Abstract
Epitopes depending on three-dimensional folding of proteins have during recent years been acknowledged to be main targets for many autoantibodies. However, a detailed resolution of conformation-dependent epitopes has to date not been achieved in spite of its importance for understanding the complex interaction between an autoantigen and the immune system. In analysis of immunodominant epitopes of the U1-70K protein, the major autoantigen recognized by human ribonucleoprotein (RNP)-positive sera, we have used diversely mutated recombinant Drosophila melanogaster 70K proteins as antigens in assays for human anti-RNP antibodies. Thus, the contribution of individual amino acids to antigenicity could be assayed with the overall structure of the major antigenic domain preserved, and analysis of how antigenicity can be reconstituted rather than obliterated was enabled. Our results reveal that amino acid residue 125 is situated at a crucial position for recognition by human anti-RNP autoantibodies and that flanking residues at positions 119–126 also appear to be of utmost importance for recognition. These results are discussed in relation to structural models of RNA-binding domains, and tertiary structure modeling indicates that the residues 119–126 are situated at easily accessible positions in the end of an α-helix in the RNA binding region. This study identifies a major conformation-dependent epitope of the U1-70K protein and demonstrates the significance of individual amino acids in conformational epitopes. Using this model, we believe it will be possible to analyze other immunodominant regions in which protein conformation has a strong impact.
The development of autoantibodies is a common feature in autoimmune diseases. Consequently, the analysis of B-cell epitopes has remained an important issue for over a decade because it could suggest which domains of the protein are attractive for the immune system and thus could improve our understanding of the functional contact between the autoantigen and the autoantibody, as well as the mechanisms resulting in self proteins acting as autoimmune targets.
Interactions between autoantigens and autoantibodies have to date mainly been analyzed by truncated subclones or synthetic peptides mimicking the proposed target epitopes. However, recent findings and reviews have implied that most autoepitopes may be conformation-dependent (1–3), thus stressing the need to develop procedures to analyze in detail which functional domain of the autoantigen could be immunodominant.
The RNA-binding U1 small nuclear ribonucleoprotein (RNP)-associated 70K protein is the major target for autoantibodies in sera from patients with mixed connective tissue disease and can also be recognized by autoantibodies in sera from patients with systemic lupus erythematosus. The cell biological role of the 70K protein is a participant in RNA-splicing, and it has been identified as a member of the large family of RNA-binding proteins denoted “RNP-80” (4–6). Several laboratories have analyzed the epitopes of the 70K protein to increase the understanding of the complex interaction between the autoantigen and the immune system, but so far the smallest fragments reported to retain antigenicity encompassed residues 99–167 or 100–156 (7–9). This region overlaps the RNA-binding region. Attempts to use smaller fragments or peptides have resulted in a substantial or total loss of antigenicity, and this has been interpreted as an indication of the importance of conformational epitopes in the 70K autoantigen.
Human nuclear autoantigens are generally well recognized across species barriers, and autoantibodies have been extensively used for the isolation and purification of low abundant intracellular splicing-related proteins from various species (10–12). Yet several studies reporting a species-specific recognition have been published (13–18). The conservation includes the U1-70K, which has been cloned from several species, e.g., frog (19), mouse (20), yeast (21), and the plant Arabidopsis thaliana (22). Even the 70K protein from the fruit fly, Drosophila melanogaster, is 68% conserved within the first amino-terminal 214 amino acids of the protein and 76% conserved in the sequences corresponding to the major antigenic region as compared with the human protein (23). The disparities are not randomly distributed but consist of occasional conservative substitutions and a few regions in which the amino acid sequences differ substantially. Using recombinant expression subclones with interchanged segments from human and Drosophila U1-70K proteins, we previously defined a smaller region of the earlier described major antigenic domain to be the prime target of the anti-70K autoantibodies (24). We thereby determined amino acid residues 99–128 to be of critical importance for the autoantigenic response, and our attention was drawn to the few disparities between the two proteins that caused such a substantial change in autoantibody response. Our previous results proposed that this was not caused by a linear sequence (17, 24) and had to be dependent on both amino acid sequence and three-dimensional folding exposure of the epitope in a proper manner. Thus, the starting point for this work was to maintain the entire antigenic region of the U1-70K protein intact in structure and analyze the importance of individual amino acids.
Materials and Methods
In Vitro Mutagenesis Procedure.
Substrate preparation.
Preparative PCRs were performed to prepare a 1,173-bp fragment of the pMAL D. melanongaster (Dm) 70K clone (17, 23) corresponding to the human immunodominant region, with the forward primer EXTMAL-1: GCCGCCAGTCCGAACAAA and the reverse primer EXTMAL-2: ATCTTCTCTCATCCGCCAAAAC. After control on 1% agarose gel, the fragment was eluted from the PCR mix with Wizard PCR preps (Promega).
