T cells into the tissue relies on a series of steps including rolling along the endothelium, activation-dependent firm adhesion to the endothelium, followed by diapedesis out of the vessel and microenvironmental localization within the tissue. The E-selectin+ postcapillary venules of the dermis are the dominant sites of T cell extravasation in the skin. The close spatial apposition between vessels in the papillary dermis and the epidermis suggests that the release of CTACK by keratinocytes could influence the influx of T cells into the skin as well as localization and retention of T cells in the epidermis; however, the precise role CTACK plays in recruiting memory T cells to the skin awaits further investigation.
The unique activity of CTACK implies the existence of a CTACK receptor potentially restricted to CLA+ T cells. A panel of transfected cells expressing the known chemokine receptors, including CCR1- 9, CXCR1, 3–5, and XCR1, responded by calcium flux or migration to their known ligands but failed to respond to CTACK (data not shown), suggesting that CTACK signals through a different chemokine receptor.
Taken together, these data suggest that CTACK may provide a skin-specific signal involved in localization of CLA+ memory T cells to skin and provides a potential target to regulate cutaneous T cell trafficking. These findings also support the hypothesis that the nonrandom recirculation of subsets of memory T cells relies not only on restricted expression of specific vascular adhesion molecules, but also on microenvironmentally restricted activation signals, including chemokines.
Acknowledgments
We thank Jim Cupp for flow cytometry, Dan Gorman for sequencing help, Terrill McClanahan for cDNA libraries, and Debra J. Gilbert for excellent technical assistance mapping mouse CTACK. We also thank Drs. Luis Llorente and Donato Alarcón-Segovia for their help in the procurement of human tissue samples. DNAX Research Institute is supported by Schering-Plough Corporation. B.H. was supported by Grant DFG HO2092/1-1 from the German Research Foundation. N.G.C. and N.A.J. received support from the National Cancer Institute, Department of Health and Human Services, under contract with Advanced BioScience Laboratories.
Abbreviations
- CLA
cutaneous lymphocyte-associated antigen
- CTACK
cutaneous T cell-attracting chemokine
- EST
expressed sequence tag
- TNF
tumor necrosis factor
- PBMC
peripheral blood mononuclear cell
- MIP
macrophage inflammatory protein
- SDF
stromal cell-derived factor
- RT-PCR
reverse transcription–PCR
Note Added in Proof
CTACK is the same chemokine reported as mouse ALP (28). In addition, two other groups will report mouse CTACK; Gerry Graham and collaborators called this chemokine Eskine, and Osamu Yoshie and collaborators will report it under the name ILC. A new systematic chemokine ligand nomenclature has been proposed, and CTACK will receive the designation CCL27. We suggest that future studies call this chemokine by this new designation (CCL27).
Footnotes
References
- 1.Butcher E C, Picker L J. Science. 1996;272:60–66. doi: 10.1126/science.272.5258.60. [DOI] [PubMed] [Google Scholar]
- 2.Picker L J, Martin R J, Trumble A, Newman L S, Collins P A, Bergstresser P R, Leung D Y. Eur J Immunol. 1994;24:1269–1277. doi: 10.1002/eji.1830240605. [DOI] [PubMed] [Google Scholar]
- 3.Picker L J. Curr Opin Immunol. 1994;6:394–406. doi: 10.1016/0952-7915(94)90118-x. [DOI] [PubMed] [Google Scholar]
