Abstract
Diatoms are important components of the biological community and food web in the aquatic environment. Here, we report the characteristics of a single-stranded RNA (ssRNA) virus (CtenRNAV01) that infects the marine diatom Chaetoceros tenuissimus Meunier (Bacillariophyceae). The ca. 31-nm virus particle is icosahedral and lacks a tail. CtenRNAV01 forms crystalline arrays occupying most of the infected host's cytoplasm. By growth experiments, the lytic cycle and the burst size were estimated to be <24 h and ∼1 × 104 infectious units per host cell, respectively. Stationary-phase C. tenuissimus cultures were shown to be more sensitive to CtenRNAV01 than logarithmic-phase cultures. The most noticeable feature of this virus is its exceptionally high yields of ∼1010 infectious units ml−1; this is much higher than those of any other algal viruses previously characterized. CtenRNAV01 has two molecules of ssRNA of approximately 8.9 and 4.3 kb and three major proteins (33.5, 31.5, and 30.0 kDa). Sequencing of the total viral genome has produced only one large contig [9,431 bases excluding the poly(A) tail], suggesting considerable overlapping between the two RNA molecules. The monophyly of CtenRNAV01 compared to another diatom-infecting virus, Rhizosolenia setigera RNA virus, was strongly supported in a maximum likelihood phylogenetic tree constructed based on the concatenated amino acid sequences of the RNA-dependent RNA polymerase domains. Although further analysis is required to determine the detailed classification and nomenclature of this virus, these data strongly suggest the existence of a diatom-infecting ssRNA virus group in natural waters.
Diatoms are the most abundant group of photosynthetic unicellular organisms; they have a very large biomass and are found in almost all aquatic and semiaquatic habitats (e.g., water, soil, and moist surfaces of rocks). They are of great ecologic importance because they are the principal primary producers in aquatic food webs and sustain the oxygen level in the atmosphere of the planet at about 21% (29). The genus Chaetoceros is highly diverse, with more than 400 species, which are considered to include the key primary photosynthetic producers that sustain the higher forms of aquatic life (17).
Since the 1970s, the significance of viruses as mortality agents for phytoplankton in aquatic systems has been highlighted. A number of algal viruses have been isolated and characterized. There used to be no direct evidence showing the relationship between diatoms and viruses, which led to the idea that the shell covering of diatoms might contribute to a reduced probability of viral infection (30). Later, however, diatoms were found to be infected by viruses similarly to other microalgae, and the first isolated diatom-infecting virus was a single-stranded RNA (ssRNA) virus lytic to a pen-shaped diatom species, Rhizosolenia setigera (R. setigera RNA virus [RsRNAV]) (12). Subsequently, another diatom-infecting virus (Chaetoceros salsugineum nuclear inclusion virus [CsNIV]) lytic to C. salsugineum was isolated (14). Its genome is distinct from those of other viruses, consisting of a covalently closed circular ssDNA (6 kb) that is partly double stranded (997 bases). A third diatom virus, Chaetoceros nuclear inclusion virus (CspNIV), infecting Chaetoceros cf. gracilis, was described by Bettarel et al. (2); however, no data concerning its genome have been reported. The discovery of these diatom-infecting viruses suggested the possibility that viruses have an impact on diatom populations as the key factors regulating the dynamics of diatom blooms in natural waters. Here, we describe the isolation and characterization of a new ssRNA virus infecting Chaetoceros tenuissimus (CtenRNAV).
MATERIALS AND METHODS
Identification of the host diatom strain.
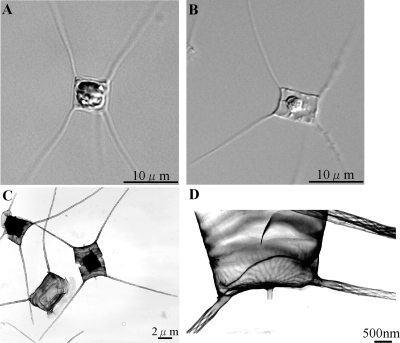
The host diatom strain used in the present study (C. tenuissimus 2-10) was isolated from the Maiko Coast, Seto Inland Sea, Japan, on 10 August 2002. It was intensively observed by both optical microscopy and transmission electron microscopy (TEM) (Fig. 1) and was identified as C. tenuissimus Meunier based on its morphology (10, 18). This planktonic diatom species is so small (<10 μm in length) that it is often confused with morphologically similar diatoms, e.g., Chaetoceros neogracilis and C. gracilis.
FIG. 1.
