SUMMARY
Nuclear pore complexes (NPCs) are 40–60 MDa protein assemblies embedded in the nuclear envelope of eukaryotic cells. NPCs exclusively mediate all transport between cytoplasm and nucleus. The nucleoporins that build the NPC are arranged in a stable core of module-like subcomplexes with 8-fold rotational symmetry. To gain insight into the intricate assembly of the NPC, we have solved the crystal structure of a protein complex between two nucleoporins, human Nup107 and Nup133. Both proteins form elongated structures that interact tightly via a compact interface in tail-to-tail fashion. Additional experiments using structure-guided mutants show that Nup107 is the critical anchor for Nup133 to the NPC, positioning Nup133 at the periphery of the NPC. Structure predictions suggest these domains to be composed of simple α-helical repeats, yet we find that they are highly individualized and non-repeating. This favors an NPC model assembled from non-redundant architectural nucleoporins.
INTRODUCTION
A membrane-enclosed nucleus that contains the genetic material is the defining feature of the eukaryotic cell. Access to the nucleus is regulated by the gatekeepers of the nucleus, large protein assemblies termed nuclear pore complexes (NPCs). They are embedded in circular openings of the nuclear envelope (NE) and control all transport into and out of the nucleus (reviewed in: (Tran and Wente, 2006; Weis, 2003)). Macromolecules larger than 20–40 kDa in mass cannot transit the NPC by passive diffusion, but are actively transported by an armada of soluble nuclear transport factors, such as the karyopherins (kaps) or importins/exportins/transportins. Kaps serve as adaptors for cargo to bind to the NPC, with transport directionality imparted by coupling cargo interaction to the Ran GTPase system (Gorlich and Kutay, 1999).
High-resolution structural data of kaps have revealed the molecular details of cargo binding and release, the regulation of the transport cycle by the small GTPase Ran, and how kaps interact with the NPC (Cook et al., 2007; Stewart, 2007; Suel et al., 2006). In contrast, X-ray crystallographic studies of the NPC itself are still in their infancy (Madrid and Weis, 2006; Suel et al., 2006). Cryo-electron tomography revealed that NPCs consist of a central framework with, on average, eight-fold rotational symmetry (Beck et al., 2007). The portion of the NPC embedded in the NE is the inner ‘spoke ring’, which is sandwiched between a cytoplasmic and a nuclear ring that anchor filamentous extensions into both compartments. The cytoplasmic filaments reach out into the cytoplasm and interact with incoming kap-cargo complexes. The nuclear filaments conjoin to a distal ring, the so-called nuclear basket of the NPC.
Proteomic studies in yeast and metazoa revealed that NPCs are assembled from a mostly homologous set of about 30 proteins, collectively termed nucleoporins (nups) (Cronshaw et al., 2002; Rout et al., 2000). In a novel approach, these proteomic studies were recently combined with a plethora of low-resolution structural information, such as distance-restraints, stoichiometric considerations and protein shape models resulting in a computer-generated assembly model of the NPC (Alber et al., 2007a; Alber et al., 2007b). In addition to the experimental tomographic structure, this assembly model now provides a framework for the relative location of the nups, and the subcomplexes they form, within the NPC.
In order to decipher the mechanism of nucleo-cytoplasmic transport in detail, we need structural resolution about an order of magnitude higher than what we currently have. Hampered by size and flexibility of the entire 40–60 MDa NPC, high-resolution structural studies have to be carried out on subassemblies (Schwartz, 2005).
The best-characterized Nup subcomplexes to date are the metazoan Nup107–160 subcomplex and its homolog in S. cerevisiae, the scNup84 subcomplex, present in 16 copies per NPC (Alber et al., 2007b). The yeast complex has a mass of ~500 kDa and contains seven proteins: Nup84, Nup85, Nup120, Nup133, the in vivo-cleaved carboxy-terminal domain of Nup145 (Nup145C), Seh1 and Sec13 (Lutzmann et al., 2002; Siniossoglou et al., 1996). Yeast strains with deletions of individual genes encoding subunits of the scNup84 complex show defects in mRNA export and clustering of NPCs within the NE (Siniossoglou et al., 1996). Isolation of the scNup84 subcomplex under native conditions or its in vitro reconstitution from recombinant nups expressed in E. coli, revealed a Y-shaped, triskelion-like morphology by negative staining electron microscopy (Lutzmann et al., 2002; Siniossoglou et al., 2000). In addition, the scNup84 subcomplex was suggested to play a role in transcription activation by providing a platform for the recruitment of transcription factors to the NPC and the nuclear periphery, a phenomenon dubbed “reverse recruitment” (Menon et al., 2005). Strikingly, members of the scNup84 subcomplex activated transcription themselves in vivo when fused to a heterologous DNA-binding domain.
The metazoan Nup107–160 subcomplex consists of Nup160 (homologous to scNup120), Nup107 (scNup84), Nup96 (scNup145C), Nup75 (scNup85), Sec13, and Seh1 (Belgareh et al., 2001; Fontoura et al., 1999; Harel et al., 2003; Vasu et al., 2001). Nup37 and Nup43 have been identified as additional members (Loiodice et al., 2004). Like their counterparts in yeast, constituents of this complex are localized on both sides of the NPC and are involved in mRNA export. Immunodepletion of the Nup107–160 subcomplex from Xenopus laevis egg extracts prevented reformation of NPCs in nuclear reconstitution assays (Harel et al., 2003; Walther et al., 2003). Depletion of single subcomplex members in HeLa cells prevented the assembly of several peripheral nucleoporins into the NPC (Boehmer et al., 2003; Harel et al., 2003; Walther et al., 2003). FRAP experiments show each Nup107–160 constituent is stably associated with the NPC during interphase (Rabut et al., 2004), further supporting that the Nup107–160 subcomplex is a crucial structural module in the central scaffold of the NPC. Moreover, in mitosis a small pool of Nup107–160 subcomplexes co-localizes with kinetochores (Belgareh et al., 2001; Loiodice et al., 2004; Zuccolo et al., 2007) and the spindle apparatus (Enninga et al., 2003; Orjalo et al., 2006) pointing toward an additional role in chromosome segregation during mitosis.
The interactions between the Nup107–160 subcomplex members appear to be very stable, since most functions are so far collectively ascribed to the entire subcomplex. For example, siRNA depletions of Nup85, Nup107 and Nup133 in metazoa have overlapping, almost identical phenotypes, i.e. they all lead to inhibition of mRNA export (Boehmer et al., 2003; Harel et al., 2003; Walther et al., 2003). Thus, to further understand the different functions of the Nup107–160 subcomplex it is highly desirable to elucidate the structure of the subcomplex members, notably to unravel the interfaces that connect the binding partners. That way, structure-guided, more specific manipulations of the Nup107–160 subcomplex would be possible allowing for dissecting the multitude of functions of this subcomplex in particular, and for the NPC in general.
Our studies were undertaken with the goal of providing insight into the assembly of nups within the Nup107–160 complex. We solved the crystal structure of the Nup107/Nup133 interaction complex, determined and analyzed the binding site, and tested structure-guided mutants. This work provides insight into the atomic details that govern nuclear pore assembly.
