Abstract
IL-7 functions as a trophic factor during T lymphocyte development by a mechanism that is partly based on the induction of Bcl-2, which protects cells from apoptosis. Here we report a mechanism by which cytokine withdrawal activates the prodeath protein Bax. On loss of IL-7 in a dependent cell line, Bax protein translocated from the cytosol to the mitochondria, where it integrated into the mitochondrial membrane. This translocation was attributable to a conformational change in the Bax protein itself. We show that a rise in intracellular pH preceded mitochondrial translocation and triggered the change in Bax conformation. Intracellular pH in the IL-7-dependent cells rose steadily to peak over pH 7.8 by 6 hr after cytokine withdrawal, paralleling the time point of Bax translocation (a similar alkalinization and Bax translocation was also observed after IL-3 withdrawal from a dependent cell line). The conformation of Bax was directly altered by pH of 7.8 or higher and was demonstrated by increased protease sensitivity, exposure of N terminus epitopes, and exposure of a hydrophobic domain in the C terminus. Eliminating charged amino acids at the C or N termini of Bax induced a conformational change similar to that induced by raising pH, implicating these residues in the pH effect. Therefore, we have shown that by either cytokine withdrawal, experimental manipulation of pH, or site-directed mutagenesis, Bax protein changes conformation, exposing membrane-seeking domains, thereby inducing mitochondrial translocation and initiating the cascade of events leading to apoptotic death.
Apoptosis, or programmed cell death, is a highly coordinated process by which cells are eliminated without inducing an inflammatory response. The development of T cells in the thymus involves a balance between expansion, survival, and apoptosis. Survival of pro-T cells depends on the trophic effect of IL-7 (1, 2), a product of the thymic epithelium. Ablation of the gene for IL-7 (3) or the IL-7 receptor α chain (IL-7Rα) (4) resulted in greatly reduced thymic cellularity, and the few T cells that developed in IL-7Rα−/− mice failed to proliferate on stimulation and underwent cell death (5). The mechanism of IL-7’s trophic effects has been partly attributed to the Bcl-2 family of related proteins, which are important intracellular mediators of apoptosis (6). Overexpression of the antiapoptotic protein Bcl-2 in IL-7Rα−/− mice partially restored T cell numbers (7), but complete restoration of a normal phenotype was not achieved (8, 9), implicating other actions of IL-7 beyond the induction of Bcl-2.
Bax is an attractive candidate for a death inducer expressed in thymocytes, because deletion of bax increased the number of thymocytes (10) and restored the thymocyte deficiency of bcl-2−/− mice (11), whereas overexpression of a bax transgene in the thymus led to decreased mature T cells (12). Bax moves from the cytosol to the mitochondria under conditions that induce cell death, for example in staurosporine-treated Cos-7 cells in which a Bax—green fluorescent protein fusion protein was overexpressed (13) or during IL-3 withdrawal in FL5.12A cells (14, 15). The mechanism of Bax translocation is unknown. Once in the mitochondria, Bax may mediate its lethal effects through a channel-forming activity (16), resulting in disruption of mitochondrial function and release of cytochrome c (17).
In the present study, we examined the role of Bax in cell death after withdrawal of IL-7. We observed that a transient rise in intracellular pH induced a conformational change in Bax that in turn induced its translocation to mitochondria. Similar phenomena were observed after IL-3 removal, suggesting this could be a general mechanism in trophic factor withdrawal.
Methods
Cells.
The IL-7-dependent cell line D1 was established from the CD4−CD8− subset sorted from p53−/− mouse thymocytes initially propagated in IL-7 and stem cell factor (SCF). The D1 cell line was maintained in RPMI 1640 (Life Technologies, Grand Island, NY) supplemented with 10% FBS (Life Technologies) and 50 ng/ml recombinant mouse IL-7 (PeproTech, Rocky Hill, NJ) (no SCF). The IL-3-dependent murine pro-B cell line FL5.12A (a kind gift from James A. McCubrey, East Carolina University, Greenville, NC) was maintained in RPMI 1640 supplemented with 10% FBS and 0.4 ng/ml recombinant mouse IL-3 (PeproTech).
Mitochondrial Protein Detection.
Lysis of D1 or FL5.12A cells was performed in isotonic buffer (200 mM mannitol/70 mM sucrose/1 mM EGTA/10 mM Hepes, pH 6.9) by Dounce homogenization. Unbroken cells, nuclei, and heavy membranes were pelleted at 1,000 × g and discarded. The mitochondrial-enriched fraction was then produced by pelleting at 12,000 × g for 20 min, washing the membrane pellet one time in isotonic buffer, followed by a final resuspension in RIPA lysis buffer (1× PBS/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS) with protease inhibitor mixture (Calbiochem). The recovered supernatant from the high-speed centrifugation represented the cytosolic fraction, whereas the high-speed pellet represented the mitochondrial fraction (14, 15).