C. tenuissimus strain 2-10. Shown are optical micrographs of an intact cell (A) and a CtenRNAV01-infected cell (B) and transmission electron micrographs of negatively stained intact cells (C and D).
Algal cultures.
The diatoms used as hosts in isolating viral pathogens were as follows: 1 strain each of Asterionella glacilis, C. salsugineum, Chaetoceros socialis, Coscinodiscus sp., Ditylum brightwellii, Eucampia zodiacus, Rhizosolenia setigera, Stephanopyxis sp., Thalassionema sp., and Thlassiosira sp.; 4 strains of Chaetoceros debilis; 2 strains of C. tenuissimus; and 13 strains of Chaetoceros sp. Each strain was grown in modified SWM3 medium (3, 5) enriched with 2 nM Na2SeO3 using a 12-h light/12-h dark cycle with ca. 110 μmol photons m−2 s−1 provided as cool white fluorescent illumination at 15°C. In the experiments described below, a clonal and axenic culture of C. tenuissimus 2-10 was used as the host for virus characterization. To test the interspecies host specificity, clonal strains of Nannochloropsis sp. (Eustigmatophyceae), Oltmannsiellopsis viridis (Chlorophyceae), Akashiwo sanguineum, Alexandrium catenella, Gymnodinium catenatum, Heterocapsa circularisquama, Heterocapsa triquetra, Karenia mikimotoi, Prorocentrum micans, Scrippsiella sp. (Dinophyceae), Chattonella verrculosa, Chattonella antiqua, Chattonella marina, Chattonella ovata, Fibrocapsa japonica, and Heterosigma akashiwo (Raphidophyceae), as well as the 29 diatom strains, were used. They were also incubated under the conditions mentioned above at either 15 or 20°C.
Virus isolation.
During June 2004, surface water samples were collected at the mouth of the Shiotsuka River in the Ariake Sound, Japan; sent to the laboratory; and stored at 4°C without fixation within 24 h of sampling. The samples were filtered through a 0.2-μm Dismic-25cs filter (Advantec) to remove eukaryotic microorganisms and most bacteria. Algal cultures (0.8 ml) were inoculated with an aliquot (0.2 ml) of the filtrates obtained from the water sample and incubated as described above. The algal cultures inoculated with modified SWM3 served as controls. Consequently, when the host growth was apparently inhibited with the filtrate, we cloned the pathogen through two cycles to extinction using the dilution procedure from the lysed culture (21, 28). The lysate in the most dilute well of the second assay was made free from bacterial contamination using filtration through a 0.1-μm polycarbonate membrane filter (Nuclepore) and transferred to an exponentially growing culture of C. tenuissimus 2-10. The resultant lysate was regarded as a clonal pathogen suspension. Serial transfers of virus from a lysed culture to an exponentially growing host culture were performed more than twice to verify its transferability. The pathogen lytic to C. tenuissimus 2-10 (later designated CtenRNAV01) was isolated from the surface water collected in June 2004 and is characterized in this paper.
TEM.
TEM was performed according to the method previously reported (26). Vigorously growing C. tenuissimus 2-10 cultures were inoculated with CtenRNAV01 at a multiplicity of infection (MOI) of 1.1 × 103. C. tenuissimus 2-10 cultures inoculated with an autoclaved pathogen served as the controls. An aliquot of the cultures was sampled at 0, 24, 48, and 72 h postinoculation (p.i.) and processed for TEM observation. Thin sections of the cells were stained with 4% uranyl acetate and 3% lead citrate and observed at 80 kV using a JEOL JEM-1010 transmission electron microscope. The algicidal pathogens negatively stained with 4% uranyl acetate were also observed using TEM.
Host range test.
The interspecies host specificity of the pathogen was tested by adding 5% (vol/vol) aliquots of fresh virus suspension to the various algal cultures mentioned above. Each culture was incubated under the conditions mentioned above; then, the growth and evidence of lysis in each algal culture were monitored by optical microscopy and compared to control cultures without the pathogen. Cultures apparently not lysed until 14 days p.i. were considered to be unsuitable hosts for the viral pathogen.
Storage test.
An exponentially growing culture of C. tenuissimus 2-10 was inoculated with the virus and incubated for 6 days. The resultant lysate was sequentially passed through 0.8- and 0.2-μm filters to remove cellular debris. The titer of the fresh lysate was estimated using the extinction dilution method (16, 21). Then, aliquots of the lysate were stored at 20, 10, 4, −20, −80, and −196°C (liquid nitrogen) in the dark without adding cryoprotectants. After 28 days of storage, the samples were retitrated using the extinction dilution method to estimate the stability of the viral pathogen at each storage temperature.
Growth experiment.
