Abstract
We describe the pathogenic interaction between a newly described gram-positive bacterium, Leucobacter chromiireducens subsp. solipictus strain TAN 31504, and the nematode Caenorhabditis elegans. TAN 31504 pathogenesis on C. elegans is exerted primarily through infection of the adult nematode uterus. TAN 31504 enters the uterus through the external vulval opening, and the ensuing uterine infection is strongly correlated with a significant reduction in host life span. Young worms can feed and develop on TAN 31504, but not preferably over the standard food source. C. elegans worms reared on TAN 31504 as the sole food source develop into thin adults with little intestinal fat stores, produce few progeny, and subsequently cannot persist on the pathogenic food source. Within 12 h of exposure, adult worms challenged with TAN 31504 alter the expression of a number of C. elegans innate immunity-related genes, including nlp-29, which encodes a neuropeptide-like protein. C. elegans worms exposed briefly to TAN 31504 develop lethal uterine infections analogous to worms exposed continuously to pathogen, suggesting that mere contact with the pathogen is sufficient for the host to become infected. TAN 31504 produces a robust biofilm, and this behavior is speculated to play a role in the virulence exerted on the nematode host. The interaction between TAN 31504 and C. elegans provides a convenient opportunity to study bacterial virulence on nematode tissues other than the intestine and may allow for the discovery of host innate immunity elicited specifically in response to vulva-uterus infection.
Leucobacter chromiireducens subsp. solipictus strain TAN 31504 was isolated in our laboratory on 15 March 2004 from a contaminated Salmonella enterica serovar Typhimurium SL1344 lawn used in a survival assay of the nematode host Caenorhabditis elegans. Worms feeding on the contaminated SL1344 lawn died significantly faster than normal. In addition, some of the exposed worms developed yellowish cyst-like structures in the uterus. We successfully isolated the contaminating organism. Based on morphological, biochemical, and chemotaxonomic characteristics of the isolate, we determined TAN 31504 to be a new subspecies of Leucobacter chromiireducens, a gram-positive bacterium belonging to the Microbacteriaceae family (29).
Although TAN 31504 is the first Leucobacter species to be identified in association with C. elegans, another member of the genus has been found within nematode-inhabited soil. Leucobacter iarius was recently isolated in association with infective juveniles of the entomopathogenic nematode Steinernema thermophilum collected from soils of New Delhi, India (40). In addition to Leucobacter, the Microbacteriaceae family includes a number of genera with species members known to be associated with nematodes (12). The characterization of “Microbacterium nematophilum” has been largely based on its interaction with C. elegans (1, 8, 14-16, 31, 32, 47). Interestingly, “M. nematophilum” was also identified as a laboratory contaminant and displays a distinctive mode of infection on C. elegans (15).
The pathogenic microbes utilized in C. elegans pathogenesis studies include many bacteria and several fungal pathogens (13). The majority of these organisms cause disease by a means correlated with accumulation of the pathogen within the intestinal lumen. At present, eight gram-positive genera include members that are pathogenic to C. elegans (13). Among this group, only “M. nematophilum” and Streptoverticillium albireticuli have been shown to affect nematode tissues other than the intestine. S. albireticuli exhibits hyphal growth on both external and internal surfaces of C. elegans and ultimately kills the nematode (33). The precise nematicidal activity exerted and specific details of the worm tissues affected by S. albireticuli have not been described. In contrast, colonization of C. elegans by “M. nematophilum” results in a nonlethal infection that is restricted to the external rectal and postanal cuticle (15). The presence of “M. nematophilum” induces swelling of the nematode hypodermal tissue beneath the infected area, resulting in a distinctive swollen tail symptom referred to as the deformed anal region (Dar) phenotype, constipation, and subsequent slow growth of the host. Interestingly, the tolerance demonstrated by C. elegans to colonization by “M. nematophilum” is not a general characteristic of the genus Caenorhabditis, as a number of species succumb to “M. nematophilum” infection during early larval development (1).