For detection of Bax, Bcl-2, or cytochrome c oxidase protein by Western blot, cell equivalent samples (10 μl containing approximately 20 μg protein) were separated by SDS/PAGE on 12% Tris–glycine gels (NOVEX, San Diego) and transferred to 0.2 μM polyvinylidene difluoride membranes (NOVEX). Blots were probed with a rabbit polyclonal antiserum specific for the amino terminal of Bax (N20, Santa Cruz Biotechnology) or to the full-length Bax protein (Δ21, Santa Cruz Biotechnology), with a hamster monoclonal antibody specific for Bcl-2 (PharMingen), or with a mouse polyclonal antibody specific for cytochrome c xidase (Research Diagnostics, Flanders, NJ), followed by the appropriate secondary antibodies conjugated to horseradish peroxidase (Santa Cruz Biotechnology) and then visualized by enhanced chemiluminescence (Pierce) following the manufacturer’s protocol.
For immunoprecipitation, cell lysates, precleared with rabbit IgG (Santa Cruz Biotechnology), were incubated with 1 μg of anti-Bax antibodies, N20 or Δ21, for 4 hr, followed by incubation with Protein A/G Plus agarose (Santa Cruz Biotechnology) for 2 hr. Antibody-agarose conjugates were washed in RIPA buffer and analyzed by Western blot, as described above.
pH Determination.
D1 or FL5.12A cells deprived of IL-7 or IL-3 for various times were treated with 1 μM BCECF-AM (Molecular Probes) for 30 min at 37°C, and intracellular pH was determined by fluorescence-activated cell sorter analysis (18), following manufacturer’s protocol. A pH calibration curve (Fig. 1 B and D Insets) was generated by preloading cells with 1 μM BCECF-AM, followed by incubation for 30 min in different pH buffers, from 6.8 to 8.0, in the presence of the permeabilizing agent nigericin (10 μM, Molecular Probes) in a high-K+ Hepes-buffered Hanks’ balanced salt solution (19).
Figure 1.
On cytokine withdrawal, Bax protein translocates to the mitochondria, and intracellular pH rises. (A) The subcellular fraction containing Bax in D1 cells was examined after IL-7 withdrawal. In the presence of IL-7, Bax was predominately cytosolic. Starting 6 hr after IL-7 withdrawal, Bax appeared in the mitochondrial fraction. Bcl-2 showed predominately a mitochondrial distribution and was not altered during the first 8 hr of IL-7 withdrawal. Cytochrome c oxidase indicates a strictly mitochondrial distribution, and in all blots serves as a control for the subcellular fractionation. (B) The effect of IL-7 withdrawal on intracellular pH in D1 cells was evaluated by using BCECF, as described in Methods. A steady rise in intracellular pH occurred, peaking at 6 hr, returning to neutral values by 8 hr after IL-7 withdrawal, and staying near neutrality for up to 30 hr. (C) The subcellular fraction containing Bax in FL5.12A cells was examined after IL-3 withdrawal. In the presence of IL-3, Bax was predominately cytosolic. However, starting 2 hr after IL-3 withdrawal, Bax appeared in the mitochondrial fraction. Cytochrome c oxidase controls for mitochondrial distribution. (D) The effect of IL-3 withdrawal on intracellular pH was evaluated by using BCECF, as described in Methods. A rapid rise in intracellular pH occurred, peaking at 2 hr after withdrawal, returning to neutral values by 5 hr after IL-3. No further rise in pH was detected up to 30 hr after IL-3 withdrawal.
In Vitro and in Vivo pH Manipulations.
To alter the pH of cell lysates, D1 cells were mechanically disrupted (as above) in isotonic buffers at pH 6.9, 7.8, or 8.1. The lysates were incubated for 15 min (to allow Bax translocation to mitochondria), then fractionated to recover mitochondria and analyzed for Bax by immunoblot, as previously described. To alter intracellular pH in living viable cells, D1 cells were permeabilized with nigericin (1 μM) in a high-K+ buffer containing Hepes (25 mM) (19) at pH 6.9, 7.8, or 8.1, for 30 min at 37°C. Cells were then disrupted, fractionated, and analyzed for cytosolic and mitochondrial Bax by immunoblot, as previously described.
Protease Digestion of Bax.