A logarithmic-phase culture and a stationary-phase culture of C. tenuissimus 2-10 (500 ml) were inoculated with the viral pathogen at MOI of 11.6 and 12.1, respectively. Host cultures inoculated with an autoclaved viral suspension served as controls. An aliquot of cell suspension was sampled from each culture every 24 h until lysis. The numbers of host cells and viral pathogens were enumerated using optical microscopy and the extinction dilution method (16, 21), respectively.
Virus purification.
Five hundred milliliters of viral lysate was sequentially passed through 0.8- and 0.2-μm polycarbonate membrane filters (Nuclepore) to remove cellular debris. Then, polyethylene glycol 6000 (Wako Co., Ltd.) was added to the filtrate to a final concentration of 10% (wt/vol), and the resultant suspension was stored at 4°C in the dark overnight. After centrifugation at 57,000 × g at 4°C for 1.5 h, the viral pellet was resuspended in 20 ml of 10 mM phosphate buffer (pH 7.2) and added to an equal volume of chloroform. After vigorous vortexing, the suspension was centrifuged at 7,500 × g, for 20 min at 4°C, the supernatant was centrifuged at 217,000 × g, for 4 h at 4°C, and then the virus particles were resuspended in 600 μl of ultrapure water.
Analysis of nucleic acids.
The viral suspension was treated with 1 mg ml−1 proteinase K (Wako Pure Chemical Industries, Ltd.) and 1% sarcosyl (International Technologies, Inc.) at 55°C for 1.5 h. Then, the nucleic acid was extracted by the standard phenol-chloroform method and ethanol precipitated, and the resultant pellet was dissolved in 100 μl of ultrapure water. Aliquots (7 μl) of the nucleic acid solution were incubated with 0.25 U μl−1 DNase I (Promega Co., Ltd.) or 0.025 μg μl−1 RNase A (Nippon Gene Co., Ltd.) in 0.01× SSC or 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M Na3 citrate, pH 7.0) at 37°C for 1 h (19, 22). Nucleic acid samples stored on ice without enzymatic treatment for 1 h served as controls. The resultant nucleic acid samples were electrophoresed on a formaldehyde-agarose gel (1.5%; 15 by 20 cm; SeaKem Gold Agarose; BMA, Inc.) at 60 V for 4 h. Nucleic acids were visualized using Sybr gold stain (Invitrogen).
Analysis of viral proteins.
The virus suspension was mixed with a fourfold volume of the sample buffer (62.5 mM Tris-HCl, 5% 2-mercaptoethanol, 2% sodium dodecyl sulfate [SDS], 20% glycerol, and 0.005% bromphenol blue) and boiled for 5 min. The proteins were separated by SDS-polyacrylamide gel electrophoresis (12.5% polyacrylamide gel; 80 by 40 by 1 mm; 150 V; 15 min) using an XV Pantera System (DRC Co., Ltd). The proteins were visualized using Coomassie brilliant blue stain. Protein molecular mass standards (DRC Co., Ltd.) ranging from 6.5 to 200 kDa were used for calibration.
Genome sequencing and phylogenetic analysis.
Sequencing of the viral genome was performed as follows. The RNA purified from the virus pellet using the RNeasy Mini Kit (Qiagen) was reverse transcribed to construct cDNAs with the cDNA Synthesis Kit M-MLV version (Takara Co., Ltd.) using random primers according to the manufacturer's recommendation. The 5′ ends of the resultant double-stranded DNA fragments were phosphorylated using T4 polynucleotide kinase (Takara Co., Ltd.); then, the resultant cDNA fragments were electrophoresed on an agarose gel, and ∼2-kb fragments were finally extracted. They were ligated into the HincII-cleaved and dephosphorylated pSTV28 plasmid vector (Takara Co., Ltd.). The ligated double-stranded DNA fragments were transformed into Escherichia coli DH10B competent cells (Invitrogen) and sequenced by the dideoxy method using an ABI 3730xl DNA analyzer (Applied Biosystems). The resultant fragmentary sequences were assembled; then, sequencing of the 5′ and 3′ ends was conducted by means of 5′ rapid amplification of cDNA ends and 3′ rapid amplification of cDNA ends, respectively, following the procedure described by Nagasaki et al. (11).