Derivation of random fragments by DNase I digestion.
Three micrograms of the substrate fragment in a 30-μl reaction mixture of 50 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, and 0.15 units of DNase I (Sigma) was digested into random fragments for 10′ at 25°C, and the reaction was stopped with EDTA to a final concentration of 10 mM. Fragments in the size range 50 to 200 bp were collected from a 1.5% agarose gel, and DNA were eluted with JetSorb gel extraction kit (Genomed, Hybaid, Middlesex, U.K.).
PCR without primers: “DNA shuffling.”
Approximately 0.7–1 μg of the DNased material was used as the DNA source in a 100-μl reaction PCR mix, but without primers. The session consisted of 1 cycle of 1′ at 94°C, 45" at 50°C, and 45" at 72°C; 40 cycles of 30" at 94°C, 30" at 50°C, and 30" 72°C; and a final incubation in 72°C for 10′. Several parallel reactions were performed, and the amplified material was pooled.
Introduction of specific point mutations.
To introduce specific point mutations with mutagenic primers, 15–40 μl from the DNA shuffling mixes were submitted to a second PCR session with 0.8 μM of one of several specific mutagenic primers in a 100-μl reaction. The PCR session consisted of 1 cycle of 1′ at 94°C, 45" at 50°C, and 45" at 72°C; 35 cycles of 45" at 94°C, 45" at 50°C, and 45" at 72°C; and final incubation in 72°C for 10′. The forward primers, with mutated residues in italics were oligo 99: ATGCCACAGGAGACCCGTTTCG, mutating the Dm glutamic acid into the corresponding human glycine, oligo 101: GAGGACGCATTTCGCACGCTGTTCA, mutating the Dm proline to the corresponding human alanine, and oligo 125: TTCGAGGTCTACGGCCCCATCAAG, mutating the Dm phenylalanine to the corresponding human valine. As reverse primer, GAAAGGGGGATGTGCTGC was used. This step produced pools of fragments of three different lengths, each containing a point mutation together with the previously produced mutations from the DNA shuffling step. To produce fragments of identical size for subsequent subcloning, these fragments were reshuffled in a 5:1 ratio with a 652-bp BsaAI/SacII fragment encoding an upstream overlapping sequence. The product mix of 15–40 μl was submitted to standard PCR in a 100-μl reaction with the forward primer GAAAACGCCCAGAAAGGTGA and the reverse primer GAAAGGGGGATGTGCTGC. A fragment of 904 bp was produced together with other minor bands. The 904-bp band was cut out, eluted, and used for subsequent subcloning.
Cloning and Expression of Recombinant Antigens.
PCR fragments were cloned into the pGEM-T-vector System 1 (Promega) according to the manufacturer’s instructions. Heterogeneous HinDIII/KpnI fragments of 667 bp were cut out with from pools of mutated clones, were subcloned into the expression vector system pMAL (New England Biolabs), and were transformed into Escherichia coli TB-1 cells, and expression was induced by isopropyl β-d-thiogalactosidase. To screen for the antigenic phenotype of the mutants, recombinant fusion proteins were tested on immunoblots. As controls, the vector-encoded fusion partner maltose binding protein (MaBP) and previously constructed human and Drosophila 70K recombinant clones (17, 25, 26) were used.
Sera.
Human anti-RNP sera from 25 anti-nuclear antibody (ANA)-positive patients previously analyzed in our laboratory (25, 27) were used. The patients, 18 women and 6 men, were attendant at the Department of Rheumatology at Huddinge University Hospital. The diagnoses of these patients were 13 mixed connective tissue disease (28), five systemic lupus erythematosus (29), one systemic lupus erythematosus and mixed connective tissue disease, one Sjögren’s syndrome (30), one juvenile chronic arthritis (31), and three unclassified connective tissue disease. Sera from five healthy individuals were used as control sera. As control for the fusion partner MaBP, commercial rabbit anti-MaBP antibodies were used (New England Biolabs).
Immunoblotting.
Immunoblotting of recombinant proteins was performed as described (25). In brief, the proteins were separated on a 10% SDS/PAGE gel and were transferred electrophoretically to a nitrocellulose membrane. After blocking with milk/BSA, the membrane was probed with primary serum at a 1:1,000 dilution. Affinity-purified alkaline phosphatase-conjugated anti-human IgG antibodies or anti-rabbit IgG antibodies (Dakopatts, Glostrup, Denmark) were used at a 1:2,000 dilution with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Sigma) as substrate.
DNA Sequencing.
Sequencing of selected mutants was performed on a 373A automated DNA sequencer (Applied Biosystems). Samples were prepared according to the instructions of the manufacturers of the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin–Elmer). Mutant clones were independently sequenced with the two sequencing primers ATGTCCGCTTTCTGGTATGC for the (+) strand and CGTTCCCTCTCGTTGTCCTC for the (−) strand, and in the majority of clones also with additional primers. The wisconsin package 9.1 (Genetics Computer Group, Madison, WI) was used for analysis and alignment.