RESULTS
Mapping the Interaction between Human Nup107 and Nup133
Sequence analysis of the 925-residue human Nup107 revealed that it consists of a ~140-residue N-terminal region, which is poorly conserved and seems to have little or no secondary structure, followed by an α-helical domain spanning approximately 750 residues (Devos et al., 2004). Nup133 is built from an N-terminal 7-bladed β-propeller, followed by a C-terminal α-helical domain (Berke et al., 2004). We have previously shown that the C-terminal α-helical domain of Nup133 interacts with Nup107 in vitro (Berke et al., 2004).
To further characterize the interaction between Nup107 and Nup133, we constructed Nup107- and Nup133-fusion proteins with either GST- or 6x-His-tag. In yeast two-hybrid screens using Nup133 as bait, the extreme C-terminus of human Nup107 was sufficient for their interaction (Belgareh et al., 2001). Accordingly, a binding assay using GST-tagged recombinant Nup107 proteins and in vitro transcribed and translated full length Nup133 showed that a C-terminal truncation mutant of Nup107, Nup1071–644, failed to bind Nup133 (Figure 1A). Consequently, GST-tagged Nup107658–925 was co-expressed with a series of N-terminally truncated constructs of Nup133’s α-helical domain in E. coli (Figures 1B and 1C) – C-terminal truncations of this domain resulted in its insolubility. Co-purification via glutathione-sepharose affinity chromatography revealed that Nup107658–925 stably interacts with the entire α-helical domain of Nup133. In addition, we defined the minimal Nup107-binding region of Nup133, comprising residues 934–1156. Further truncation of this domain to residues 987–1156 resulted in complete failure to bind Nup107 (Figure 1B).
Figure 1. Nup107 and Nup133 Interact in Tail-to-Tail Fashion.
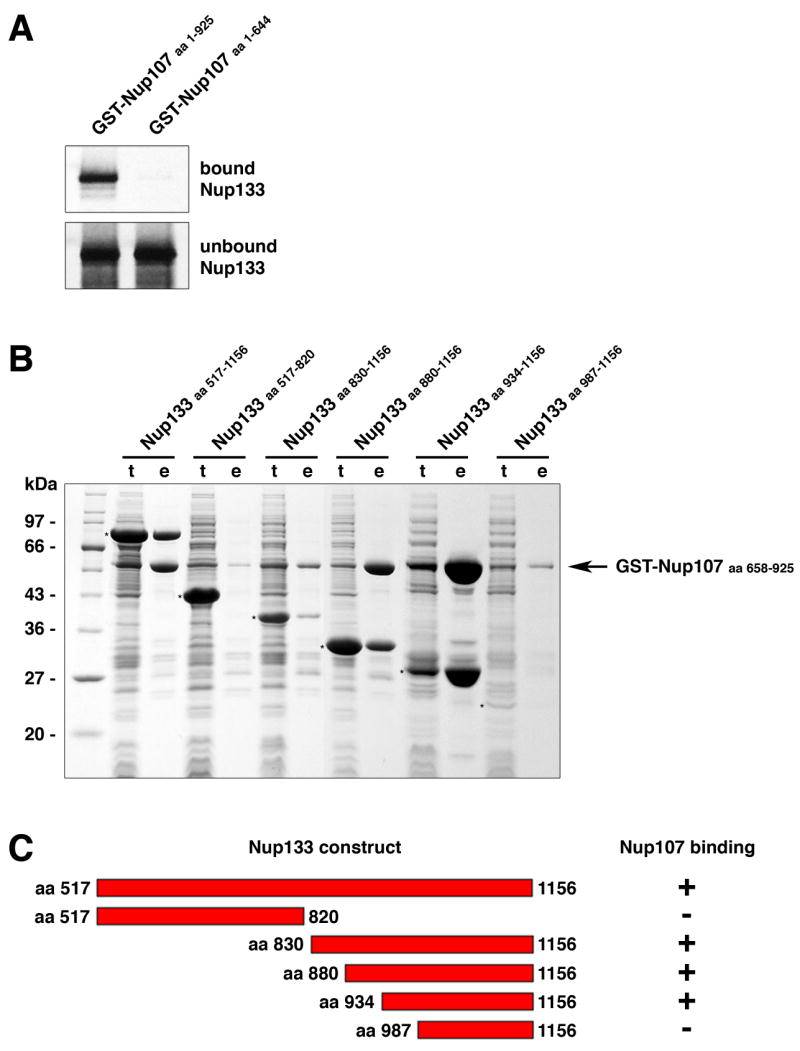
(A) In vitro binding of full length human Nup133 to recombinant GST-tagged human Nup107 visualized by autoradiography. Top and bottom panels show the bound and unbound fractions of [35S] methionine-labeled Nup133 translation product incubated with recombinant GST-Nup107 proteins immobilized on affinity resin. The C-terminus of Nup107 is critical for its interaction with Nup133. (B) Various C-terminal fragments of Nup133 (marked with asterisks) were co-expressed with GST-tagged Nup107 (aa 658–925) from separate vectors in E. coli and co-purified by glutathione sepharose affinity chromatography. Total cell lysates (t) and elutions from the GST-affinity columns (e) were separated by SDS-PAGE and analyzed by Coomassie staining. (C) Schematic representation of Nup133 constructs tested for Nup107-binding (+). A construct spanning residues 934 to 1156 of Nup133 represents the minimum domain required for the interaction with Nup107. The relative amounts of co-expressed proteins differs from clone to clone, due to pGEX- and pET-derived vectors harboring the same origin of replication. The results of the binding experiment are therefore limited to qualitative interpretation.
Structural Studies of Nup107/Nup133
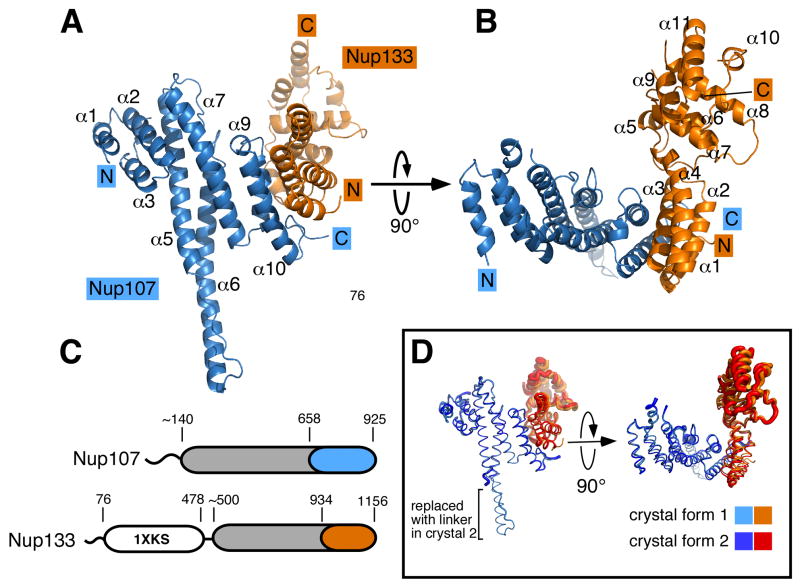
Based on our mapping experiments we constructed a bi-cistronic expression vector encoding the interacting domains of Nup107658–925 and Nup133934–1156. The protein complex was expressed and purified from E.coli lysate (Boehmer and Schwartz, 2007). After crystallization the structure of the complex was determined at a resolution of 2.55 Å (Rfree/R = 24.5%/21.4%; Figure 2; Table 1). A complex of Nup107658–925, with residues 772–801 replaced by a flexible 6-residue linker, and Nup133934–1156 was also crystallized and the structure was determined at 3.0 Å resolution (Rfree/R = 28.8%/22.7%; Figure 2C; Table 1). The crystals belong to different space groups. Both crystal forms contain one Nup107-Nup133 heterodimer in the asymmetric unit.