We identified a conserved RNA-dependent RNA polymerase (RdRp) domain in the assembled contig by using BLAST (Basic Local Alignment Search Tool) (1). The deduced amino acid sequence of the corresponding region was compared with other viruses' RdRp domains. They were automatically aligned using ClustalW (24) and manually refined. Phylogenetic trees based on the deduced amino acid sequences of the RdRp domains were constructed using the neighbor-joining (NJ) and maximum likelihood (ML) methods with the Jones-Taylor-Thornton matrix (JTT model) packaged in the Phylip 3.65 program (6). Amino acid sequences used for comparison in the analyses were as follows, with the database accession numbers (referring to the NCBI database unless otherwise stated): Aichi virus, AB010145; bovine enteric calicivirus, AJ011099; bean pod mottle virus, NC_003496; black queen cell virus, NC_003784; CtenRNAV, DDBJ accession number AB375474; cowpea severe mosaic virus, M83830; cricket paralysis virus, NC_003924; Drosophila C virus, NC_001834; deformed wing virus, NC_004830; H. akashiwo RNA virus (HaRNAV), NC_005281; Norwalk virus, M87661; human poliovirus 1 Mahoney, V01149; parsnip yellow fleck virus, D14066; RsRNAV, AB243297; rice turgo spherical virus, AAA66056; Sacbrood virus, NC_002066; Schizochytrium single-stranded RNA virus, BAE47143; Triatoma virus, NC_003783; and Taura syndrome virus, NC_003005.
Nucleotide sequence accession number.
The CtenRNAV01 contig sequence has been deposited at DDBJ under accession no. AB375474.
RESULTS AND DISCUSSION
Isolation and morphological features of the virus.
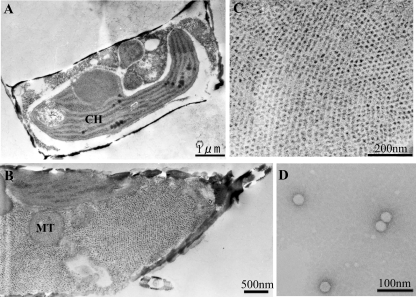
A clonal pathogen causing lysis of C. tenuissimus strain 2-10 was isolated from Ariake Sound. The virus was filterable through a 0.1-μm polycarbonate membrane filter and was serially transferable to either logarithmic-phase or stationary-phase cultures of C. tenuissimus 2-10 (data not shown). Host cells lysed by the pathogen became pale, as did the host culture, presumably due to the loss or degradation of photosynthetic pigments (Fig. 1). TEM observations using thin sections of healthy C. tenuissimus 2-10 cells from control cultures showed the normal cytoplasmic organization with the frustules that is typical of diatoms; no symptoms of viral infection were detected (Fig. 2A). In contrast, at 48 h p.i. with virus, most of the cytoplasm area was occupied with small virus-like particles (VLPs) forming crystalline arrays (Fig. 2B and C); occasionally, the VLPs were also found in an unidentified organelle (data not shown). Free VLPs were also observed in the culture lysate by using negative staining; the virus was icosahedral, 31 ± 2 nm in diameter (n = 34), and lacked a tail or an outer membrane (Fig. 2D). Because (i) the virus was transferable to a fresh algal culture, (ii) VLPs were observed in the lysed culture, and (iii) the VLPs were not found in healthy cultures, Koch's postulates were fulfilled, and we concluded the VLPs were a pathogenic virus lytic to C. tenuissimus. Considering the genome type of this virus (see below), we designated the strain CtenRNAV01.
FIG. 2.
Transmission electron micrographs of C. tenuissimus 2-10 and CtenRNAV01; thin sections of an intact C. tenuissimus cell (A) and a CtenRNAV01-infected cell (B) at 48 h p.i., higher magnification of the CtenRNAV01-infected cell's cytoplasm at 48 h p.i. (C), and negatively stained CtenRNAV01 particles (D). CH, chloroplast; MT, mitochondrion.
Host range.
The host range of CtenRNAV01 was tested using 45 phytoplankton strains, including 29 diatom strains isolated from the coastal waters of western Japan (see Materials and Methods). CtenRNAV01 was lytic only to C. tenuissimus, showing the high host specificity of the virus, which is a common feature of most viruses infecting eukaryotic microalgae.
Stability.
CtenRNAV01 suspension samples containing 7.02 × 1010 virus particles were stored at 20, 10, 4, −20, −80, and −196°C in the dark; the titers after 28 days were 3.01 × 109, 3.01 × 109, 3.85 × 109, 2.26 × 1010, 2.08 × 1010, and 9.82 × 1010 infectious units ml−1 (relative titer, 4 to 140%), respectively. Hence, CtenRNAV01 was shown to be very stable under any storage conditions without the addition of cryoprotectant. This is similar to other ssRNA viruses infecting microalgae, e.g., RsRNAV and Heterocapsa circularisquama RNA virus (HcRNAV) (12, 27).