Enzyme-Linked Immunosorbent Assay (ELISA).
ELISA was performed as described (17). In brief, medium binding ELISA plates (Costar) were coated overnight at 4°C with 1 μg of recombinant protein/well in 0.05 M carbonate buffer (pH 9.5) and were blocked with 1% BSA before incubation for 2 h with primary antibody. Bound antibodies were detected by using alkaline phosphatase conjugated anti-human IgG antibodies (Dakopatts) diluted 1:2,000 and the substrate p-nitrophenylphosphate (Sigma). The reaction was stopped with 1 M NaOH, and optical density (OD) read at 405 nm. The sera were tested in triplicates and at 1:1,000 dilution. For MaBP in the pMAL vector system, the mean background reaction was 0.040 in OD with a standard deviation of 0.038.
Preincubation Experiments.
Sera diluted 1:1,000 were incubated with 25 μg of recombinant antigen in 2.5 ml TPBS/BSA for 1.5 h before analysis in ELISA as described above.
Statistical Analysis.
To compare the reaction of the mutants with the reaction against the human clone, an ANOVA followed by a Scheffe post hoc test was used.
Tertiary Structure Modeling.
The Swiss-Model Protein Modeling Server (Glaxo Wellcome Experimental Research S.A., Geneva) was used to select optimal homology model structures (refs. 32–35; Swiss-Model at www.expasy.ch/swissmod/SWISS-MODEL.html). A search for tertiary structure templates was performed by the ExPASy Molecular Biology Server (Glaxo Wellcome Research and Development S.A., Geneva) (36). swisspdbviewer 2.6 (ref. 37; www.expasy.ch/spdbv/mainpage.htm) was used for investigating and analyzing the derived structure.
Results
To define the major epitopes of the U1-70K protein recognized by anti-RNP sera, we chose the approach of inducing mutations throughout the immunodominant region of the similar but less antigenic Dm U1-70K protein. Random mutations were introduced by DNA shuffling (38–40), and specific point mutagenesis was used to induce mutations at three residues estimated by three-dimensional modeling to be at possible key positions.
Selection of Clones.
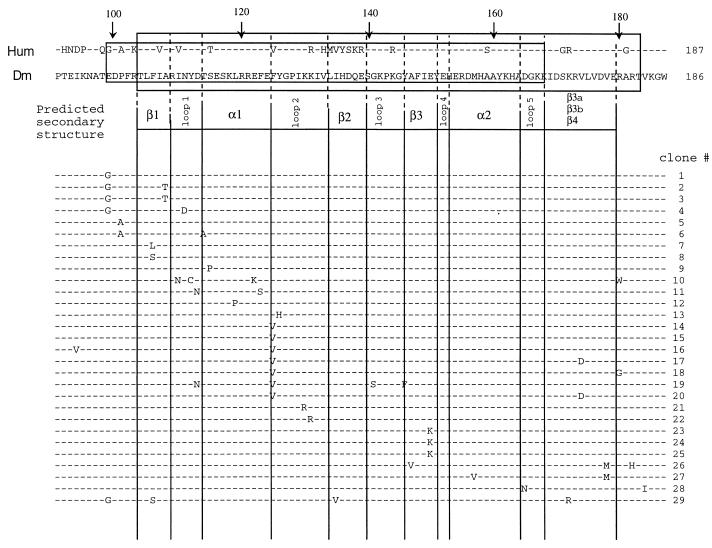
To analyze the mutational impact on the antigenicity of the immunodominant region, the mutants were subcloned into the pMAL expression vector, and 254 expression clones were screened by immunoblotting for “phenotypic” changes in antigenicity. For this, we used two patient sera previously established as low-reactive against Dm 70K (17), but with high recognition of human U1-70K. Seventy clones expressing recombinant proteins of apparently higher, lower, or equal antigenicity as the original pMAL Dm 70K clone were selected for DNA sequencing. Eighteen of the 70 clones contained the nonmutated wild-type Drosophila DNA sequence, which was in concordance with their appearance on immunoblotting. Apart from the specifically induced point mutations at amino acid residue positions 99, 101, and 125, spontaneous mutations throughout the major antigenic region were observed as expected. Of the 70 sequenced clones, 28 clones with different phenotypes, as determined by recognition of RNP-positive sera, were selected for protein purification and subsequent quantification of their antigenic properties by ELISA (Fig. 1). Mutations, denoted by their corresponding amino acid residues between amino acids 91–187, are depicted in Fig. 1, but also mutations outside this region were observed. The antigenic properties did not seem to be affected in clones containing only mutations outside of the major antigenic region (data not included).
Figure 1.