Figure 2. The Nup107/Nup133 Complex Structure.
(A) Overall structure of the Nup107658–925/Nup133934–1156 interaction complex. Nup107 is colored in blue, Nup133 in orange. Both proteins are entirely α-helical and interact via a compact interface, mainly involving two helices from each binding partner. Secondary structure elements in Nup107 are labeled (helices α4 and α8 of Nup107 are obscured and labels omitted for clarity). Nup107 forms an irregular stack of helices, with helix 6 protruding noticeably from the domain core. (B) Overall structure rotated by 90° compared to (A). Helices in Nup133 are numbered. Nup133 consists of two connected domains, an interacting 4-helix bundle and the C-terminal domain. (C) Schematic domain organization of Nup107 and Nup133. Important residue positions are indicated. α-helical domains are marked in grey outside of the crystallized regions which are colored in blue and orange, respectively. The β-propeller domain (PDB code 1XKS) is colored white. (D) Superposition of the Nup107658–925/Nup133934–1156 interaction complex crystallized in two different crystal forms, shown as tubes. In the protein construct used in crystal form 2 the helix 6 protrusion is replaced by a short flexible linker. Tube thickness proportional to temperature factors, indicating flexibility. The C-terminal helical domain of Nup133 is significantly more flexible than the rest of the protein and shows rigid body movement.
Table I.
Data collection and refinement statistics
| Data set | Nup107 658–925 Nup133 934–1156 | Nup107 658–925dF Nup133 934–1156 | SeMet |
|---|---|---|---|
| Data collection | |||
| Wavelength | 0.9792 | 0.9792 | 0.9792 |
| Space group | C2 | P212121 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 51.7, 127.9, 152.7 | 47.6, 65.5, 193.2 | 51.9, 128.8, 153.4 |
| abg(°) | 90.0, 97.36, 90.0 | 90.0, 90.0, 90.0 | 90.0, 97.65, 90.0 |
| No. of unique reflections | 32243 | 12599 | 39961 |
| Resolution (Å) | 40-2.55 (2.64-2.55) | 50-3.0 (3.11-3.0) | 40-2.9 (3.0-2.9) |
| Rsyma (%) | 10.5 (48.7) | 9.8 (52.4) | 6.7 (21.4) |
| Completeness (%) | 99.4 (98.8) | 97.8 (97.3) | 92.0 (52.5) |
| Redundancy | 4.2 (3.1) | 4.8 (4.3) | 3.4 (1.8) |
| I/s(I) | 16.1 (2.0) | 16.1 (2.0) | 17.8 (2.7) |
|
| |||
| Refinement | |||
|
| |||
| Resolution (Å) | 40-2.55 | 46–3.0 | |
| No. of unique reflections | 32190 | 12547 | |
| Rwork b/Rfree c (%) | 21.9/24.8 | 22.7/28.8 | |
|
| |||
| No. atoms | |||
| Protein | 3353 | 3249 | |
| Water | 131 | 34 | |
|
| |||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.012 | |
| Bond angles (deg) | 1.0 | 1.3 | |
|
| |||
| B-factors (Å2) | |||
| Protein | |||
| Nup107 | 54.1 | 64.3 | |
| Nup133D1/Nup133D2d | 70.5/144.7 | 77.0/114.4 | |
| Water | 59.3 | 63.6 | |
|
| |||
| Ramachandran | |||
| Most favored regions (%) | 95.4 | 91.6 | |
| Add. allowed regions (%) | 4.6 | 8.4 | |
| Outliers (%) | 0.0 | 0.0 | |
Rsym = Σ|Ii − <Ii>|/ΣIi, where Ii is the intensity of the ith observation and <Ii> is the mean intensity of the reflection.
Rwork = Σ(||Fobs| − |Fcalc||/Σ|Fobs|)
Rfree = R value for a randomly selected subset (10%) of the data that were not used for minimization of the crystallographic residual.
Highest resolution shell is shown in parenthesis.
Average B-factors are reported separately for the two domains of Nup133. D1 denotes the Nup107 interaction domain (residues 934–1008), D2 the C-terminal domain (residues 1028–1156)
Overall Architecture of the Nup107–133 complex
As illustrated in Figure 2, both proteins, Nup107658–925 and Nup133934–1156, are built from α-helices. Nup107658–925 and Nup133934–1156 form distinct domains that bind via a compact interface resulting in an L-shaped overall arrangement. Nup107658–925 forms an elongated domain. The helices are stacked perpendicularly to the long domain axis. The long axis of the domain measures about 60 Å, whereas its other two dimensions are ~ 25 Å. The domain is organized around a core composed of 3 helical hairpins stacked on top of each other. One helix, α6, protrudes noticeably from the helical core. The very C-terminus of Nup107 interacts with Nup133. Nup133934–1156 consists of two separate helical domains that are connected with moderate flexibility. In accordance with our biochemical data (Figure 1), the interacting domain of Nup133 comprises residues 934–1008 and forms a rigid block of four helices. The C-terminus, residues 1027–1134, adopts a compact, second domain that is closely tethered to the interacting domain.
Structure of Nup107658–925
Nup107658–925 contains 10 helices. With the exception of the short helices α4 and α9, located within connecting loops, the helices are arranged as a stack in consecutive and antiparallel order. The domain is structured around a core of three helical hairpins (α2/3, α5/6, α7/8). The helices vary considerably in length and the twist between each helical pair ranges from ~ 0º to ~ 50º. Helices α1 on the N-terminal side and α10 on the C-terminal end flank the core and close the domain (Figure 2A). Loops linking helical segments are generally short and well ordered. The only disordered region is a 15aa stretch between helices α4 and α5, omitted from the model. The irregular twist between consecutive helix pairs results in the absence of the typically observed overall features of helical repeat proteins (Kobe and Kajava, 2000). The domain is straight rather than curved and superhelicity is not observed. Within the Nup107658–925 domain α6 is markedly extended and protrudes like a finger from the core domain. With a length of 30Å the finger (residues 769 – 802) is as long as the core domain is wide. The finger is very well ordered in the crystal structure, including the extended loop that connects the fingertip back to the domain. The extended loop and finger form a common hydrophobic core, which helps explain the rigidity of this structural element.
Structure of Nup133934–1156
Nup133934–1156 folds into 11 helices, arranged in two blocks. Helices α1–α4 stack as two helical hairpins and form a rigid entity. Helices α6–α11 form the second helical block comprising the very C-terminal domain of Nup133. The latter domain has temperature factors averaging twofold higher than the rest of the complex (Table 1) resulting in electron density of limited quality. Helices α6–α11 form a domain of defined, compact structure. We examined whether poor crystal packing might be the cause for weak electron density within the C-terminal Nup133 domain. A modified protein complex, replacing the finger domain of Nup107 with a short flexible linker, was crystallized. Clipping the finger was based on the rational that this modification might trigger crystallization in a different space group and so it did. The combination of both crystal structures allowed us to build the C-terminal domain of Nup133 confidently, even though the absence of strong crystal contacts involving the Nup133 C-terminus was also observed in the second crystal form. A superposition of both structures shows that the two helical blocks of Nup133 exhibit a slight rigid body movement relative to each other (Figure 2C). It is suggestive that the tether between the C-terminal domain of Nup133 and the remainder of the protein inherently allows for restrained movement. The tether is composed of 15 residues forming an ordered loop structure, including the short helix α5. This connector module is ordered well enough to suggest that the two domains are linked through a continuous hydrophobic core. Thus, both crystal structures suggest that the two Nup133 domains can move independently, albeit only in restrained fashion.