Lysates of D1 cells were treated with either proteinase K (Life Technologies), 1, 10, or 100 μg/ml, for 5 min and digestion stopped with 10 mM PMSF (Sigma) or with trypsin (Calbiochem), 0.3, 3, or 30 μg/ml for 5 min and the reaction stopped with 30× excess aprotinin (Sigma). Bax produced by in vitro translation (described below) was treated with either proteinase K, 0, 1 or 10 μg/ml, for 15 min and digestion stopped with 10 mM PMSF, or with trypsin, 0, 0.5, or 5 μg/ml for 15 min and the reaction stopped with 30× excess aprotinin.
Triton-X114 Phase Partitioning.
To examine the hydrophobicity of Bax, Triton X-114 phase partitioning was used (20, 21). Briefly, cytosolic extracts prepared in isotonic buffers at either pH 6.9, 7.8, or 8.1 were treated with 2% (vol/vol) Triton X-114 (Sigma), and the hydrophobic detergent phase was partitioned by incubation at 37°C followed by high-speed centrifugation. Bax was immunoprecipitated from the detergent phase and analyzed by immunoblot, as previously described.
Site-Directed Mutagenesis of Bax.
Site-directed mutagenesis was performed to substitute Leu for Lys-189 and Lys-190 by using the PCR with mutated primers. For wild-type (WT) Bax, the primer pair used was: 5′-TTC ATG GAC GGG TCC GGG GAG C-3′ and 5′-TTA TCA GCC CAT CTT CTT CCA GAT GGT-3′. For the C-terminal leucine mutant (MUT) Bax, the primer pair used was 5′-TTC ATG GAC GGG TCC GGG GAG C-3′ and 5′-TTA TCA GCC CAT GAG GAG CCA GAT GGT-3′. All PCR constructs were verified by dideoxy-sequencing protocols. PCR products for WT-Lys–189Lys-190 and MUT-Leu-189–Leu-190 were cloned in TA vectors (Invitrogen), and protein was expressed by using the TnT T7-coupled rabbit reticulocyte lysate system (Promega), following manufacturer’s guidelines. The translation products were radiolabeled with [35S]methionine (10 mCi/ml, Amersham), electrophoresed on a 12% Tris–glycine gel (NOVEX), fixed, dried, and placed on film for overnight exposure.
Site-directed mutagenesis to substitute Ala for Asp-2 and Glu-6 was performed by using PCR with mutated primers, as above. For WT-Bax, the same primer pair as above was used. For the N-terminal alanine MUT-Bax, the primer pair used was 5′-TTC ATG GCC GGG TCC GGG GCG C-3′ and 5′-TTA TCA GCC CAT CTT CTT CCA GAT GGT-3′. All PCR constructs were verified by dideoxy-sequencing protocols. WT-Asp-2–Glu-6 and MUT-Ala-2–Ala-6 PCR products were cloned in PCR-script vectors (Stratagene), then expressed and translated as above.
Results
To study how IL-7 maintains cell survival, we established a thymocyte cell line, D1, that depends on exogenous IL-7. This is, to our knowledge, the first use of such a cell line to study the molecular mechanisms underlying death induced by withdrawal of this cytokine. On loss of IL-7 signaling in D1 cells, death occurs in 30% of cells by 24 hr, 70% by 32 hr, and >90% by 48 hr, with no survival beyond 72 hr. On IL-7 withdrawal, transcripts for the antiapoptotic proteins Bcl-2 and Bcl-XL sharply declined after 2 hr, but transcripts for the death proteins Bax, Bak, and Bad persisted in the absence of IL-7 (data not shown). To determine whether Bax was involved in cell death in the IL-7-dependent D1 cells, we pretreated these cells with Bax antisense oligodeoxynucleotides and found that they were protected from withdrawal-induced death (data not shown). Therefore, we examined the relationship of Bax to the cell death process.
Bax Translocation Coincides with an Intracellular pH Rise.
D1 cells were deprived of IL-7 for various times, and subcellular protein fractions were made and analyzed. As shown in Fig. 1A, in the presence of IL-7 (time 0 hr), Bax was found in the cytosolic fraction, whereas very little was seen in the mitochondrial fraction. However, 6 hr after IL-7 withdrawal, a large increase in inserted Bax protein was found in the mitochondrial fraction. Although bcl-2 mRNA levels declined 2 hr after IL-7 withdrawal, Bcl-2 protein persisted in the mitochondrial membrane fractions (Fig. 1A). Thus Bcl-2 may protect mitochondria from Bax-induced damage, because loss of Bcl-2 by 24 hr coincided with the onset of cell death (data not shown). Cytochrome c oxidase (Fig. 1A) is shown as a positive control for mitochondrial protein.