Summary of 28 Dm 70K mutants used for ELISA quantification. Mutated residues in each clone are indicated. The amino acid alignment of amino terminal part of the human (Hum) and Dm 70K proteins is according to Mancebo et al. (23). Dashes denote identity between human and Drosophila sequence. The major antigenic region according to Netter et al. (7) is shaded in gray, and the RNA binding region is boxed. The predicted secondary structure of the 70K RNA binding domain is based on the Swiss-Model tertiary derivation (www.expasy.ch/swissmod/SWISS-MODEL.html) and the crystal structure of the hnRNP A protein (52).
Analysis of Antibody Response Against Mutant Clones.
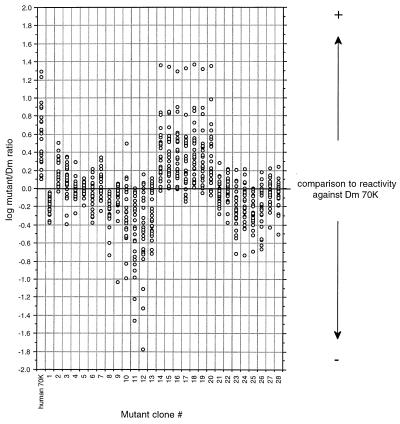
Initially, mutant clones 1–28 were subjected to ELISA analysis with 25 patient sera and three normal sera. The clones against which the patient sera gave the highest response were also tested against two additional normal sera. All normal serum responses were below optical density (OD) at 0.1. Fig. 2 is a scatter diagram in logarithmic scale, demonstrating the optical densities of the mutant clones. Compared with the response against the original Dm 70K protein, antigenicity was reconstituted by the recombinant 70K proteins produced by clones 14–20, containing a substitution of the Dm phenylalanine at position 125 with the corresponding human valine. As a contrast, the shuffling-produced mutation from a conserved tyrosine to histidine in the neighboring residue 126 in clone 13 caused a dramatic loss of antigenicity. Other clones demonstrating a large loss of antigenicity were 10–12, whose common feature was mutations between residues 119 and 124. None of the clones containing mutations in the first part of the immunodominant region (clones 1–9) demonstrated a significant change in antigenicity, even though some heterogeneity in the responses was observed.
Figure 2.
Scatter diagram in logarithmic scale depicting the response against the mutated Dm 70K clones relative to the response against the pure Dm 70K protein for 25 RNP-positive sera. Each column represents one clone, and each circle in this column represents the optical density of one patient serum against this clone. The value 0.0 on the y axis indicates that the response against the mutated protein is the same as the response against the wild-type Dm 70K protein. As a reference, the human/Dm ratio is depicted at the left in the diagram.
In conclusion, clones 10–20 demonstrated the largest change in antigenicity. As a common component, these clones contained mutations between amino acid residues 119–126. Clones 14–20 all contained the same mutation at amino acid position 125, from the Dm residue phenylalanine to the human valine, and exhibited a large antigenic increase, whereas clones 10–13 showed a substantial decrease. These clones all contained “nonhuman” mutations.
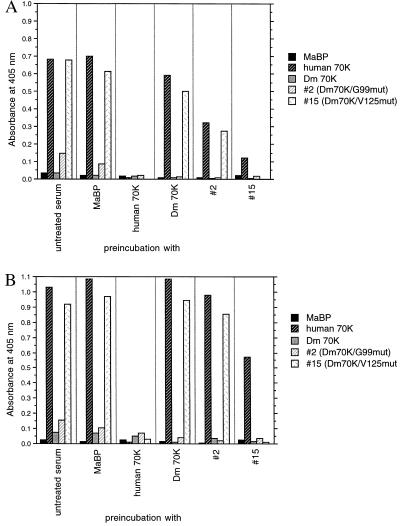
Preincubation Experiments.
To examine the specificity against the mutant clones, we incubated six representative high titer RNP-positive sera with a selection of recombinant proteins before ELISA. Preincubation experiments were performed with the recombinant protein produced by clone 15 containing the mutation to human valine at position 125 (hence termed Dm 70K/V125 mut) and were compared with preincubations with the fusion partner MaBP, human 70K, Dm 70K, and clone 2 (Dm 70K/G99 mut). Preincubation with the Dm 70K/V125 mut protein reduced the response against the human 70K protein, and these results imply that the response against the Dm70K/V125 mut is to a large extent specific and that the recombinant proteins can specifically adsorb the antibodies. Fig. 3 illustrates that the remaining response against human 70K for patient serum B is 18% after preincubation with Dm 70K/V125 mut (Fig. 3A) and that the remaining response of serum P is 56% (Fig. 3B). The preincubation of the four other sera produced intermediate results compared with these two patient sera.
Figure 3.