The gram-negative bacteria Yersinia pestis and Y. pseudotuberculosis and the fungal pathogens Duddingtonia flagrans and Drechmeria coniospora also exert virulence on nonintestinal nematode tissues. D. flagrans is a nematode-trapping fungus that successfully preys on C. elegans (27). D. flagrans trap cells adhere to the cuticle of C. elegans and are effective at capturing the nematode. D. coniospora adheres as spores to the external cuticle in the head, vulval, and tail regions of C. elegans (9, 19). From there, the spores grow into mycelia that penetrate and spread rapidly throughout the worm body, inevitably killing the host. Y. pestis and Y. pseudotuberculosis adhere as a robust biofilm to the nematode head that ultimately covers the worm mouth and inhibits feeding of the host (10). More recently, a biofilm-deficient strain of Y. pestis was reported to cause disease in C. elegans through virulence mechanisms dependent on intestinal accumulation of the pathogen (41).
Here, we have investigated TAN 31504 and C. elegans in the context of a host-pathogen interaction. We present findings that place TAN 31504 within the small set of C. elegans pathogens that display unusual modes of infection on the nematode host. We show that TAN 31504 is pathogenic to C. elegans, primarily through infection of the uterus. In addition, we show that C. elegans exposure to TAN 31504 alters nematode physiology, feeding behavior, and host gene expression.
MATERIALS AND METHODS
Nematodes.
C. elegans Bristol strain N2, vulvaless strain MT3232 lin-10(n1390) (22), vulvaless strain CB1309 lin-2(e1309) (18), and temperature-sensitive germ line proliferation-defective strain SS104 glp-4(bn2) (5) were kindly provided by the Caenorhabditis Genetics Center (CGC), University of Minnesota, Minneapolis. The pnlp-29::gfp reporter strain (9, 30) was generously provided by Jonathan J. Ewbank (Centre d'Immunologie de Marseille-Luminy, France). Nematodes were propagated by standard techniques on nematode growth medium (NGM) utilizing the standard food source strain, Escherichia coli OP50-1 (7).
Bacterial strains.
E. coli OP50-1 was provided by the CGC. S. enterica serovar Typhimurium SL1344 and the green fluorescent protein (GFP)-expressing derivative SM022 have been described previously (17, 45). The type strain of L. chromiireducens subsp. chromiireducens (LMG 22506) was obtained from the BCCM/LGM (Belgian Coordinated Collections of Micro-organisms collection of bacteria, Laboratorium voor Microbiologie Universiteit Gent). E. faecalis V583 and P. aeruginosa PA14 have been described previously (35, 36). L. chromiireducens subsp. solipictus can be obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) as strain DSM 18340 or from the American Type Culture Collection (ATCC) as strain ATCC BAA-1336.
Bacterial growth conditions.
All strains were maintained on Luria-Bertani (LB) medium (1.5% agar). The LB medium for OP50-1, SL1344, SM022, V583, and PA14 was supplemented with the following, respectively: 100 μg ml−1 streptomycin, 300 μg ml−1 streptomycin, 50 μg ml−1 kanamycin, 20 μg ml−1 gentamicin, and 100 μg ml−1 rifampin. TAN 31504 and LMG 22506 were grown without selection. Overnight growth of TAN 31504 and LMG 22506 in liquid culture was done at 25°C in LB medium without antibiotics. The following were cultured in liquid media aerated overnight at 37°C in the indicated media: OP50-1 was grown in LB medium; PA14 was grown in King's B; SM022 was grown in LB medium containing 50 μg ml−1 kanamycin; V583 was grown in brain heart infusion (BHI) medium. SL1344 liquid overnight cultures were grown standing at 37°C in high-salt (1.5%) LB medium. The cdc-25.1 RNA interference (RNAi) clone from the Ahringer RNAi feeding library was maintained on and cultured overnight in LB medium plus 100 μg ml−1 ampicillin at 37°C (20). The cdc-25.1 RNAi plates were made as previously described (38).