Signals from the IL-7 receptor therefore maintained Bax in a soluble cytosolic form, whereas withdrawal of IL-7 led to translocation of Bax to mitochondrial membranes. To determine how IL-7 receptor regulates this process, several possible mechanisms were evaluated. Unlike the mechanism by which another death protein, Bad, is regulated (22), we could not detect phosphorylated forms of Bax (data not shown). The addition of the caspase inhibitor Z-VAD-FMK at 100 μM did not interfere with Bax translocation (data not shown), nor was any cleavage product of Bax detected (23) (Fig. 1A); this suggests that, unlike the death protein Bid, which participates in Fas-induced death (24), proteolytic cleavage of Bax did not occur on IL-7 withdrawal. To examine whether cytosolic Bax associated with a partner protein or formed a homodimer with itself (15) by which it could be retained in the cytosol (or escorted to the mitochondria), protein chemical crosslinking with disuccinimidyl suberate was used. Only the monomeric form of Bax protein could be detected in the cytosolic protein fraction, whereas in mitochondria, Bax could be found both as a monomer and in multimers crosslinked to Bcl-2 or itself (data not shown). Therefore, Bax was not found crosslinked to another protein in the cytosol prior to its mitochondrial translocation.
We next sought a mechanism by which the conformation of Bax protein could change, inducing mitochondrial translocation, and evaluated whether alterations in intracellular pH could account for this. As shown in Fig. 1B, IL-7 withdrawal led to a rapid and steady intracellular alkalinization, rising to a peak above pH 8.0 at 6 hr after withdrawal and returning to more neutral pH levels by 8 hr after withdrawal. Intracellular pH remained near neutrality from 8 to 30 hr after withdrawal. The transient and early alkalinization could be reversed by readdition of IL-7 (after 3 hr of withdrawal), which restored neutral intracellular pH (not shown). Treatment with Z-VAD or cycloheximide did not prevent intracellular alkalinization, suggesting that the rise in intracellular pH did not require the activation of caspases or de novo protein synthesis (data not shown). Pro-T cells (isolated from day 15 fetal thymuses), which depend on the trophic action of IL-7 (6), also underwent alkalinization, increasing intracellular pH 0.2–0.3 units within a few hours of IL-7 deprivation (data not shown).
To determine whether transient alkalinization was unique to IL-7 withdrawal or occurred more generally during apoptosis, we examined the pro-B cell line, FL5.12A, which undergoes cell death on IL-3 withdrawal. Within 2 hr of IL-3 deprivation, Bax protein was found in the mitochondrial fraction (Fig. 1C). In parallel, we observed that within 2 hr a transient alkalinization also occurred (Fig. 1D). This alkalinization lasted several hours, followed by a return to neutral pH. The use of staurosporine to induce apoptosis in the IL-7-dependent D1 cells also caused a rapid intracellular alkalinization with 30 min of treatment (the same time point at which Bax translocation was detected), followed by acidification and death by 8 hr (data not shown).
Acidification has been reported to occur during apoptosis in various cell lines (25, 26). Acidification also eventually occurred in D1 cells, but only after 30 hr of IL-7 deprivation, when the cells had started to demonstrate the morphological changes of cell death, such as annexin staining (data not shown). Therefore, trophic factor withdrawal may generally be associated with an early and transient alkalinization, followed closely by Bax protein translocation to the mitochondria, and later a terminal acidification occurring during the final effector stages of apoptosis.
High pH Induces the Mitochondrial Translocation of Bax.
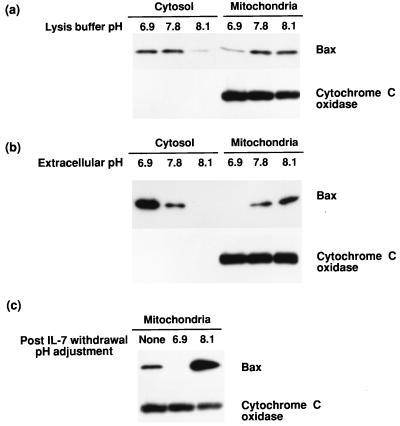
In D1 cells, because the peak alkalization time point of 6 hr coincided with the time of Bax translocation to mitochondria, we asked whether high pH triggered this movement. To test whether high pH could induce Bax insertion into mitochondria, D1 cells were disrupted in isotonic buffers of different pH (ranging from pH 6.6 to 8.1), and the homogenates were briefly incubated to allow Bax translocation. As illustrated in Fig. 2A, exposure to cell lysis buffer of pH of 7.8 or higher induced translocation of Bax to mitochondria. A second method was also used to assess the effect of pH on Bax translocation. Whole cells were permeabilized with nigericin, adjusting the intracellular pH to that of the extracellular buffers. As shown in Fig. 2B, Bax translocated to mitochondria at pH 7.8 or above and became correspondingly depleted from the cytosolic fraction. Therefore, two methods of raising pH (Fig. 2 A and B) mimicked the effect of IL-7 withdrawal on Bax translocation to mitochondria. Conversely, preventing the pH rise blocked Bax translocation after IL-7 withdrawal, as shown in Fig. 2C. We also tested the effects of acidic pH on the induction of Bax translocation and found that very little protein could be recovered from cell lysates incubated at pH 6.5 or lower, likely resulting from the effects of protein aggregation or activation of proteolytic digestion.