Effect of preincubation with MaBP, human 70K, Dm 70K, and mutant clones 2 (Dm70K/G99 mut) and 15 (Dm70K/V125 mut). The sera were preincubated with the appropriate protein (specified under the bars) before ELISA-determination of the reactivity of both untreated and preincubated sera with the various recombinant 70K proteins (specified within each bar). (A) Serum P. (B) Serum B.
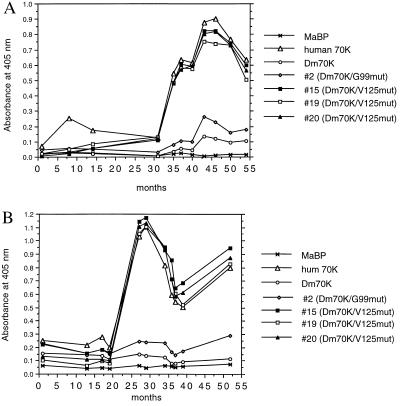
Analysis of the Reactivity with Mutated Dm 70K Proteins in Two Longitudinally Collected Serum Series.
To examine how the autoantibody response to Dm 70K/V125 mut was related longitudinally to the human 70K, we tested two RNP-positive sera exhibiting changes in autoantibody levels with time. The responses against MaBP, human 70K, Dm 70K, clone 2 (Dm70K/G99 mut), and clones 15, 19, and 20 (all Dm70K/V125 mut) (Fig. 4) were compared, and we observed that the response against Dm 70K/V125 mut clones mirrored the response against the human 70K protein. Recognition of the residue 99 mutation in clone 2 (Dm70K/G99 mut) was distinctly lower but not as low as that of Dm 70K, indicating that a small fraction of the species-specific antibodies are directed against epitopes containing this amino acid residue. The observation that one patient serum (illustrated in Fig. 4B) gave a higher response with Dm70K/V125 mut compared with the human 70K protein was consistent and reproducible for this patient serum.
Figure 4.
Longitudinal studies of human patient serum reactivity against MaBP, human 70K, Dm 70K, and mutant clones 2 (Dm70K/G99 mut), 15, 19, and 20 (all Dm70K/V125 mut) as determined by ELISA. (A) Serum O. (B) Serum S.
Statistical Analysis.
We compared the reaction of RNP-positive sera against the human clone with the reaction against the mutants described in Fig. 2 and hence divided the clones into the following groups: group 99, clones 1–4; group 102, clones 5–6; group 108, clones 2–3; group 120X, clones 10–13; and group 125, clones 14–20. All groups except the 125-group could significantly be distinguished from the human clone (data not included).
Tertiary Structure Modeling.
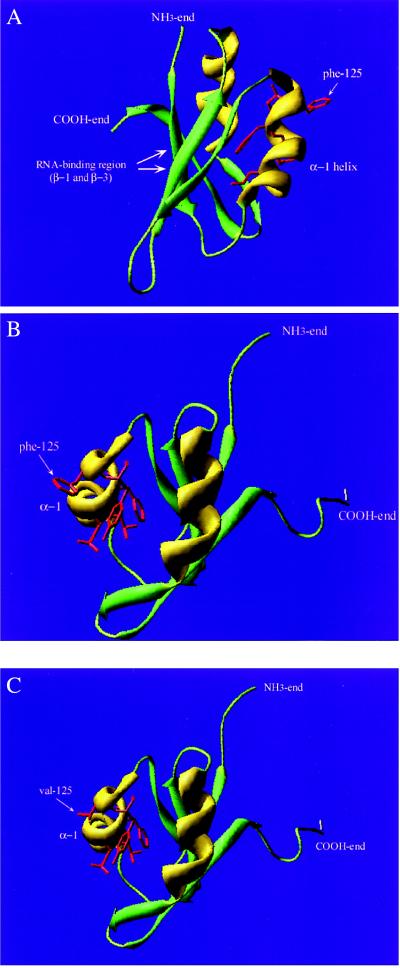
The tertiary structure of the U1-70K protein has not been determined, but results from x-ray crystallography and NMR spectroscopy of highly homologous proteins in the same large family of RNA binding proteins containing a prevalent RNA recognition motif have been reported (4, 5, 41–43). Every protein structure so far determined in this group, the hnRNP C (45), Drosophila Sxl (46), U1A (47–50), and hnRNP A1 (51, 52), have exhibited the same tertiary structure built of β-sheets and α-helices in secondary domains, wrapped in the characteristic folding order βαββαβ with connecting loops. This commonly shared RNA-binding domain has been described as a minimal folding unit. To investigate whether this could be used to model a putative structure for the U1-70K protein, the amino acid sequence from amino acid position 95–190 of the human U1-70K protein was sent to the Swiss-Model Automated Protein Modeling Server (www.expasy.ch/swissmod/SWISS-MODEL.html) for modeling by homology. The modeling server selected the hnRNP A protein (52) and the Dm sex-lethal protein (Sxl) (45) from the same RNA-binding family as the 70K protein as the best homologues for deriving a putative tertiary structure of the RNA binding region. In this model, the conformational epitope at residues 119–126 would be situated in the end of α-helix 1 (Fig. 5).