Nucleic acids.
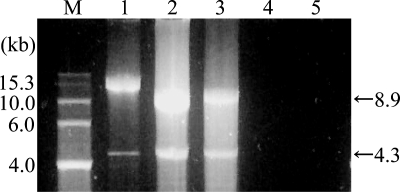
Denaturing gel electrophoresis showed that the major nucleic acids isolated from CtenRNAV01 virions were 8.9 and 4.3 kb. The electrophoretic bands were sensitive to RNase A both under high- and low-salt concentrations, but not to DNase I (Fig. 3). These data indicated that both bands were ssRNA. There are other viruses whose genomes consist of two species of linear positive-sense ssRNA (e.g., Sadwavirus and Cheravirus) (8, 9), in which each RNA molecule codes for a different product and both are essential for infection. Sequencing of the total viral genome produced only one contig sequence with a poly(A) tail; it was 9,431 bases long, excluding the poly(A) tail. Because only the 3′ end was shown to be polyadenylated, the larger band is unlikely to be circular. Based on these observations, there may be a considerable overlap between the two RNA molecules; further analysis is essential to clarify the detailed genome structure of this virus.
FIG. 3.
Nucleic acids of CtenRNAV01 with no treatment (lane 2) and treatment with DNase I (lane 3) or RNase A in low- and high-salt buffers (lanes 4 and 5, respectively). Samples were electrophoresed in a formaldehyde agarose gel with RNA molecular size markers (lane M) and Sendai virus genome RNA (15.3 kb) (lane 1).
Proteins.
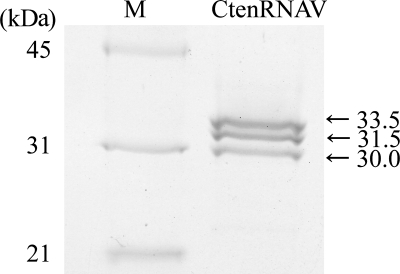
The sizes and numbers of structural proteins of the CtenRNAV01 virions were estimated using SDS-polyacrylamide gel electrophoresis. CtenRNAV01 had three major polypeptides of 33.5, 31.5, and 30.0 kDa (Fig. 4). The number of major proteins of CtenRNAV01 was the same as that of the ssRNA diatom virus RsRNAV, whose band masses were 41.5, 41.0, and 29.5 kDa (12).
FIG. 4.
Major proteins of CtenRNAV01.
Viral replication.
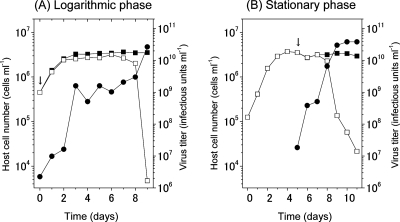
When a logarithmic culture of C. tenuissimus was inoculated with CtenRNAV01 at a high MOI, the host culture exhibited a growth curve similar to the control culture (without virus inoculation) until 7 days p.i.; however, the virus titer of the infected culture showed a significant increase (∼3 orders of magnitude) within the initial 3 days p.i. Then, at 8 to 9 days p.i., the host culture showed a drastic decrease in cell numbers, and the virus titer reached ∼1010 infectious units ml−1 at 9 days p.i. (Fig. 5A). Considering the initial rapid increase in the virus titer following virus inoculation, the lytic cycle of CtenRNAV01 was estimated to be less than 24 h. The host cell numbers, however, did not show significant decreases until after 8 days p.i., in spite of the remarkably high MOI; this was repeatedly observed in several preliminary tests (data not shown). When a stationary-phase host culture was inoculated with CtenRNAV01 at a high MOI, the host culture also showed a growth curve similar to that of the control culture until 3 days p.i., and the virus titer showed a significant increase (∼3 orders of magnitude) within the initial 3 days p.i.; then, at 3 to 6 days p.i., the host cells showed a rapid decrease in number, and the virus yield reached ∼1010 infectious units ml−1 at 6 days p.i. (Fig. 5B). The rapid increase in virus titer following inoculation of the stationary-phase host culture also suggests that the lytic cycle is shorter than 24 h. We also observed significant accumulation of CtenRNAV01 genome-size RNA (the 8.9-kb band) in the host culture within 24 h p.i., as observed by extracting the total RNA from the host cell pellet (data not shown). We found the time from virus inoculation to the sudden crash of the host culture was shorter in stationary-phase culture than in logarithmic-phase culture (Fig. 5). This suggests higher sensitivity of stationary-phase cells than logarithmically growing cells to CtenRNAV01 infection. This is contrary to the other microalgal host-virus systems, in which logarithmically growing cells are more sensitive to virus infection (12, 15). Considering that viruses utilize the biosynthetic functions of hosts, it seems rational that host cells in the vigorously growing (logarithmic) phase are more suitable for viral growth because of their high biosynthesis activity; hence, future interpretation of these unexpected results will be of great interest.