Interaction between Nup107 and Nup133
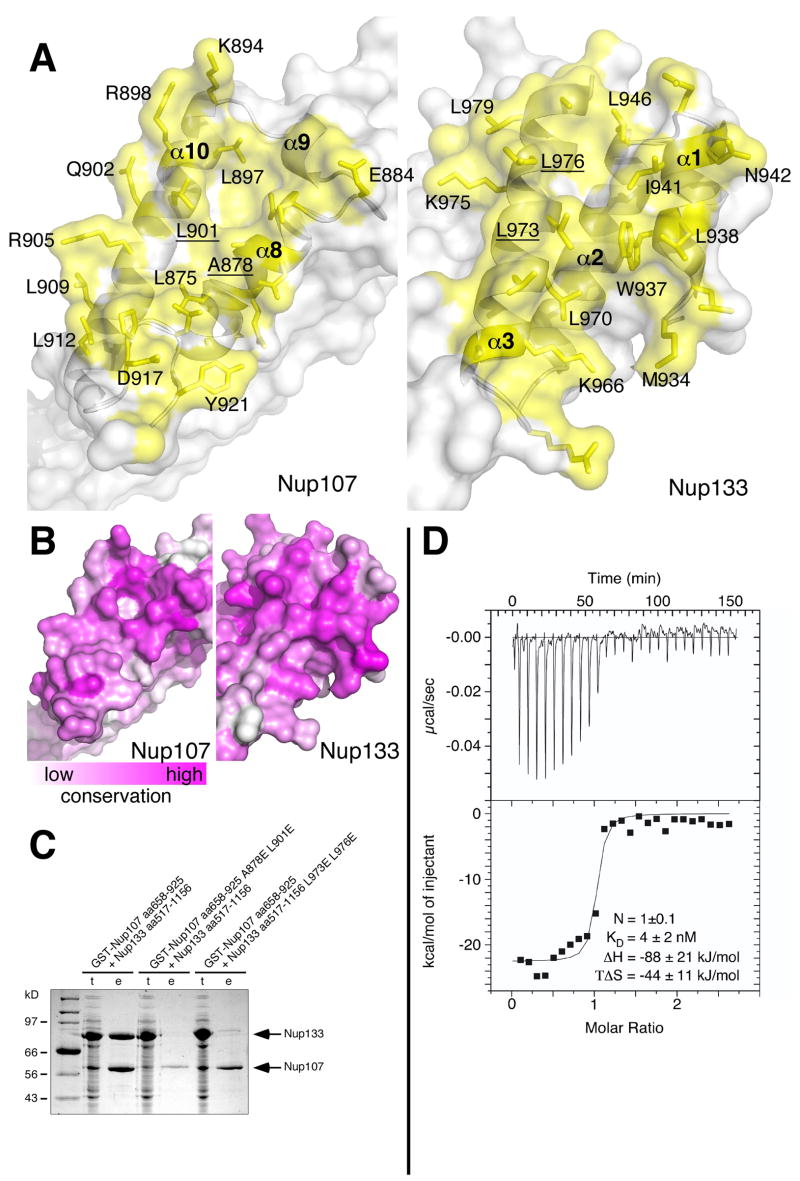
Nup107 and Nup133 bind via a compact interface of 1180 Å2 (Figure 3A). The interacting residues of Nup107 are located within its very C-terminal region (aa 875–925) and predominantly belong to helices α8 and α10, and the terminal loop of the protein. The Nup133 interface mostly consists of residues of helices α1 and α3. The interface is distinctly hydrophobic in its core. Residues L875, A878, L897, L901, P918, L919 and Y921 form a continuous van der Waals surface on the Nup107 side, highly complementary in shape to the surface of W937, L938, I941, L946, A965, L970, L973, L976, L979 and A980 displayed on Nup133 (Figure 3A). No water molecules are found as part of the interface, confirming the exquisite shape complementarity. At the periphery of the ‘dry’ interface numerous hydrogen bonds and salt bridges are observed that strengthen the interaction. The contacting helices of Nup107 and Nup133 are aligned in a ~ 45º angle, with the C-terminal loop of Nup107 wedged into the interface. As a result, the contact area is distinctly uneven with pronounced pockets and protrusions, characteristics typical for intramolecular interactions such as those between consecutive stacked helical pairs within Nup107. The size of these intramolecular interfaces measures about 1000 ± 100 Å2, quite similar to the Nup107/133 contact surface. The intramolecular interfaces are hydrophobic in the interior and are also flanked by residues forming a peripheral network of polar and ionic interactions.
Figure 3. The Nup107/Nup133 Interaction.
(A) Open book view of the Nup107/Nup133 interface, Nup107 contact side shown in the left panel, Nup133 contact side in the right panel. Protein surface is shown half-transparent, contact region colored yellow. The contact area measures 1180 Å2. Interface residues are labeled. The interface is ‘dry’ and entirely hydrophobic in its core. (B) Conservation of the binding interface across eukaryotes. (C) Structure-based mutants disrupt the interaction between Nup107 and Nup133. Wildtype and mutant Nup133 (aa 517–1156) was co-expressed with wildtype and mutant GST-tagged Nup107 (aa 658–925) from separate vectors in E. coli and co-purified by glutathione sepharose affinity chromatography. Total cell lysates (t) and elutions from the GST-affinity columns (e) were separated by SDS-PAGE and analyzed by Coomassie staining. Both double mutations, Nup107 (A878E L901E) and Nup133 (L973E L976E), are sufficient to abolish the interaction of Nup107 and Nup133. Regarding relative amount of expressed proteins see comment in legend to Figure 1. (D) Isothermal titration calorimetry. The titration curve of Nup107658–925 against Nup133934–1156 demonstrates a binding stoichiometry of 1.0 ± 0.1. 8 μl aliquots of Nup107 (13.1μM) were successively injected into 1.38 ml of a 0.75 μM solution of Nup133. The dissociation constant KD, binding enthalpy ΔH and entropy TΔS were derived by curve fitting using the single set of independent sites model. The binding reaction is enthalpically driven.
Next, we examined the sequence conservation of the interface. Mapping the sequence conservation comparing eukaryotes spanning the entire tree of life (Ciccarelli et al., 2006) on the surfaces of Nup107 and Nup133 shows that the interface is very well conserved in character (Figure 3B and Supplementary Figures 1+2). It is in fact the best-conserved surface region of both Nup107 and Nup133.
To characterize the interface further, isothermal titration calorimetry (ITC) was performed to measure the binding energy. We determined an equilibrium dissociation constant KD of 4 ± 2 nM (Figure 3D), confirming the very strong interaction between both proteins as suggested from the structure.