Figure 5.
Tertiary folding model of the immunodominant region of the U1-70K protein. β-sheets are presented in green, α-helices in yellow, and loops in light green. The NH3 and COOH ends of the fragment are depicted. (A) The immunodominant region with the RNA-binding β1- and β3-sheets as well as the α-helix-1 are depicted. Side chains of mutated amino acid residues in the 119–126 conformation-dependent epitope are presented in red (residues 119, 122, 123, 125, and 126). The arrow points to the amino acid residue at position 125. (B) The structure model viewed from the opposite direction. The 119–126 epitope is depicted with the Dm residue phenylalanine at position 125. (C) The epitope depicted with the human valine at amino acid residue 125.
Discussion
A major restraint in the detailed analysis of autoepitopes has been that they are often conformation-dependent, and thus have individual amino acids constituting an epitope been hard to pinpoint. This has been a major problem in the case of the U1-70K protein, and a detailed resolution has never been attained.
We have developed a procedure to retain the major antigenic region structurally intact and yet analyze the impact of individual amino acids. Combining the DNA shuffle method (38, 39) in which general homologous recombination could be performed in vitro, and specific point mutagenesis (40), we generated a number of both randomly derived and specifically altered mutant clones. To our knowledge, DNA shuffling has not previously been modified to reconstitute antigenicity and thus to overcome some of the difficulties in the analysis of conformation-dependent epitopes.
Two specifically mutated residues in the beginning of the immunodominant region did not appear to influence the autoimmune recognition, and a randomly derived mutation at position 108 caused a slight but nonsignificant increase in antigenicity. In contrast, a mutation at position 125 caused a considerable change. All mutant clones containing this specific point mutation from the Dm phenylalanine to the human valine at amino acid position 125 resulted in a substantial increase in antigenic response, whatever other random mutations throughout the major antigenic region they contained in total. Other “shuffle”-produced mutations at amino acid residues 119–126, mutated from either conserved residues to a substitution present in neither the Dm nor the human protein or from a Dm specific residue to another nonhuman amino acid residue, gave strongly reduced antigenic responses. Nonconservative substitutions in other areas did not have the same dramatic effects. Statistical analysis supported the hypothesis that the reaction against the human clone and the reaction against the 125-mutants to a large extent were identical.
Additional experiments further validated the importance of the 125 substitution. After preincubation with the Dm 70K/V125 mut protein, anti-RNP sera poorly recognized the pure human clone, demonstrating that a major part of the antibody response against this clone is directed against a human-specific epitope. Results obtained by using series of anti-RNP positive sera collected over time also indicated that a major part of the reaction against the pure human clone and the Dm 70K/V125 mut clone were identical.
We also attempted to relate the conformational epitope to a three-dimensional position on the U1-70K protein. Using the closely related human hnRNP A1 and Dm Sxl proteins for modeling the three-dimensional structure of the U1-70K protein, one can observe that the region around residues 119–126 is situated in the end of α-helix 1 (Figs. 1 and 5). These residues face away from the residues in the RNA-binding β1 and β3 sheets, and, by using this model, the amino acid residue at position 125 should be an easily accessible B-cell epitope (Fig. 5 B and C). We have previously reported that not only the amino acid residue sequence is important, but that the epitope has to be kept in place by the α-helix and β-sheet (17, 24). By now demonstrating that valine-125 is part of a major human epitope, it is not surprising that a switch to phenylalanine (Fig. 5) induces a disarrangement of the epitope surface. Both amino acids involved are hydrophobic and contain nonpolar side chains, but phenylalanine is larger and contains a bulky aromatic side chain. Valine, however, is one of the smallest amino acids containing a less complex carbon side chain. Thus, the tertiary structure derivation supports the theory of the region 119–126 as an autoantigenic, conformational epitope of the U1-70K protein. The positions of the key amino acids that constitute this major epitope also make it possible to understand why previous approaches have failed to identify smaller fragments than 56–67 amino acid residues as being antigenic. A truncation from either end would affect protein conformation and would cause the compressed helix-loop-β-sheet structure to unfold, thus obscuring the originally prominent and crucial valine at position 125.
Our findings might have clinical implications because all tested sera recognized the identified epitope. This demonstrates a remarkable homogeneity of the otherwise heterogeneous autoantigenic B-cell response and might also instigate therapeutic approaches of inducing tolerance.
In conclusion, this study indicates possibilities of mapping conformational epitopes, and we propose the approach to reconstitute antigenicity in a related but poorly antigenic protein. With the current pace of gene identification, more examples of related proteins should become available from families of related proteins within a species or proteins for intraspecies comparisons.