Figure 2.
Bax translocation to mitochondria is induced by high pH. The effect of alkaline pH on Bax translocation to mitochondria was examined by using two methods (A and B), and movement to the mitochondria was inhibited by pH neutralization (C). (A) D1 cells were mechanically disrupted in isotonic buffers at pH 6.9, 7.8, or 8.1. At pH 7.8 or higher, an increase in mitochondrial Bax was detected. The blot for cytochrome c oxidase was included as a control. (B) D1 cells were permeabilized with nigericen to alter intracellular pH. At pH 7.8 or greater, mitochondrial Bax protein levels increased, whereas cytosolic Bax decreased. The blot for cytochrome c oxidase was included as a control. (C) IL-7 was withdrawn from D1 cells for 5 hr and the cells permeabilized with nigericen to maintain the intracellular pH at 6.9 or 8.1 during the last 3 hr of withdrawal. Neutralization of the pH after IL-7 withdrawal prevented the mitochondrial translocation of Bax.
High pH Induces a Conformational Change in Bax Protein.
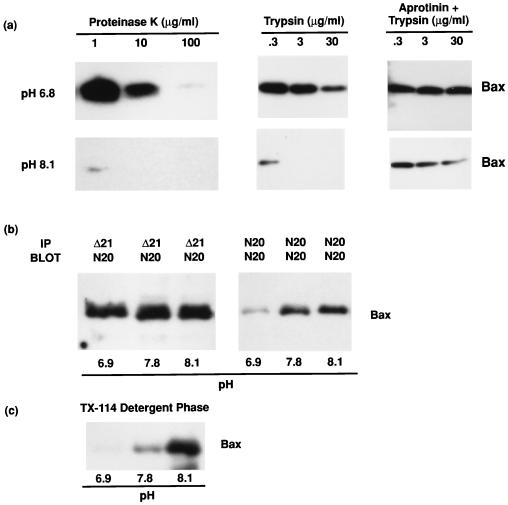
Because high pH could induce Bax translocation from a hydrophilic compartment to a hydrophobic one, we examined the possibility that pH controlled Bax conformation. Three criteria of Bax conformational change were used: protease susceptibility, exposure of cryptic epitopes, and hydrophobicity. Cytosols prepared at different pH values were treated with proteinase K or trypsin and, as a control, trypsin digests were also pretreated with the trypsin inhibitor aprotinin. As shown in Fig. 3A, Bax was much more susceptible to proteolytic digestion above pH 7.8, consistent with a more “open” conformation at high pH. Control experiments showed that digestion of BSA by these enzymes showed little variation over this pH range (not shown). In Fig. 3B, cytosol at different pH values was treated with either a Bax antiserum (N20) that recognizes epitopes in a small region of the N terminus of Bax or with an antiserum (Δ21) that recognizes epitopes on most of the Bax protein. The N terminus-specific antibody immunoprecipitated more Bax protein at high than at low pH, suggesting that at neutral pH the N terminus is hidden, whereas it becomes more exposed at high pH. As a control, antiserum against the full-length protein could immunoprecipitate Bax equally well at different pHs (Fig. 3B). To assess whether an increase in the hydrophobicity of Bax occurred at high pH (resulting from exposure of the hydrophobic transmembrane domain in the C terminus), cytosol prepared in buffers of different pH was partitioned into Triton X-114 detergent. As shown in Fig. 3C, more hydrophobic protein was detected at high pH than at neutral pH, indicated by the increasing recovery of Bax in the detergent phase, consistent with high pH inducing exposure of the hydrophobic C terminus of Bax.
Figure 3.
Demonstration of a pH-induced conformational change in cytosolic Bax. To determine whether high pH induced a conformational change in Bax, assays were performed to examine three parameters: sensitivity to proteolytic cleavage, exposure of cryptic epitopes, and hydrophobicity. (A) To examine Bax sensitivity to proteolytic cleavage, cytosolic extracts from D1 cells were prepared in isotonic buffers at either pH 6.9, 7.8, or 8.1, as in Fig. 2B, and digested with either proteinase K or trypsin, as described in Methods. Bax protein was analyzed by immunoblot with anti-Bax (N20). High pH rendered Bax much more susceptible to proteolytic digestion. (B) To examine exposure of cryptic epitopes in Bax, cytosolic extracts were prepared in isotonic buffer at either pH 6.9, 7.8, or 8.1 as in Fig. 2B. Immunoprecipitation reactions were then performed with anti-Bax N20, an antiserum produced against the N terminus 20 aa, or with anti- Bax Δ21, an antiserum produced against full-length Bax protein. Bax was then analyzed by immunoblot with anti-Bax N20. High pH induced exposure of the N-terminal epitopes recognized by N20. (C) To examine the hydrophobicity of Bax, Triton X-114 phase partitioning was used (20, 21). Bax was immunoprecipitated from the detergent phase and analyzed by immunoblot, as previously described. High pH increased the hydrophobicity of Bax.