Structure-based mutants disrupt Nup107–133 complex formation
Based on the crystal structure of the Nup107–133 complex and analysis of its interface we designed point mutations to disrupt the interface. Two residues on each of the protein interaction surfaces were chosen whose aliphatic side chains are deeply buried in the hydrophobic interface, but would be solvent-exposed in the uncomplexed proteins. The results of in vitro binding assays with non-binding-mutants Nup133517–1156L973E L976E and Nup107658–925A878E L901E are shown in Figure 3C. Wildtype and mutant Nup133 and Nup107 proteins were co-expressed in E. coli and co-purification examined via glutathione-sepharose affinity chromatography. Both double mutants, Nup133517–1156L973E L976E and Nup107658–925A878E L901E, strongly inhibited complex formation. This is a direct result of the manipulation of the binding interface. The structure-based point mutations did not affect protein folding of either Nup107 or Nup133 (Supplementary Figure S3).
The N-terminal part of Nup107’s α-helical domain anchors Nup107 and Nup133 in the NPC
We have previously shown that Nup133 is incorporated into the NPC via its C-terminal α-helical domain (Berke et al., 2004) and not the N-terminal β-propeller. Since the interaction-interface with Nup107 represents only a relatively small part of this domain, we examined whether Nup107-interaction is indeed the crucial determinant for the integration of Nup133 into the NPC. HeLa cells transfected with EGFP-tagged full-length Nup133 L973E L976E, which does not bind Nup107, failed to incorporate the fusion protein into the NPC and exhibited a diffuse cytoplasmic staining (Figure 5A).
Figure 5. Nup107 and Nup133 are Anchored to the NPC via the N-terminus of Nup107.
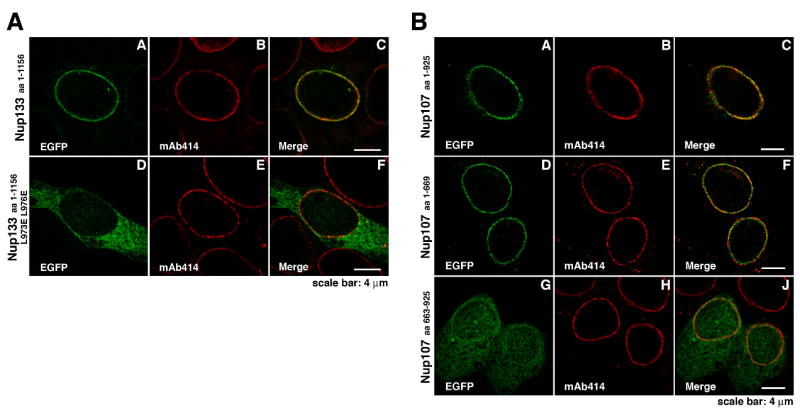
(A) Localization of EGFP-tagged Nup133 proteins in HeLa cells. Cells, co-stained with the NPC marker mAb414, were analyzed 48 h after transfection. In contrast to full length Nup133 (A–C), showing punctate nuclear rim staining, the non-Nup107-binding mutant Nup133 (L973E L976E) is dispersed over the cytoplasm and largely excluded from the nucleus and nuclear envelope (D–F). (B) Localization of EGFP-tagged Nup107 proteins in HeLa cells. Cells, co-stained with mAb414, were analyzed 48 h after transfection. EGFP-tagged Nup107 lacking the last ~250 residues shows punctate nuclear rim staining that overlaps with the mAb414 signal (D–F), similar to full length Nup107 (A–C). In contrast, the Nup133-binding C-terminus of Nup107 shows a dispersed EGFP-signal, with only a slight concentration at the nuclear envelope (G-J).
Transfection of HeLa cells with various EGFP-tagged Nup107 proteins revealed that Nup107 (aa 1–669) assembled into the NPC and exhibited wildtype-like punctate nuclear rim staining (Figure 5B). EGFP-tagged Nup107 (aa 140–669) missing the poorly conserved and non-helical N-terminus of Nup107 showed a similar behavior, whereas further truncation of this domain abolished NPC-targeting completely (data not shown). The Nup133-binding C-terminus of Nup107 comprising residues 663 to 925 was dispersed throughout the cell and only weakly concentrated at the NE (Figure 5B). These data indicate that both Nup107 and Nup133 are integrated into the Nup107–160 subcomplex and the NPC via the N-terminal part of Nup107’s α-helical domain. This also strongly suggests that the binding interface characterized in the crystal structure is the exclusive interaction site between both proteins.
Conservation of Nup107 and Nup133
Even though the inventory list of nucleoporins is largely identical in all eukaryotes, their sequence conservation is rather weak (Vasu and Forbes, 2001). Nup107 and Nup133 are no exception to this observation and thus it is interesting to now analyze sequence conservation in a structural context. Comparing Nup107 sequences across the entire spectrum of eukaryotes (Ciccarelli et al., 2006), we observe that the best conserved residues are either located in the hydrophobic core or participate in binding to Nup133 (Supplementary Figure 1). The helical elements are all reasonably well conserved in length and they are present in all species, suggesting that the overall architecture of the crystallized domain is maintained throughout eukaryotic evolution. However, we note one possibly important exception (Figure 5). The distinctive extension of helix α6 observed in this structure of human Nup107 is present in all metazoa, but it is absent in Saccharomyces cerevisiae and, strikingly, all budding yeast species analyzed. It appears that the finger module was lost early after budding yeast split off from their last common ancestor with other eukaryotes 300–400 million years ago (Dujon, 2006). Budding yeasts are distinct from other eukaryotes in that they divide by closed mitosis, meaning their nuclear envelope and with it the NPCs remain largely intact. In contrast, most other organisms, specifically metazoa, undergo open mitosis with concomitant disassembly of the NPC (Prunuske and Ullman, 2006). Hence, the finger module might play a direct role in NPC disassembly or another mitotic function of Nup107.
The sequence conservation of Nup133 is equally weak as observed in Nup107 (Supplementary Figure 2). Sequence identity across eukaryotes is scarce, yet the well-conserved residues shape the hydrophobic core or are involved in the binding interface. Both, the interaction domain and the C-terminal domain appear to be present in all species.
DISCUSSION
The NPC is one of the largest known macromolecular assemblies in the cell, consequently its structure determination is a formidable challenge. Based on proteomic studies that lead to the nucleoporin inventory of ~30 proteins, symmetry restraints obtained from electron microscopic analysis, pull-down experiments performed under many different conditions, and computational methods to classify domain topologies of nucleoporins, the picture of the NPC as a highly modular machine has emerged (Alber et al., 2007b). The modularity has opened up a vista to elucidate the high-resolution structure of the NPC, namely by solving crystal structures of individual nucleoporins and their subcomplexes and combining this information with cryo-electron microscopic and computational methods to eventually obtain a quasi-atomic model of the assembled structure (Schwartz, 2005). The architectural core of the NPC, excluding the FG-repeat containing unstructured fiber meshwork that fills the central channel of the NPC, is estimated to be less than 40 MDa in size (Alber et al., 2007b). The observation that a significant number of nucleoporins are dynamically attached to the NPC means that the stable scaffold structure contains only a subset of nucleoporins, reducing its size even further (Rabut et al., 2004). A prominent working hypothesis in the field is that the Nup107–160 complex forms a cytoplasmic and a nucleoplasmic ring of eight copies each, situated at the boundaries of the NPC core and coating the nuclear envelope. Sandwiched between these outer rings is an inner ring of a second architectural subcomplex minimally containing the large non-FG nucleoporins Nup205, Nup188 and Nup93 (Schwartz, 2005). The position of a third stable NPC subcomplex containing the essential membrane-spanning Ndc1 nucleoporin is likely proximal to the Nup205 subcomplex but still awaits more detailed characterization (Madrid et al., 2006; Mansfeld et al., 2006; Stavru et al., 2006). Tertiary structure classification of all constituents of the ring-forming subcomplexes strongly predicts them to be predominantly composed of either an α-helical repeat domain, a β-propeller domain, or a tandem arrangement of both (Berke et al., 2004; Devos et al., 2006; Schwartz, 2005). Such simplicity could be a sign of redundancy within the NPC proper. In fact, it has been proposed that the entire inventory of architectural nucleoporins in extant eukaryotes is the result of gene duplication in the past (Alber et al., 2007b).