FIG. 5.
Changes in cell numbers of C. tenuissimus 2-10 without (closed squares) or with (open squares) virus inoculation and virus titers (closed circles). Virus inoculation (arrows) was performed at day 0 (logarithmic phase) (A) or day 4 (stationary phase) (B), where the MOI was set at 11.6 and 12.1, respectively.
The burst sizes of CtenRNAV01 were estimated to be 1.18 × 104 and 1.10 × 104 infectious units cell−1, respectively, when virus inoculation was performed in exponential- and stationary-phase cultures; the calculations were based on the changes in host cell numbers and virus titers at 8 to 9 and 3 to 4 days p.i., respectively. These burst sizes are higher than those of the other diatom viruses, ∼103 infectious units cell−1 for RsRNAV and ∼102 infectious units cell−1 for CsNIV (12, 14). A typical thin section of the CtenRNAV-infected cell contained ∼3,000 particles. Assuming the virus particles' aggregation is three dimensional, each cell would contain ∼1.5 × 105 virus particles. Considering their possible aggregation and partial deficiency, the above estimated burst size (∼1 × 104 infectious units cell−1) appears reasonable.
BLAST searching and phylogenetic analysis.
By using ORF finder (http://www.ncbi.nlm.nih.gov/projects/gorf/), two open reading frames (ORFs) were identified in the 9.4-kb contig mentioned above. By using BLAST, the upstream and downstream ORFs (ORF-1 and ORF-2) were predicted to be a replicase polyprotein gene and a structural polyprotein gene, respectively. The significant BLAST hits (E value < 1E−60) of ORF-1 were to the replicase polyprotein of RsRNAV (E value = 0.0) and hypothetical proteins of marine RNA virus JP-A (2E−109) and JP-B (6E−68); those of ORF-2 were to the capsid polyprotein of RsRNAV (5E−134), a hypothetical protein of marine RNA virus JP-B (4E−87), the capsid polyprotein of HaRNAV (3E−80), and HP of marine RNA virus JP-B (3E−67) (4, 7, 20). Here, we note that JP-A and JP-B are previously unknown RNA viruses whose genomes were assembled from reverse-transcribed whole-genome shotgun libraries originated from British Columbian waters (Canada) (4). The significant BLAST hits may support the hypothesis of Culley et al. (4) that both JP viruses likely have protist hosts.
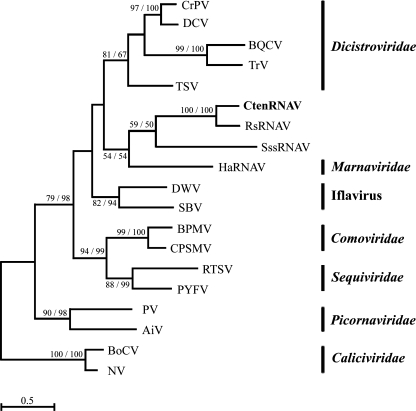
The RdRp domain was identified in ORF-1 and analyzed as follows. Both the NJ and ML methods were used to assess the phylogenetic relationships among positive-sense ssRNA viruses, including CtenRNAV. Similar topologies were obtained using the two methods; hence, only the ML phylogenetic tree is shown in Fig. 6. The monophyly of CtenRNAV01 and RsRNAV was supported with a bootstrap value of 100% (Fig. 6). This suggests the existence of a diatom-infecting ssRNA virus clade within the positive-sense ssRNA viruses. The tree also suggests lower relatedness of the diatom-infecting ssRNA viruses with those infectious to the other marine stramenopile organisms: Schizochytrium single-stranded RNA virus, infecting a fungoid protist, Sicyoidochytrium minutum (previously reported as Schizochytrium sp.) (23), and HaRNAV, infecting a bloom-forming raphidophyte, H. akashiwo (7).
FIG. 6.
ML tree based on deduced amino acid sequences of the RdRp whole domain. ML bootstrap values (percent) from 100 samples are shown at the nodes, followed by bootstrap values (percent) based on the NJ analysis of 100 samples. The ML distance scale bar is shown. CrPV, cricket paralysis virus; DCV, Drosophila C virus; BQCV, black queen cell virus; TrV, Triatoma virus; TSV, Taura syndrome virus; SssRNAV, Schizochytrium single-stranded RNA virus; DWV, deformed wing virus; SBV, Sacbrood virus; BPMV, bean pod mottle virus; CPSMV, cowpea severe mosaic virus; RTSV, rice turgo spherical virus; PYFV, parsnip yellow fleck virus; PV, human poliovirus 1 Mahoney; AiV, Aichi virus; BoCV, bovine enteric calicivirus; NV, Norwalk virus.