Site-Directed Mutagenesis of Bax Termini Mimics High pH Effects and Abolishes pH Dependency.
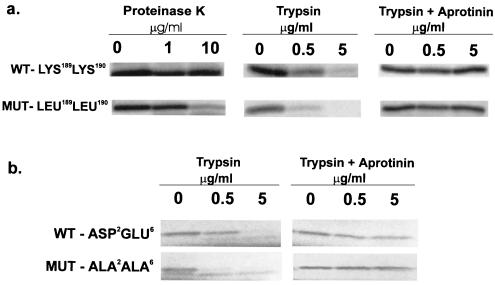
The results shown in Fig. 3 A–C suggest a model of Bax changing from a “closed” conformation at neutral pH, with its N and C termini hidden, to an “open” conformation at alkaline pH, with its N and C termini exposed. To test this model for Bax translocation to the mitochondria, site-directed mutagenesis was used to create a Bax mutant in which the positively charged C-terminal amino acids, Lys-189 and Lys-190, were exchanged for uncharged leucines. If the model of termini interacting via charged amino acids is correct, this mutant Bax should mimic high pH, assume the “open” conformation and, therefore, be protease sensitive. Fig. 4A shows in vitro-translated Bax, lacking the positively charged residues at the C terminus, increased sensitivity to digestion by either proteinase K or trypsin. Another Bax mutant in which the negatively charged N-terminal amino acids Asp-2 and Glu-6 were exchanged for uncharged alanines also showed increased protease sensitivity (Fig. 4B). Therefore, loss of charged amino acids at either terminus resulted in enhanced proteolytic digestion.
Figure 4.
Charged amino acids in the C terminus or N terminus make Bax protease resistant. (A). Positively charged amino acids in the C-terminal domain of Bax are required to maintain a conformation that is resistant to protease digestion. Site-directed mutagenesis, to replace Lys-189 and Lys-190 with Leu-189 and Leu-190, was performed by using PCR with mutated primers. Bax PCR products were cloned in TA cloning vectors and protein expressed in a coupled transcription/translation procedure incorporating [35S]methionine. Proteinase K and trypsin digestions were performed as described in Methods. (B) Negatively charged amino acids in the N-terminal domain of Bax are required to maintain a conformation that is resistant to protease digestion. Site-directed mutagenesis, to replace Asp-2 and Glu-6 for Ala-2 and Ala-6, was performed by using PCR with mutated primers. Bax PCR products were cloned in PCR-Script cloning vectors and protein expressed in a coupled in vitro transcription/translation procedure incorporating [35S]methionine. Trypsin digestion was performed as described in Methods.
Discussion
This study identifies a mechanism of mitochondrial targeting of the death protein Bax after loss of trophic stimuli. After IL-7 or IL-3 withdrawal from dependent cell lines, a transient intracellular alkalinization occurred. We show that this cytosolic alkalinization altered the conformation of Bax, exposing membrane-targeting domains, resulting in Bax translocation to the mitochondrial membrane. Site-directed mutagenesis of Bax implicated charged amino acids in the N and C termini as the pH-sensitive sites.
Cytosolic acidification had been reported to occur during apoptosis in various cell lines, such as an IL-2-dependent cytotoxic T lymphocyte line (25) or an IL-3-dependent myeloid cell line, BAF3 (26). However the cytosolic acidification described in those reports, as well as in our own findings, is a late event briefly preceding or coinciding with annexin staining, DNA fragmentation, and the activation of proteases, all terminal events of apoptosis. In accord with our results, mature thymocytes were reported to undergo cytosolic alkalinization during the process of corticosteroid-induced apoptosis (19, 27).
Recent studies support the concept that Bax changes conformation during initiation of cell death (28). Bax bound to mitochondria has been shown to expose the N terminus, and deletion of the N terminus resulted in constitutive mitochondrial translocation (14, 29). The C terminus of Bax has also been proposed to control its subcellular localization (30).