The Nup107/133 interaction complex reported here, together with the published structures of other architectural nucleoporins now allows us to evaluate the modeling attempts on solid experimental basis. The predictions for propeller domains have been matched experimentally in all cases where crystal structures were obtained, including the N-terminal domains of hNup133, yNup159, and its human homolog hNup214 (Berke et al., 2004; Napetschnig et al., 2007; Weirich et al., 2004). β-Propellers are a very common architectural fold and can be found in the cell in many different contexts (Fulop and Jones, 1999; Paoli, 2001; Pons et al., 2003). Thus, knowledge of the fold does not add much to the understanding of the functional role of a protein. Strikingly, functionally significant features of hNup133 and yNup159 have been attributed to loops outside the basic β-propeller fold (Drin et al., 2007; Weirich et al., 2004). We now have several crystal structures of supposedlyα-helical repeat containing proteins of the NPC scaffold. Nup107 and Nup133 are reported here, Nic96/Nup93 as well as yNup145C were published recently (Hsia et al., 2007; Jeudy and Schwartz, 2007; Schrader et al., 2008). α-Helical repeats or solenoids are characterized by a regular arrangement of two or three helical elements that stack on top of each other to form a superhelix. Due to the regular displacement of consecutive stack elements around the superhelical axis the overall structure can be described with parameters like curvature, handedness and helical twist (Kobe and Kajava, 2000), also familiar from the description of stacked base pairs in nucleic acid helices. All known members of the karyopherin family of nuclear transport factors adopt this fold type (Cook et al., 2007; Suel et al., 2006). Flexibility is inherent in these structures and is functionally important (Conti et al., 2006). Neither Nup107 nor Nup133 form a similarly regular repeat structure. Instead, each one of these proteins adopts its own, unique architecture. The same observation was made with the α-helical domains of Nic96/Nup93 and yNup145C (Hsia et al., 2007; Jeudy and Schwartz, 2007; Schrader et al., 2008). Karyopherin gain flexibility mostly from slight twist movements that individual helical hairpins perform against each other, producing pronounced effects over the length of the protein. In the Nup107/133 complex structure there are no indications that would point to similar pliability. An important observation is that the α-helical regions in nucleoporins are not necessarily forming a single contiguous domain. In Nup133, the α-helical region is at least divided in three tethered subdomains, the C-terminal helical bundle, the Nup107 interaction domain, and the N-terminally preceding domain that is not part of our structure. This latter domain seems flexibly attached to the Nup107 interaction domain as assessed by limited proteolysis (not shown). Rather than bending motion, we envision the possibility of restrained tilting at the hinges connecting domain units. The crystallized Nup107 region is one rigid unit without intermittent gaps. It remains to be determined whether the complete helical region of Nup107 is equally fragmented as Nup133 or whether it forms one continuous domain.
Based on both biochemical and structural data now at hand, architectural nucleoporins appear to have very specific functions and are not generally redundant. The combination of structural and phylogenetic data supports this view most obviously. The sequence conservation of nucleoporins with structural roles is weak and does not, as we now know, lend itself well to structure prediction. From the available crystal structures we have learned that the weak sequence conservation is a consequence of the domain architectures involved, which tolerate sequence alterations remarkably well. However, a structure-based sequence alignment reveals phyla-specific alterations, like the absence of the finger domain of Nup107 in budding yeasts (Figure 6). Similarly, plants appear to have a characteristic insertion of about 100 residues between helices α6 and α7, potentially pointing to a specific function (data not shown).
Figure 6. The Nup107 α6-Finger is Absent in Budding Yeast.
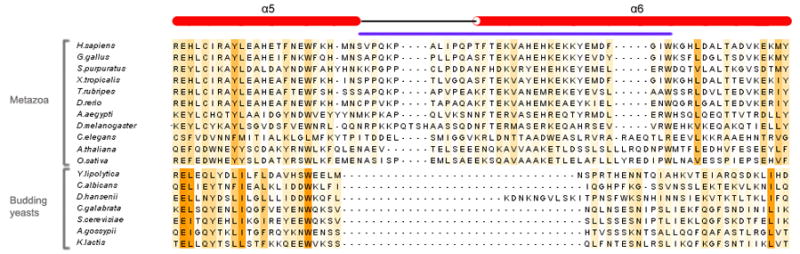
Alignment of the Nup107 amino acid sequence folding into the protruding helix α6 ‘finger’. Sequences representative of the entire phylogenetic tree of eukaryotes are shown. The finger is present in metazoa, but noticeably absent in all budding yeasts analyzed. Maximally diverged budding yeast species are shown, indicating that the finger was lost early after this clade split off from its last common ancestor with other eukaryotes ~300–400 Mya (Dujon, 2006).
Redundancy in an assembly requires protein-protein interactions to be promiscuous, rather weak and non-obligate. Nup107 and Nup133 however interact in specific manner with very high affinity as determined in this study. The stability of binary contacts between other architectural nucleoporins is also well documented (Lutzmann et al., 2002; Lutzmann et al., 2005). Even for the two structural nucleoporins with highest sequence similarity and obvious common ancestry, the β-propeller proteins Sec13 and Seh1, it does not appear likely that they can functionally replace each other. In contrast, redundancy is integral to the function of the FG-repeat containing, unstructured fibrils that make up the central transport channel of the NPC. The deletion of FG-regions of different nucleoporins show similar effects in vivo (Strawn et al., 2004). And, maybe most strikingly, an engineered gel made up of only one FG-repeat segment (although there are hundreds of variants in the NPC) has properties that were suggested in broad measure to be comparable with the natural meshwork (Frey and Gorlich, 2007). Also, the coiled-coil containing nucleoporins might associate in dynamic fashion with various binding partners. However, the biochemical data available on stable, architectural nucleoporins does not support similar exchangeability. How else then does the architectural scaffold accomplish flexibility, as the experimental data suggests (Beck et al., 2007; Tran and Wente, 2006)? First, domain boundaries of large nucleoporins are easy to conceive as possible hinge regions allowing flexibility. Second, our study demonstrates that the predicted α-helical domains are not necessarily composed of one continuous entity, but that they can be built from several smaller domains that move with at least restrained motility. Occurrence of such hinge mobility is further supported by the fact that Nup107–160 subcomplex adopts an extended, Y-shaped structure composed of binary protein units forming the arms (Lutzmann et al., 2002). Hinge motility would be much more difficult to conceive in a globular assembly with contact areas involving more than two components.