For survival assays and phenotypic analysis: NGM lawns of OP50-1, TAN 31504, and LMG 22506 were generated from 25-fold-concentrated liquid overnight cultures; LB lawns of OP50-1, TAN 31504, LMG 22506, and SM022 and BHI lawns of V583 were prepared as previously described, with the exception of overnight incubation at 25°C (38). Freshly inoculated LB plates supplemented with 0.5% cholesterol, having no prior incubation before use, were used in the developmental growth rate and fat-staining assessments. LB agar supplemented with 0.5% cholesterol was also used for generational and progeny production assessments of worms performed on TAN 31504 and LMG 22506 lawns. Addition of 0.5% cholesterol to media used for nematode growth-related studies fulfills the requirement of exogenous sterols for C. elegans development (11).
In mixed-lawn assays, the mixed inoculums of TAN 31504 and OP50-1 were generated by combining various amounts of the two overnight cultures prior to plating. After overnight incubation at 25°C, two test plates representative of each mixed-lawn condition were subjected to CFU counts in order to determine the proportion of each lawn constituent. LB plates with 15 μg ml−1 gentamicin and growth at 25°C for 2 days were used to select for the TAN 31504 CFU count, since TAN 31504 is resistant to gentamicin and OP50-1 is sensitive to gentamicin. LB plates with 100 μg ml−1 streptomycin and growth at 37°C overnight were used to select for the OP50-1 CFU count, since TAN 31504 grows extremely poorly at 37°C and produces no visible colonies after overnight incubation.
LB lawns generated for biofilm characterization were each produced at 25°C from 100 μl of overnight culture spread onto 9-cm2 petri plates of plain LB medium and incubated for 24 h. Bacterial lawns generated for the assessment of host gene expression by quantitative real-time PCR (qRT-PCR) were prepared as follows. Petri plates (9 cm2) of the following media were inoculated with 100 μl of the corresponding overnight culture for the strains listed and incubated at the specified temperatures for the time indicated prior to use: TAN 31504 and LMG 22506, plain LB medium, 25°C overnight; PA14, plain slow-killing plates (42), 37°C overnight; SL1344, plain LB medium, 37°C for 4 h. For OP50-1 plates, 100 μl of a 25-fold-concentrated liquid overnight culture was spread onto NGM plates and incubated at room temperature (∼23°C) 12 to 24 h prior to use.
Production of Glp worms.
Wild-type N2 and lin-10(n1390) worms were made sterile by a feeding RNAi method utilizing the cdc-25.1 RNAi clone (38, 43). The cdc-25.1 gene encodes a dual-specificity phosphatase that is required for germ line proliferation (2). RNAi to cdc-25.1 during oogenesis has been found to produce, in the following generation, sterile germ line proliferation-deficient (Glp) adults that behave similarly to sterile glp-4(bn2) worms (38). Unless otherwise indicated, 1-day-old Glp adults reared on the cdc-25.1 RNAi clone lawns were used in experiments involving Glp worms.
Survival analysis and infection conditions.
Approximately 100 to 120 1-day-old Glp adults were apportioned between three and four bacterial lawns generated as described above for each experiment conducted. For the brief-exposure assays, distinct worm populations of the initial infection condition were maintained when shifted to the subsequent infection condition. The worms were incubated at 25°C, unless otherwise indicated. Worms were assessed for survival essentially as described previously (38). The resulting data were analyzed with StatView, version 5.0.1, software (SAS Institute, Inc.) and plotted using the statistical test of Kaplan-Meier (nonparametric survival analysis). The host life span and associated pathogen virulence were considered significantly different when the P value of the log rank (Mantel-Cox) test was <0.05. All experiments were replicated in a comparable fashion and found to produce findings similar to the data presented.
Microscopes.