Implications.
CtenRNAV is the second ssRNA virus described that infects diatoms. The most remarkable feature of this virus is its distinctively high yield of ∼1010 infectious units ml−1. Generally, the highest yield found in other viruses infecting microalgae is between 107 and 108 ml−1; thus, the yield of CtenRNAV01 is 2 to 3 orders of magnitude higher. The high concentration of the host cell in a culture (∼106 cells ml−1) and the burst size of CtenRNAV01 (∼104 infectious units cell−1) may reasonably contribute to the high yield. This feature may make this virus suitable for experiments in which large numbers of virions are necessary.
There is insufficient data to determine the roles of ssRNA viruses in marine environments. However, studies of the bloom-forming dinoflagellate H. circularisquama and its infectious ssRNA virus, HcRNAV, provide a rational explanation for the ecological relationship between them. HcRNAV affects the dynamics (especially bloom termination) of host algae and may be preserved in sediments in the absence of its host population (13, 25). The ecology of C. tenuissimus and its viruses is now under examination; some preliminary results of our recent field survey show the existence of virus-like agents that are lytic to C. tenuissimus in both waters and sediments of western Japan, and they increase during blooms of Chaetoceros spp. (Y. Tomaru, unpublished data). This may indicate an intimate ecological relationship between the host diatom and its viruses, as was observed with H. circularisquama and HcRNAV. Further study will be required to determine the ecological importance of CtenRNAV as an agent affecting the dynamics of C. tenuissimus in natural environments.
Acknowledgments
This work was supported by grants-in-aid for scientific research (A) (2) (no. 16208019) from the Ministry of Education, Science and Culture of Japan; the Fisheries Agency of Japan; and the Ministry of Agriculture, Forestry and Fisheries of Japan.
Thanks are also due to H. Mizumoto (University of Tokyo, Japan) for technical assistance.
Footnotes
Published ahead of print on 9 May 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bettarel, Y., J. Kan, K. Wang, S. Cooney, K. Williamson, F. Chen, E. Wommack, and W. Coats. 2005. Isolation and characterisation of a small nuclear inclusion virus infecting the diatom Chaetoceros cf. gracilis. Aquat. Microb. Ecol. 40:103-114. [Google Scholar]
- 3.Chen, L. C. M., T. Edelstein, and J. McLachlan. 1969. Bonnemaisonia hamifera Hariot in nature and in culture. J. Phycol. 5:211-220. [DOI] [PubMed] [Google Scholar]
- 4.Culley, A. I., A. S. Lang, and C. A. Suttle. 2007. The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol. J. 4:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh, K., and I. Imai. 1987. Raphidophyceae, p. 122-130. In Japan Fisheries Resource Conservation Association (ed.), A guide for studies of red tide organisms. Shuwa, Tokyo, Japan. (In Japanese.)
- 6.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 7.Lang, A. S., A. I. Culley, and C. A. Suttle. 2004. Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo. Virology 320:206-217. [DOI] [PubMed] [Google Scholar]
- 8.Le Gall, O., T. Iwanami, A. V. Karasev, T. Jones, K. Lehto, H. Sanfacon, J. Wellink, T. Wetzel, and N. Yoshikawa. 2005. Genus Cheravirus, p. 803-805. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier, London, United Kingdom.
- 9.Le Gall, O., T. Iwanami, A. V. Karasev, T. Jones, K. Lehto, H. Sanfacon, J. Wellink, T. Wetzel, and N. Yoshikawa. 2005. Genus Sadwavirus, p. 799-802. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier, London, United Kingdom.