Acknowledgments
We are grateful to Drs. Ulf Nyman, Stephen Mount, and Ricardo Mancebo for originally providing the human and Drosophila melanogaster cDNA clones, respectively, Dr. Eva Hedfors for the gift of human RNP-sera, Jan Kowalski for statistical help, and Dr. Robert Harris for critical reading. This work was supported by the Swedish Medical Research Council, the Swedish Society for Medicine, the Swedish Rheumatism Association, the Swedish Foundation for Strategic Research, the Jubileum Foundation of Disease Insurance, and the Foundations of King Gustaf V:s 80-year, Lundström, Golje, Axel & Margaret Ax:son Johnson, Magn. Bergvall, Wallström, Prof. Nanna Svartz’, Åke Wiberg, and Bengt Dahlin.
Abbreviations
- Dm
Drosophila melanogaster
- RNP
ribonucleoprotein
- MaBP
maltose binding protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Casiano C A, Tan E M. Int Arch Allergy Immunol. 1996;111:308–313. doi: 10.1159/000237385. [DOI] [PubMed] [Google Scholar]
- 2.Hassfeld W, Steiner G, Studnicka-Benke A, Skriner K, Graninger W, Fischer I, Smolen J S. Arthritis Rheum. 1995;38:777–785. doi: 10.1002/art.1780380610. [DOI] [PubMed] [Google Scholar]
- 3.Tan E M, Muro Y, Pollard K M. Clin Exp Rheum Suppl. 1994;11:S27–S31. [PubMed] [Google Scholar]
- 4.Burd C G, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 5.Kenan D J, Query C C, Keene J D. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 6.Query C C, Keene J D. Cell. 1987;51:211–220. doi: 10.1016/0092-8674(87)90148-6. [DOI] [PubMed] [Google Scholar]
- 7.Netter H J, Guldner H H, Szostecki C, Will H. Scand J Immunol. 1990;32:163–176. doi: 10.1111/j.1365-3083.1990.tb02906.x. [DOI] [PubMed] [Google Scholar]
- 8.Cram D S, Fisicaro N, Coppel R L, Whittingham S, Harrison L C. J Immunol. 1990;145:630–635. [PubMed] [Google Scholar]
- 9.Klein Gunnewiek J M T, van de Putte L B A, van Venrooij W J. Clin Exp Rheumatol. 1997;15:549–560. [PubMed] [Google Scholar]
- 10.Tan E M. J Clin Invest. 1989;84:1–6. doi: 10.1172/JCI114127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balczon R. Proc Soc Exp Biol Med. 1993;204:138–154. doi: 10.3181/00379727-204-43645. [DOI] [PubMed] [Google Scholar]
- 12.von Mühlen C A, Tan E M. Semin Arthritis Rheum. 1995;24:323–358. doi: 10.1016/s0049-0172(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 13.Keech C L, Gordon T, McCluskey J. Clin Exp Immunol. 1996;104:255–263. doi: 10.1046/j.1365-2249.1996.16726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruijn G J M, Thijssen J P H, Smith P R, Williams D G, van Venrooij W J. Eur J Biochem. 1995;232:611–619. doi: 10.1111/j.1432-1033.1995.611zz.x. [DOI] [PubMed] [Google Scholar]
- 15.Reichlin M, Rader M, Harley J B. Clin Exp Immunol. 1989;76:373–377. [PMC free article] [PubMed] [Google Scholar]
- 16.Semsei I, Tröster H, Bartsch H, Schwemmle M, Igloi G L, Bachmann M. Gene. 1993;126:265–268. doi: 10.1016/0378-1119(93)90378-g. [DOI] [PubMed] [Google Scholar]
- 17.Welin Henriksson E, Pettersson I. J Autoimmun. 1996;9:551–559. doi: 10.1006/jaut.1996.0074. [DOI] [PubMed] [Google Scholar]
- 18.Weng Y M, McNeilage J, Topfer F, McCluskey J, Gordon T. J Clin Invest. 1993;92:1104–1108. doi: 10.1172/JCI116617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Etzerodt M, Vignali R, Ciliberto G, Scherly D, Mattaj I W, Philipson L. EMBO J. 1988;7:4311–4321. doi: 10.1002/j.1460-2075.1988.tb03330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornig H, Fischer U, Costas M, Rauh A, Lührmann R. Eur J Biochem. 1989;182:45–50. doi: 10.1111/j.1432-1033.1989.tb14798.x. [DOI] [PubMed] [Google Scholar]
- 21.Smith V, Barrell B G. EMBO J. 1991;10:2627–2634. doi: 10.1002/j.1460-2075.1991.tb07805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy A S, Czernik A J, An G, Poovaiah B W. Biochim Biophys Acta. 1992;1171:88–92. doi: 10.1016/0167-4781(92)90143-n. [DOI] [PubMed] [Google Scholar]