This structural study sheds light on the assembly of nucleoporins. A surprisingly small, yet very stable contact area is formed between the C-terminal regions of Nup107 and Nup133. The strong interaction, highlighted by the low equilibrium dissociation constant KD of 4 ± 2 nM as directly measured by isothermal titration calorimetry, implies that both proteins are stably associated. In higher eukaryotes the NPC disassembles during mitosis, but which connections have to be broken? The stability of the Nup107/Nup133 interface suggests that these two proteins are permanently associated. Accordingly, it was shown that phosphorylation, which fosters the mitotic disassembly of the NPC into its various subcomplexes, occurs exclusively at the N-termini of Nup107 and Nup133 (Glavy et al., 2007) consistent with the Nup107/Nup133 complex being maintained. The interaction between S. cerevisiae Nup133 and the yeast homolog of Nup107, scNup84, has not been readily detectable in vivo (Siniossoglou et al., 1996; Siniossoglou et al., 2000), leading to the notion their interaction might be rather weak (Lutzmann et al., 2002). Similarly, in a series of pull-down experiments of the Nup84 subcomplex using wash buffers of various stringency, Nup133 was found to be less stably associated with the Nup84 complex than other components (Alber et al., 2007a). The structure explains this apparent antagonism: in both studies by Siniossoglou et al. as well as in the Alber et al. paper the Nup84 complex was isolated via protein A fused to the C-terminus of Nup84, which very likely interferes with the Nup133 interaction. This observation further supports the view that Nup84/Nup107 is the sole interaction partner of Nup133 within the Y-shaped complex and that their interaction is exclusively mediated by the C-terminal binding sites revealed in the present study.
We show that Nup133 is anchored to the NPC via Nup107 in tail-to-tail fashion. Nup107 and Nup133 both form elongated domains that interact in a stark angle. These observations point to a largely extended overall structure of the Nup107/Nup133 complex. This is consistent with electronmicroscopic pictures of the assembled heptameric yeast Nup84 complex, where Nup133/Nup84 form the long arm of the Y-shaped assembly (Lutzmann et al., 2002). Further, Nup107 integrates into the NPC via a separate N-terminal interaction site that is independent of the Nup133 binding interface (Figure 5B). Based on our in vivo labeling experiments we place Nup133 at the periphery of the NPC, with Nup107 acting as the chain-link to the nonameric Nup107–160 complex. This is in accordance with the relative localization of Nup133 and Nup84/107 in the assembly model by Rout and colleagues (Alber et al., 2007b), where both proteins are localized at the periphery of a so-called membrane-coating scaffold complex. A radically different model has been proposed recently. Based on crystal contacts between hSec13/yNup145C heterodimers it is hypothesized that the NPC is built from concentric layers, like the skins of an onion (Hsia et al., 2007). In this model the Y-shaped complex forms the outermost, membrane-coating skin, followed by a layer of ‘adaptor nups’, which surround FG-repeat containing ‘channel nups’. This ‘onion-model’ suggests that the Y-shaped complex forms four eight-membered rings that are stacked on top of each other and span the entire NPC from the cytoplasmic to the nucleoplasmic side, which stands in stark contrast to the preeminent model with two peripheral rings of 8 complexes each sandwiching a central core ring built from a second set of scaffold nucleoporins (likely containing Nup93, Nup205 and Nup188). Our in vivo experiments do not support the model by Hsia et al.. With four adjacent rings, we would not expect Nup133 to be detached from the NPC by the exclusive disruption of the Nup107 interface with two surface point mutations, since Nup133 would be expected to be integrated into its layer by additional contacts. Furthermore, if Nup133 was part of a membrane-spanning pole, mislocalization of Nup133 (L973E L976E) should have a strong effect on the overall NPC assembly, which we do not observe.
The Nup107–160 complex is a major component of the NPC scaffold. Its extended, Y-shaped structure suggests that the NPC scaffold is porous rather than compact. Such an architecture could potentially generate peripheral transport channels in addition to the central pore as has been postulated before (Hinshaw et al., 1992; Shahin et al., 2001). It could also perhaps facilitate lateral opening of the NPC to allow the passage of membrane-anchored proteins on their journey from the outer to the inner nuclear membrane (King et al., 2006). With the knowledge obtained in this study, we can now engineer structure-based protein probes to address these issues in a sensible way. We will surely see more atomic structures of nucleoporin complexes in the near future. Together with electron-microscopic, computational and molecular biological methods this will lead us to much-needed in-depth understanding of the still largely enigmatic NPC.
EXPERIMENTAL PROCEDURES
Summary of Plasmid Construction, Protein Expression, Purification and Structure Determination
For protein interaction studies, full-length or fragments of human Nup107 were fused to GST. 6x-His N-terminal fusions of human Nup133 fragments were co-expressed with GST-Nup107 fusions in E.coli and co-purified by affinity chromatography. Protein complexes for crystallization were expressed in E.coli from bi-cistronic vectors, with Nup107 N-terminally tagged with GST (Nup107658–925/Nup133934–1156) or 6x-His (Nup107658–925dF/Nup133934–1156). The affinity-tags were removed before crystallization. Structures were determined by SAD (Nup107658–925/Nup133934–1156) or molecular replacement (Nup107658–925dF/Nup133934–1156) and refined as summarized in Table 1. Full details of plasmid construction, protein expression, crystallization, and structure determination are provided in the Supplemental Data.
Protein-Protein Interaction Experiments
GST pulldown assays were performed using recombinantly expressed GST-tagged full length Nup1071–925 or Nup1071–644 as bait by capturing GST-Nup107 as previously described (Berke et al., 2004). Full length Nup1331–1156 was in vitro transcribed and translated in the presence of [35S] methionine by a coupled reticulocyte lysate transcription/translation system (TNT T7, Promega). Bound and unbound fractions were separated by SDS-PAGE and analyzed by autoradiography.
For experiments mapping and analyzing the interaction site of Nup133 and Nup107 GST-tagged wildtype Nup107 and mutant Nup107658–925 were co-expressed with C-terminal fragments of wildtype and mutant Nup133. Bacterial cell pellets were resuspended in PBS supplemented with 1 mM DTT. Then, cells were lysed using a french press and supplemented with 200 mM PMSF. After centrifugation at 15000×g for 20 min at 4 °C, the supernatant was incubated with glutathione sepharose (4 Fast Flow, GE Healthcare) for 30 min at 4 °C. The resin was washed 3 times with PBS in batch, added to a disposable column (Pierce), and eluted with 4 column volumes of glutathione elution buffer (50 mM Tris-HCl pH 8.0, 10 mM reduced glutathione). Total cell lysate and elution were separated by SDS-PAGE and visualized by Coomassie Blue staining. Due to the use of pGEX- and pET-derived vectors harboring the same origin of replication, the relative protein amounts of the GST- and the 6xHis-fusion proteins differ from clone to clone.
Isothermal titration calorimetry
His-tagged hNup107658–925 and hNup133934–1156 were separately expressed and chromatographically purified to homogeneity. The proteins were dialysed against buffer containing 10 mM potassium phosphate pH 8.0, 150 mM NaCl, 0.1 mM EDTA, 0.1 mM TCEP. Concentrations were determined spectrophotometrically at 278 nm. ITC was performed using a VP-ITC microcalorimeter (MicroCal, Northhampton, MA). Titrations were performed at 25 ºC by injecting 5–10 μl aliquots of hNup107658–925 into the ITC cell containing 1.43 ml of hNup133934–1156. Binding stoichiometry, enthalpy and entropy as well as the equilibrium dissociation constant was determined by using the “single set of independent sites” model of molecular association (MicroCal Origin 2.9; MicroCal). The experiment was repeated four times.