- 4.Picker L J, Michie S A, Rott L S, Butcher E C. Am J Pathol. 1990;136:1053–1068. [PMC free article] [PubMed] [Google Scholar]
- 5.Santamaria Babi L F, Perez Soler M T, Hauser C, Blaser K. Immunol Res. 1995;14:317–324. doi: 10.1007/BF02935627. [DOI] [PubMed] [Google Scholar]
- 6.Berg E L, Yoshino T, Rott L S, Robinson M K, Warnock R A, Kishimoto T K, Picker L J, Butcher E C. J Exp Med. 1991;174:1461–1466. doi: 10.1084/jem.174.6.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groves R W, Allen M H, Barker J N, Haskard D O, MacDonald D M. Br J Dermatol. 1991;124:117–123. doi: 10.1111/j.1365-2133.1991.tb00419.x. [DOI] [PubMed] [Google Scholar]
- 8.Catalina M D, Estess P, Siegelman M H. Blood. 1999;93:580–589. [PubMed] [Google Scholar]
- 9.Springer T A. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 10.Zlotnik A, Morales J, Hedrick J A. Crit Rev Immunol. 1999;19:1–47. [PubMed] [Google Scholar]
- 11.Baggiolini M. Nature (London) 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 12.Kelner G S, Kennedy J, Bacon K B, Kleyensteuber S, Largaespada D A, Jenkins N A, Copeland N G, Bazan J F, Moore K W, Schall T J, et al. Science. 1994;266:1395–1399. doi: 10.1126/science.7973732. [DOI] [PubMed] [Google Scholar]
- 13.Gilliland G, Perrin S, Blanchard K, Bunn H F. Proc Natl Acad Sci USA. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platzer C, Richter G, Uberla K, Muller W, Blocker H, Diamantstein T, Blankenstein T. Eur J Immunol. 1992;22:1179–1184. doi: 10.1002/eji.1830220511. [DOI] [PubMed] [Google Scholar]
- 15.Hedrick J A, Helms A, Vicari A, Zlotnik A. Blood. 1998;91:4242–4247. [PubMed] [Google Scholar]
- 16.Larsen C G, Anderson A O, Oppenheim J J, Matsushima K. Immunology. 1989;68:31–36. [PMC free article] [PubMed] [Google Scholar]
- 17.Goebeler M, Yoshimura T, Toksoy A, Ritter U, Brocker E B, Gillitzer R. J Invest Dermatol. 1997;108:445–451. doi: 10.1111/1523-1747.ep12289711. [DOI] [PubMed] [Google Scholar]
- 18.Schroder J M. J Invest Dermatol. 1995;105, Suppl. 1:20S–24S. [PubMed] [Google Scholar]
- 19.Rollins B J. Blood. 1997;90:909–928. [PubMed] [Google Scholar]
- 20.Nagira M, Imai T, Hieshima K, Kusuda J, Ridanpaa M, Takagi S, Nishimura M, Kakizaki M, Nomiyama H, Yoshie O. J Biol Chem. 1997;272:19518–19524. doi: 10.1074/jbc.272.31.19518. [DOI] [PubMed] [Google Scholar]
- 21.Hromas R, Kim C H, Klemsz M, Krathwohl M, Fife K, Cooper S, Schnizlein-Bick C, Broxmeyer H E. J Immunol. 1997;159:2554–2558. [PubMed] [Google Scholar]
- 22.Hedrick J A, Zlotnik A. J Immunol. 1997;159:1589–1593. [PubMed] [Google Scholar]
- 23.Bleul C C, Fuhlbrigge R C, Casasnovas J M, Aiuti A, Springer T A. J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bos J D, de Boer O J, Tibosch E, Das P K, Pals S T. Arch Dermatol Res. 1993;285:179–183. doi: 10.1007/BF00372006. [DOI] [PubMed] [Google Scholar]
- 25.Picker L J, Kishimoto T K, Smith C W, Warnock R A, Butcher E C. Nature (London) 1991;349:796–799. doi: 10.1038/349796a0. [DOI] [PubMed] [Google Scholar]
- 26.De Boer O J, Horst E, Pals S T, Bos J D, Das P K. Immunology. 1994;81:359–365. [PMC free article] [PubMed] [Google Scholar]
- 27.Tedder T F, Steeber D A, Chen A, Engel P. FASEB J. 1995;9:866–873. [PubMed] [Google Scholar]
- 28.Hromas R, Broxmeyer H E, Kim C, Christopherson K, 2nd, Hou Y H. Biochem Biophys Res Commun. 1999;258:737–740. doi: 10.1006/bbrc.1999.0507. [DOI] [PubMed] [Google Scholar]