On the basis of our findings relating pH and Bax, we propose the following model. At neutral pH, in the presence of trophic factors, negative residues in the N terminus could interact with positive residues in the C terminus, concealing the hydrophobic transmembrane region. Then at high pH, after trophic factor withdrawal, the net charge at the termini would change, resulting in reduced attraction between the termini, exposing the membrane-seeking regions. The candidate negative residues in the N terminus are Asp-2 and Glu 6—all these negative residues fall within the first 19-aa segment which, on deletion, renders Bax membrane seeking (14). The candidate positive residues, occurring just before the end of the C terminus, are Lys-189 and Lys-190. We show that by mutating these charged amino acids, either at the C or N termini, we can mimic the effects of high pH during IL-7 withdrawal. These results lend support to our proposed model of the pH effect on Bax conformation, and also, in showing a direct effect of pH on Bax, argue against a role for intermediary proteins in Bax translocation induced by alkaline pH.
It is reasonable to propose that a rise in intracellular pH could alter the conformation of a protein by acting on charged side groups of amino acids. Although the pK of lysine as a free amino acid is 10.5, its environment within a protein can reduce it by 2 or more pH units (31). Adjacent positive charges, as occurs in the two adjacent lysines in the Bax C terminus, have the potential to lower their pKs from 10.5 to the range observed (in Fig. 3) and affect Bax conformation. Other pH reactive proteins have similar arrangements of charged amino acids at critical structural sites; for example, the inhibitor protein (IF1) of the mitochondrial F0F1 ATP synthase complex is a pH-dependent protein (32). The activity of IF1 decreases with increasing pH, with maximal loss occurring at pH 8. This pH dependency is lost when three lysines residues (Lys-46, Lys-47, Lys-58) are exchanged with uncharged alanines, providing another case in which conformational change affecting function occurs at alkaline pH values and is mediated by adjacent charged residues.
In conclusion, we have shown that during withdrawal of IL-7 in a dependent cell line, a transient rise in intracellular pH promotes the mitochondrial translocation of Bax by altering charged interactions between the N and C termini, exposing the hydrophobic transmembrane domain. There may be additional effects of pH on other charged regions of Bax, for example in the putative membrane penetrating BH1 region formed by α-5 and α-6 helices of Bax (33), whereby high pH could facilitate mitochondrial insertion. It remains to be determined how cessation of signaling from the IL-7 receptor causes a rise in intracellular pH. Possible mechanisms include decreased glucose transport resulting in decreased lactic acid production (as well as ATP synthesis) or deregulation of the Na+/H+ exchanger (34), the Na+/HCO3−/CO32− cotransporter, or the Cl−/HCO3− exchanger (35), all of which have been implicated in thymocyte apoptosis (19).
Acknowledgments
We thank P. Henckart for suggesting use of Triton X-114, J. McCubrey for the FL5.12A line, R.Youle for helpful discussions, and T. Sayers, C. Zacharchuk, and J. Oppenheim for comments on the manuscript.
Abbreviation
- WT
wild type
References
- 1.Watson J D, Morrissey P J, Namen A E, Conlon P J, Widmer M B. J Immunol. 1989;143:1215–1222. [PubMed] [Google Scholar]
- 2.Hofmeister R, Khaled A R, Benbernou N, Rajnavolgyi E, Muegge K, Durum S K. Cytokine Growth Factor Rev. 1999;10:41–60. doi: 10.1016/s1359-6101(98)00025-2. [DOI] [PubMed] [Google Scholar]
- 3.von-Freeden-Jeffry U, Vieira P, Lucian L A, McNeil T, Burdach S E, Murray R. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peschon J J, Morrissey P J, Grabstein K H, Ramsdell F J, Maraskovsky E, Gliniak B C, Park L S, Ziegler S F, Williams D E, Ware C B, et al. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maraskovsky E, Teepe M, Morrissey P J, Braddy S, Miller R E, Lynch D H, Peschon J J. J Immunol. 1996;157:5315–5323. [PubMed] [Google Scholar]
- 6.Kim K, Lee C K, Sayers T J, Muegge K, Durum S K. J Immunol. 1998;160:5735–5741. [PubMed] [Google Scholar]