- 10.Meunier, A. 1913. Microplankton de la Mer Flamande, 1 ére partie: le genre “Chaetoceros” Ehr. Mem. Mus. R. Hist. Nat. Belg. 7:1-49. [Google Scholar]
- 11.Nagasaki, K., Y. Shirai, Y. Takao, H. Mizumoto, K. Nishida, and Y. Tomaru. 2005. Comparison of genome sequences of single-stranded RNA viruses infecting the bivalve-killing dinoflagellate Heterocapsa circularisquama. Appl. Environ. Microbiol. 71:8888-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagasaki, K., Y. Tomaru, N. Katanozaka, Y. Shirai, K. Nishida, S. Itakura, and M. Yamaguchi. 2004. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl. Environ. Microbiol. 70:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagasaki, K., Y. Tomaru, K. Nakanishi, N. Katanozaka, and M. Yamaguchi. 2004. Dynamics of Heterocapsa circularisquama (Dinophyceae) and its viruses in Ago Bay, Japan. Aquat. Microb. Ecol. 34:219-226. [Google Scholar]
- 14.Nagasaki, K., Y. Tomaru, Y. Takao, K. Nishida, Y. Shirai, H. Suzuki, and T. Nagumo. 2005. Previously unknown virus infects marine diatom. Appl. Environ. Microbiol. 71:3528-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagasaki, K., and M. Yamaguchi. 1998. Effect of temperature on the algicidal activity and stability of HaV (Heterosigma akashiwo virus). Aquat. Microb. Ecol. 15:211-216. [Google Scholar]
- 16.Nagasaki, K., and M. Yamaguchi. 1997. Isolation of a virus infectious to the harmful bloom causing microalga Heterosigma akashiwo (Raphidophyceae). Aquat. Microb. Ecol. 13:135-140. [Google Scholar]
- 17.Rines, J. B. E., and P. E. Hargraves. 1988. The Chaetoceros Ehrenberg (Bacillariophyceae) flora of Narragansett Bay, Rhode Island, USA. Bibl. Phycol. 79:1-196. [Google Scholar]
- 18.Sar, E. A., D. U. Hernandez-Becerril, and I. Sunesen. 2002. A morphological study of Chaetoceros tenuissimus Meunier, a little-known planktonic diatom, with a discussion of the section Simplicia, subgenus Hyalochaete. Diatom Res. 17:327-335. [Google Scholar]
- 19.Semancik, J. S., A. K. Vidaver, and J. L. Van Etten. 1973. Characterization of segmented double-helical RNA from bacteriophage phi6. J. Mol. Biol. 78:617-625. [DOI] [PubMed] [Google Scholar]
- 20.Shirai, Y., Y. Takao, H. Mizumoto, Y. Tomaru, D. Honda, and K. Nagasaki. 2006. Genomic and phylogenetic analysis of a single-stranded RNA virus infecting Rhizosolenia setigera (Stramenopiles: Bacillariophyceae). J. Mar. Biol. Assoc. U.K. 86:475-483. [Google Scholar]
- 21.Suttle, C. A. 1993. Enumeration and isolation of viruses, p. 121-137. In P. F. Kemp, E. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, FL.
- 22.Tai, V., J. E. Lawrence, A. S. Lang, A. M. Chan, A. I. Culley, and C. A. Suttle. 2003. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J. Phycol. 39:343-352. [Google Scholar]
- 23.Takao, Y., K. Nagasaki, K. Mise, T. Okuno, and D. Honda. 2005. Isolation and characterization of a novel single-stranded RNA virus infectious to a marine fungoid protist, Schizochytrium sp. (Thraustochytriaceae, Labyrinthulea). Appl. Environ. Microbiol. 71:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomaru, Y., N. Hata, T. Masuda, M. Tsuji, K. Igata, Y. Masuda, T. Yamatogi, M. Sakaguchi, and K. Nagasaki. 2007. Ecological dynamics of the bivalve-killing dinoflagellate Heterocapsa circularisquama and its infectious viruses in different locations of western Japan. Environ. Microbiol. 9:1376-1383. [DOI] [PubMed] [Google Scholar]
- 26.Tomaru, Y., N. Katanozaka, K. Nishida, Y. Shirai, K. Tarutani, M. Yamaguchi, and K. Nagasaki. 2004. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat. Microb. Ecol. 34:207-218. [Google Scholar]
- 27.Tomaru, Y., H. Tanabe, S. Yamanaka, and K. Nagasaki. 2005. Effects of temperature and light on stability of microalgal viruses, HaV, HcV and HcRNAV. Plankton Biol. Ecol. 52:1-6. [Google Scholar]
- 28.Tomaru, Y., K. Tarutani, M. Yamaguchi, and K. Nagasaki. 2004. Quantitative and qualitative impacts of viral infection on Heterosigma akashiwo (Raphidophyceae) population during a bloom in Hiroshima Bay, Japan. Aquat. Microb. Ecol. 34:227-238. [Google Scholar]
- 29.Werner, D. 1977. Introduction with a note on taxonomy, p. 1-23. In D. Werner (ed.), The biology of diatoms, vol. 13. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 30.Zingone, A. 1995. The role of viruses in the dynamics of phytoplankton blooms. Giorn. Bot. It. 129:415-423. [Google Scholar]