Bright-field, Nomarski differential interference contrast (DIC), and fluorescent microscope images were obtained using Leica ×10 and ×20 HC PL Fluotar and ×40 HCX PL Fluotar objectives on a Leica DM RXA2 upright microscope. The fluorescent micrographs were generated utilizing Leica filter cube N3 (red, bandpass filter, 600/40 nm), Leica filter cube I3 (green-red, longpass filter, 515 nm), and Chroma Technology Corp. filter 31019 (green, bandpass filter, 505/40 nm). Images were captured using a Leica DC500 camera and Leica Firecam software version 1.2. A Leica MZFLIII stereoscope utilizing the same camera and software mentioned was used to produce the bright-field image at ×80 magnification. Image overlays were generated using Adobe Photoshop cs version 8.0 on a Macintosh PowerBook G4 using Mac OS X, version 10.4.11.
Dye penetration method.
To ensure that only the external structures of the worms are exposed to the dye, infected Glp worms were paralyzed in 100 mM levamisole in 1× M9 salts. Paralyzed worms were then treated with 10 mg ml−1 of the 10-kDa tetramethylrhodamine dextran dye in the levamisole solution for 15 min in the dark; paralyzed worms do not exhibit pharyngeal pumping and therefore do not take up the dye orally. For oral exposure, worms were treated with the same concentration of dye in 1× M9 salts for 15 min in the dark. The level of bacteria carried over with the worms into the 1× M9 dye solution was sufficient to maintain feeding/pharyngeal pumping and allowed passage of the dye into the digestive tract of the treated worms. After external and oral exposure to dye, worms were rinsed gently with the levamisole solution three times prior to visualization. All worms subjected to microscopy on the Leica DM RXA2 upright microscope were paralyzed with levamisole prior to visualization.
Food choice assessments.
The design of the unbiased- and biased-food-choice plates was derived from that presented by B. B. Shtonda and L. Avery (39). Ten microliters of the appropriate overnight culture was spotted or spread onto 5-cm2 petri plates of LB medium plus cholesterol in the arrangements shown in Fig. 4G. The plates were incubated overnight at 25°C prior to use. The pnlp-29::gfp reporter strain was used in place of N2 because the GFP reporter in this otherwise wild-type strain, which is expressed at a considerable level throughout the larval stages of development, aided tremendously in the detection of young worms during the assays. TAN 31504 lawns express a yellow pigment that makes visualization of young worms within the lawns extremely difficult, and it is essentially impossible to find all of the young worms contained in the lawns without the aid of a fluorescent marker. The use of a fluorescent strain also allowed assessment of the same plates at all time points in the experiment.
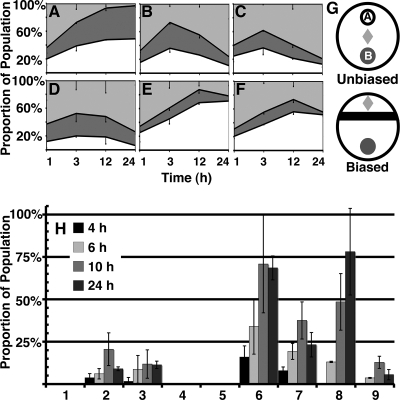
FIG. 4.