- 7.Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman I L. Cell. 1997;89:1033–1041. doi: 10.1016/s0092-8674(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 8.Di Santo J P, Rodewald H R. Curr Opin Immunol. 1998;10:196–207. doi: 10.1016/s0952-7915(98)80249-5. [DOI] [PubMed] [Google Scholar]
- 9.Blom B, Spits H, Krimpenfort P. In: The Role of the Common Gamma Chain of the IL-2, IL-4, IL-7 and IL-15 Receptors in Development of Lymphocytes. Smit Sibinga S, Das P, Loewenberg D, editors. Dordrecht, the Netherlands: Kluwer; 1997. pp. 3–11. [Google Scholar]
- 10.Knudson C M, Tung K S, Tourtellotte W G, Brown G A, Korsmeyer S J. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- 11.Knudson C M, Korsmeyer S J. Nat Genet. 1997;16:358–363. doi: 10.1038/ng0897-358. [DOI] [PubMed] [Google Scholar]
- 12.Williams O, Norton T, Halligey M, Kioussis D, Brady H J M. J Exp Med. 1998;188:1125–1133. doi: 10.1084/jem.188.6.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolter K G, Hsu Y T, Smith C L, Nechushtan A, Xi X G, Youle R J. J Cell Biol. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goping I S, Gross A, Lavoie J N, Nguyen M, Jemmerson R, Roth K, Korsmeyer S J, Shore G C. J Cell Biol. 1998;143:207–215. doi: 10.1083/jcb.143.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross A, Jockel J, Wei M C, Korsmeyer S J. EMBO J. 1998;17:3878–3885. doi: 10.1093/emboj/17.14.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antonsson B, Conti F, Ciavatta A, Montessuit S, Lewis S, Martinou I, Bernasconi L, Bernard A, Mermod J J, Mazzei G, et al. Science. 1997;277:370–372. doi: 10.1126/science.277.5324.370. [DOI] [PubMed] [Google Scholar]
- 17.Eskes R, Antonsson B, Osen-Sand A, Montessuit S, Richter C, Sadoul R, Mazzei G, Nichols A, Martinou J C. J Cell Biol. 1998;143:217–224. doi: 10.1083/jcb.143.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franck P, Petitipain N, Cherlet M, Dardennes M, Maachi F, Schutz B, Poisson L, Nabet P. J Biotechnol. 1996;46:187–195. doi: 10.1016/0168-1656(95)00189-1. [DOI] [PubMed] [Google Scholar]
- 19.Tsao N, Lei H Y. J Immunol. 1996;157:1107–1116. [PubMed] [Google Scholar]
- 20.Brusca J S, Radolf J D. Methods Enzymol. 1994;228:182–193. doi: 10.1016/0076-6879(94)28019-3. [DOI] [PubMed] [Google Scholar]
- 21.Pryde J G. Methods Mol Biol. 1998;88:23–33. doi: 10.1385/0-89603-487-9:23. [DOI] [PubMed] [Google Scholar]
- 22.del-Peso L, Gonzalez G M, Page C, Herrera R, Nunez G. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 23.Wood D E, Thomas A, Devi L A, Berman Y, Beavis R C, Reed J C, Newcomb E W. Oncogene. 1998;17:1069–1078. doi: 10.1038/sj.onc.1202034. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Zhu H, Xu C J, Yuan J. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Eastman A. J Biol Chem. 1995;270:3203–3211. doi: 10.1074/jbc.270.7.3203. [DOI] [PubMed] [Google Scholar]
- 26.Furlong I J, Ascaso R, Lopez R A, Collins M K. J Cell Sci. 1997;110(Pt 5):653–661. doi: 10.1242/jcs.110.5.653. [DOI] [PubMed] [Google Scholar]
- 27.Dai H Y, Tsao N, Leung W C, Lei H Y. Radiat Res. 1998;150:183–189. [PubMed] [Google Scholar]
- 28.Hsu Y T, Youle R J. J Biol Chem. 1997;272:13829–13834. doi: 10.1074/jbc.272.21.13829. [DOI] [PubMed] [Google Scholar]
- 29.Desagher S, Osen-Sand A, Nichols A, Eskes R, Montessuit S, Lauper S, Maundrell K, Antonsson B, Martinou J C. J Cell Biol. 1999;144:891–901. doi: 10.1083/jcb.144.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nechushtan A, Smith C L, Hsu Y T, Youle R J. EMBO J. 1999;18:2330–2341. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehninger A L, Nelson D L, Cox M M. Principles of Biochemistry. New York: Worth; 1993. , chap. 8. [Google Scholar]
- 32.Papa S, Zanotti F, Cocco T, Perrucci C, Candita C, Minuto M. Eur J Biochem. 1996;240:461–467. doi: 10.1111/j.1432-1033.1996.0461h.x. [DOI] [PubMed] [Google Scholar]
- 33.Schlesinger P H, Gross A, Yin X M, Yamamoto K, Saito M, Waksman G, Korsmeyer S J. Proc Natl Acad Sci USA. 1997;94:11357–11362. doi: 10.1073/pnas.94.21.11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu W H, Loh T T. Biochim Biophys Acta. 1995;1269:122–128. doi: 10.1016/0167-4889(95)00102-x. [DOI] [PubMed] [Google Scholar]
- 35.Restrepo D, Kozody D J, Spinelli L J, Knauf P A. J Gen Physiol. 1988;92:489–507. doi: 10.1085/jgp.92.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]