- 23.Mancebo R, Lo P C, Mount S M. Mol Cell Biol. 1990;10:2492–2502. doi: 10.1128/mcb.10.6.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welin Henriksson E, Pettersson I. J Autoimmun. 1997;10:559–568. doi: 10.1006/jaut.1997.0163. [DOI] [PubMed] [Google Scholar]
- 25.Nyman U, Lundberg I, Hedfors E, Pettersson I. Clin Exp Immunol. 1990;81:52–58. doi: 10.1111/j.1365-2249.1990.tb05290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welin Henriksson E, Hansson H, Karlsson-Parra A, Pettersson I. Vet Immunol Immunopathol. 1998;61:157–170. doi: 10.1016/s0165-2427(97)00142-6. [DOI] [PubMed] [Google Scholar]
- 27.Pettersson I, Wang G, Smith E I, Wigzell H, Hedfors E, Horn J, Sharp G C. Arthritis Rheum. 1986;29:986–996. doi: 10.1002/art.1780290807. [DOI] [PubMed] [Google Scholar]
- 28.Alarcón-Segovia D, Cardiel M H. J Rheumatol. 1989;16:328–334. [PubMed] [Google Scholar]
- 29.Tan E M, Cohen A S, Fries J F, Masi A T, McShane D J, Rothfield N F, Schaller J G, Talal N, Winchester R J. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 30.Manthorpe R, Oxholm P, Prause J U, Schiodt M. Scand J Rheum Suppl. 1986;61:19–21. [PubMed] [Google Scholar]
- 31.Brewer E J J, Bass J, Baum J, Cassidy J T, Fink C, Jacobs J, Hanson V, Levinson J E, Schaller J, Stillman J S. Arthritis Rheum. 1977;20:195–199. [PubMed] [Google Scholar]
- 32.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Peitsch M C. Bio/Technology. 1995;13:658–660. [Google Scholar]
- 34.Peitsch M C. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 35.Peitsch M C, Jongeneel C V. Int Immunol. 1993;5:233–238. doi: 10.1093/intimm/5.2.233. [DOI] [PubMed] [Google Scholar]
- 36.Appel R D, Bairoch A, Hochstrasser D F. Trends Biochem Sci. 1994;19:258–260. doi: 10.1016/0968-0004(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 37.Peitsch M C, Wilkins M R, Tonella L, Sanchez J C, Appel R D, Hochstrasser D F. Electrophoresis. 1997;18:498–501. doi: 10.1002/elps.1150180326. [DOI] [PubMed] [Google Scholar]
- 38.Stemmer W P C. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 39.Stemmer W P C. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crameri A, Stemmer W P C. BioTechniques. 1995;18:194–196. [PubMed] [Google Scholar]
- 41.Birney E, Kumar S, Krainer A R. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hodgkin J, Plasterk R H, Waterston R H. Science. 1995;270:410–414. doi: 10.1126/science.270.5235.410. [DOI] [PubMed] [Google Scholar]
- 43.Nagai K, Oubridge C, Ito N, Avis J, Evans P. Trends Biochem Sci. 1995;20:235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]
- 44.Wittekind M, Gorlach M, Friedrichs M, Dreyfuss G, Mueller L. Biochemistry. 1992;31:6254–6265. doi: 10.1021/bi00142a013. [DOI] [PubMed] [Google Scholar]
- 45.Lee A L, Kanaar R, Rio D C, Wemmer D E. Biochemistry. 1994;33:13775–13786. doi: 10.1021/bi00250a031. [DOI] [PubMed] [Google Scholar]
- 46.Avis J M, Allain F H, Howe P W, Varani G, Nagai K, Neuhaus D. J Mol Biol. 1996;257:398–411. doi: 10.1006/jmbi.1996.0171. [DOI] [PubMed] [Google Scholar]
- 47.Hoffman D W, Query C C, Golden B L, White S W, Keene J D. Proc Natl Acad Sci USA. 1991;88:2495–2499. doi: 10.1073/pnas.88.6.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu J, Hall K B. J Mol Biol. 1995;247:739–752. doi: 10.1006/jmbi.1995.0177. [DOI] [PubMed] [Google Scholar]
- 49.Nagai K, Oubridge C, Jessen T H, Li J, Evans P R. Nature (London) 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 50.Oubridge C, Ito N, Evans P R, Teo C-H, Nagai K. Nature (London) 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 51.Garrett D S, Lodi P J, Shamoo Y, Williams K R, Clore G M, Gronenborn A M. Biochemistry. 1994;33:2852–2858. doi: 10.1021/bi00176a015. [DOI] [PubMed] [Google Scholar]
- 52.Shamoo Y, Krueger U, Rice L M, Williams K R, Steitz T A. Nat Struct Biol. 1997;4:215–222. doi: 10.1038/nsb0397-215. [DOI] [PubMed] [Google Scholar]