C. elegans will feed on TAN 31504, but not preferably to LMG 22506 or OP50-1. (A to F) Unbiased food choice of L1 larvae on single-food-source (A to C) and choice-test (D to F) plates. Please refer to panel G (top) for food arrangement details. The food source strains tested were E. coli OP50-1, L. chromiireducens subsp. solipictus TAN 31504, and L. chromiireducens subsp. chromiireducens LMG 22506. The 100% stacked area graphs represent the percentage of worms found within lawn A (white), lawn B (dark gray), and off the lawns (light gray) over time. Error bars of the standard error of the mean are shown. (A) OP50-1 constitutes both lawns A and B. A comparably similar number of worms occupied each lawn at all time points, with few worms remaining off the lawn by 12 h and 24 h. (B) TAN 31504 constitutes both lawns A and B. A similar number of worms to that in panel A reached each lawn by 1 h and 3 h. A notable movement of the worms off the lawns was observed at 12 h, with the trend continuing at 24 h. (C) LMG 22506 constitutes both lawns A and B. A similar number of worms to that in panels A and B reached each lawn by 1 h. Although more worms moved into the lawns after 1 h, a slight decrease in the overall number of worms occupying the lawns at 3 h compared to panels A and B was observed. Movement of the worms off the lawns began at 12 h and continued to 24 h, as was seen in panel B. (D) More worms went to LMG 22506 (lawn B) than to TAN 31504 (lawn A) by 1 h and 3 h. The fewest worms occupied these lawns together at 3 h over all other 3-h conditions assessed. Movement of the worms off both lawns occurred at 12 h and 24 h, as in panels B and C. (E and F) Notably more worms went to OP50-1 (lawn A) over TAN 31504 (E; lawn B) and LMG 22506 (F; lawn B) at all time points. Movement of the worms off the TAN 31504 and LMG 22506 lawns was evident by 24 h. (G, top) Unbiased-food-choice arrangement of food source lawns for panels A to F. Lawn A is depicted in white, and lawn B is depicted in dark gray. Constituents of lawns A and B are indicated above. The light gray diamond represents the position on the plate where the L1 larvae were placed. (G, bottom) Biased-food-choice arrangement of food source lawns for (H). The biased-food-source lawn is depicted in black, and the unbiased-food-source lawn is shown in dark gray. The light gray diamond represents the position on the plate where the L1 larvae were placed. (H) Biased food choice of L1 larvae. Please refer to panel G (bottom) for food arrangement details. Columns 1 to 9 represent the following combinations of food source lawns listed as biased lawn/unbiased lawn: 1, OP50-1/OP50-1; 2, TAN 31504/TAN 31504; 3, LMG 22506/LMG 22506; 4, OP50-1/TAN 31504; 5, OP50-1/LMG 22506; 6, TAN 31504/OP50-1; 7, TAN 31504/LMG 22506; 8, LMG 22506/OP50-1; 9, LMG 22506/TAN 31504. The numbers of worms found within the unbiased lawns at the indicated time points are represented as the percentages of the total populations. The error bars of the standard error of the mean are shown. Worms biased with an OP50-1 lawn did not venture into the unbiased lawns during the 24-h assessment. Worms moved beyond the TAN 31505-biased lawns as early as 4 h for all corresponding tests and were seen to accumulate in the largest numbers within unbiased lawns at 10 h. The largest number of worms that traveled beyond the TAN 31504-biased lawns moved into OP50-1. A smaller proportion of these worms moved into LMG 22506, and the fewest worms moved back into TAN 31504 of the unbiased lawn. The largest proportions, but relatively small overall percentages, of worms that ventured beyond the LMG 22506-biased lawns into the TAN 31504- and LMG 22506-unbiased lawns were observed at 10 h. The majority of the LMG 22506-biased population moved into OP50-1 by 24 h, with most of the worms reaching the unbiased OP50-1 lawn by 12 h.
Between 2,800 to 3,200 and 1,000 to 1,100 worms in two replicate assays (repeat assays were performed on a separate occasion) were equally dispersed onto at least four plates of each of the unbiased- and biased-food-choice conditions described in Fig. 4. The numbers of worms on the lawns were recorded at the indicated times by either real-time visual assessment (1-h unbiased and 4-h biased time points only) or image capture of fluorescent micrographs generated of the food choice plates. The numbers of worms in and off the lawns of the later time points were counted subsequent to completion of the experiment. Worms found on the walls of assay plates and worms that appeared dead within the area of initial placement after 24 h were not counted. The data generated from the two independent assays of each of the unbiased- and biased-food-choice tests were combined and are shown in Fig. 4.
Developmental progress and fat staining.