Transfection of HeLa cells and immunofluorescence microscopy
For transient transfection of DNA plasmids coding for EGFP-tagged wildtype and mutant Nup107 and Nup133 proteins, HeLa cells were grown on coverslips to a confluency of 40–60% and transfected using Effectene reagent (Qiagen) according to the manufacturer’s instructions. 24–48 h after transfection, cells were fixed/permeabilized in −20ºC-cold 100% Methanol for 5 min. After blocking with 2% BSA/PBS for 2 h, cells were incubated with mAb414 antibody (Covance Research Products) diluted 1:2000 in 1% BSA/PBS for 1 h. Cells were washed three times with PBS for 5 min and then incubated with CY5-conjugated donkey α-mouse (1:200) IgG antibodies (Jackson ImmunoResearch) diluted in 1% BSA/PBS for 30 min. Cells were washed again three times with PBS for 5 min and mounted in ProLong Gold antifade reagent (Molecular Probes). Samples were analyzed with a Leica TCS SP spectral confocal microscope or a DeltaVision image restoration microscope (Applied Precision/Olympus), and images were processed in Adobe Photoshop CS.
Supplementary Material
Figure 4. Comparison to α-Helical Repeat Proteins.
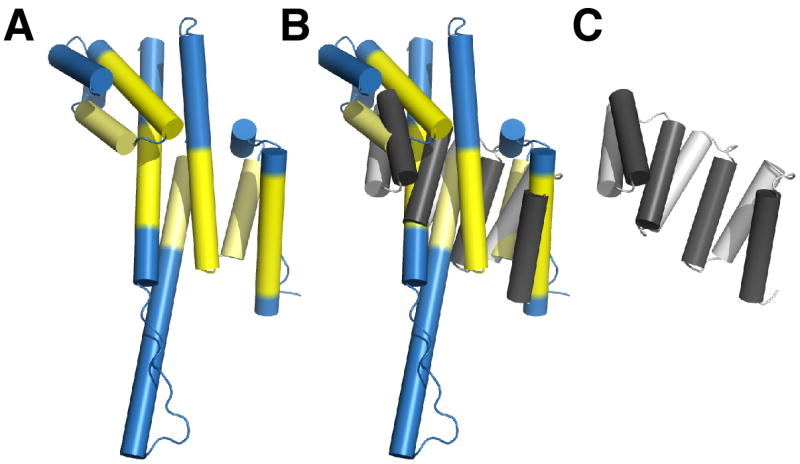
Superposition of Nup107 (panel A) and the consensus α-helical TPR-repeat protein (PDB code 2FO7) (panel C) is shown in (B). RMS deviation 4.0 Å over 92 equivalent Cα positions (indicated in yellow). The similarity between Nup107 and αhelical repeat proteins is limited, in contrast to current computer predictions. No significant match with α-helical repeats was found for Nup133 (not shown).
Acknowledgments
We thank members of the Schwartz laboratory for many helpful discussions; Günter Blobel for support during the initial phase of this project; and the staff of the NE-CAT beamlines 24ID-C/-E, especially Kanagalaghatta Rajashankar, at Argonne National Laboratory for excellent assistance with data collection. This work was support by NIH Grant 5R01 GM77537 (T.S.) and a Pew Scholar Award (T.S.).
Footnotes
Accession Numbers
The atomic coordinates of the Nup107/133 structures described here have been deposited with the Protein Data Bank (PDB) under the accession codes 3CQC and 3CQG.
This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2–3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Boehmer T, Blobel G, Schwartz TU. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol. 2004;167:591–597. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci USA. 2003;100:981–985. doi: 10.1073/pnas.252749899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T, Schwartz TU. Purification, crystallization and preliminary X-ray analysis of a Nup107-Nup133 heterodimeric nucleoporin complex. Acta Crystallogr F. 2007;63:816–818. doi: 10.1107/S1744309107040523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Conti E, Muller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Enninga J, Levay A, Fontoura BM. Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol Cell Biol. 2003;23:7271–7284. doi: 10.1128/MCB.23.20.7271-7284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura BM, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Fulop V, Jones DT. Beta propellers: structural rigidity and functional diversity. Curr Opin Struct Biol. 1999;9:715–721. doi: 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- Glavy JS, Krutchinsky AN, Cristea IM, Berke IC, Boehmer T, Blobel G, Chait BT. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107–160 subcomplex. Proc Natl Acad Sci USA. 2007;104:3811–3816. doi: 10.1073/pnas.0700058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeudy S, Schwartz TU. Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J Biol Chem. 2007;282:34904–34912. doi: 10.1074/jbc.M705479200. [DOI] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. When protein folding is simplified to protein coiling: the continuum of solenoid protein structures. Trends Biochem Sci. 2000;25:509–515. doi: 10.1016/s0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Kunze R, Stangl K, Stelter P, Toth KF, Bottcher B, Hurt E. Reconstitution of Nup157 and Nup145N into the Nup84 complex. J Biol Chem. 2005;280:18442–18451. doi: 10.1074/jbc.M412787200. [DOI] [PubMed] [Google Scholar]
- Madrid AS, Mancuso J, Cande WZ, Weis K. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J Cell Biol. 2006;173:361–371. doi: 10.1083/jcb.200506199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid AS, Weis K. Nuclear transport is becoming crystal clear. Chromosoma. 2006;115:98–109. doi: 10.1007/s00412-005-0043-3. [DOI] [PubMed] [Google Scholar]
- Mansfeld J, Guttinger S, Hawryluk-Gara LA, Pante N, Mall M, Galy V, Haselmann U, Muhlhausser P, Wozniak RW, Mattaj IW, et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell. 2006;22:93–103. doi: 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci USA. 2005;102:5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napetschnig J, Blobel G, Hoelz A. Crystal structure of the N-terminal domain of the human protooncogene Nup214/CAN. Proc Natl Acad Sci USA. 2007;104:1783–1788. doi: 10.1073/pnas.0610828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ. The Nup107–160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoli M. Protein folds propelled by diversity. Prog Biophys Mol Biol. 2001;76:103–130. doi: 10.1016/s0079-6107(01)00007-4. [DOI] [PubMed] [Google Scholar]
- Pons T, Gomez R, Chinea G, Valencia A. Beta-propellers: associated functions and their role in human diseases. Curr Med Chem. 2003;10:505–524. doi: 10.2174/0929867033368204. [DOI] [PubMed] [Google Scholar]
- Prunuske AJ, Ullman KS. The nuclear envelope: form and reformation. Curr Opin Cell Biol. 2006;18:108–116. doi: 10.1016/j.ceb.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol Cell. 2008;29:46–55. doi: 10.1016/j.molcel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol. 2005;15:221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Shahin V, Danker T, Enss K, Ossig R, Oberleithner H. Evidence for Ca2+- and ATP-sensitive peripheral channels in nuclear pore complexes. FASEB J. 2001;15:1895–1901. doi: 10.1096/fj.00-0838com. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Stavru F, Hulsmann BB, Spang A, Hartmann E, Cordes VC, Gorlich D. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J Cell Biol. 2006;173:509–519. doi: 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- Suel KE, Cansizoglu AE, Chook YM. Atomic resolution structures in nuclear transport. Methods. 2006;39:342–355. doi: 10.1016/j.ymeth.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu SK, Forbes DJ. Nuclear pores and nuclear assembly. Curr Opin Cell Biol. 2001;13:363–375. doi: 10.1016/s0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, et al. The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Berger JM, Weis K. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol Cell. 2004;16:749–760. doi: 10.1016/j.molcel.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, Doye V. The human Nup107–160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.