Sterile eggs from gravid adults reared at 25°C were obtained using an alkaline bleach solution (38). Approximately 200 to 250 eggs were dropped onto each of two freshly seeded plates (LB plus cholesterol) of the three bacterial strains assessed, and 50 to 60 eggs were dropped onto each of three previously prepared Nile red plates of the TAN 31504 (generated on LB agar supplemented with cholesterol) and standard food source lawns. Nile red fat staining was performed as described previously (3). The Nile red plates were incubated in the dark at 25°C and images of late L4 larvae/young adults were taken of 87-h-old and 43-h-old worms grown on the Nile red-containing TAN 31504 and standard food sources, respectively. The plates assessed for progeny development rate were incubated at 25°C, and the numbers of gravid adults, young adults, L4 larvae, and L3-or-younger larvae were recorded at 24-h intervals for each condition. These experiments were repeated twice with similar numbers of worms and produced comparable results.
Biofilm characterization.
Bacterial lawns were generated as described above. Five milliliters of water was dispensed slowly by gravity onto the center of the 24-h-old lawns or plain LB plates using a 5-ml serological pipette. After a 5-min incubation at room temperature, images of the water-treated plates were captured. The water sample from each plate was collected directly, by pouring, into test tubes after the 5-min room temperature incubation, and a picture was taken. The optical density at 600 nm of each sample was determined using a Beckman Coulter DU 800 spectrophotometer. TAN 31504 lawns were submerged under 25 ml of water for 15 min prior to image capture and lawn disruption. The submerged lawns were disrupted by 2 to 3 min of manual agitation. The images shown in Fig. S5 in the supplemental material were captured using a Sony DSC-F717 digital camera. The data described are representative of multiple repeated observations.
Host gene expression.
Synchronized populations of wild-type young adult worms (approximately 44-h-old worms) were generated at 25°C on the standard food source from sterilized eggs. Worms were collected from the food source lawns in 1× M9 salts at room temperature. Worms were washed twice with the salts before the large population was distributed between four and six plates per bacterial strain tested. Worms were exposed to the pathogens and the standard food source control for 12 h at 25°C. Worms were then collected from the assay plates in 1× M9 salts and washed with the salts an additional two to three times prior to suspension in TRIzol reagent from Invitrogen. The samples were flash frozen in liquid nitrogen and stored at −80°C until processing. These experiments were repeated two to four times for each infection condition tested.
RNA from whole worms was extracted according to the protocol of R. D. Burdine and M. J. Stern (http://www.wormbase.org; WormBase release WS188, WBPaper00015272). qRT-PCRs were prepared with a Bio-Rad iScript one-step RT-PCR kit with Sybr green according to the manufacturer's protocol. The qRT-PCRs were performed on a Bio-Rad iCycler iQ RT-PCR detection system, and the resulting cycle threshold (CT) data were generated using the system's software version 3.0 for Windows.
The panel of infection-responsive genes assessed was constructed in our lab (S. S. Slutz, E. A. Evans, and M.-W. Tan, unpublished data). Primer sequences used for each gene are available upon request. The panel of 96 genes includes 4 normalization controls represented by 3 genes (ama-1, pan-actin, and F44B9.5). Data from two to four biological replicates of each infection condition assessed were successfully collected on different occasions for 91 of the 96 genes; three replicates of the standard food source condition, three replicates of the PA14 condition, two replicates of the SL1344 condition, and four replicates of the TAN 31504 condition. Each biological replicate was normalized separately to its four internal controls. Student's t tests, with two-tailed distributions, were performed between the replicates of the standard growth condition and the replicates from each of the infection conditions tested, as well as between the replicates of one infection condition to another. The expression of a gene within a particular profile is reported to be significantly different in expression from the other profiles when the corresponding P values were <0.05. The statistical tests were performed with Microsoft Excel 2004 for Mac version 11.3.7. The four arrays of 46 genes presented in the dendrogram (see Fig. 6) were constructed from the averaged control-normalized CT values generated from the two to four biological replicates. The gene expression sets of the averaged arrays were normalized to their median CT values and clustered hierarchically using the average linkage algorithm of Cluster 3.0 for Mac OS X (0.1). The median-normalized CT values of the preclustered arrays and the P values generated from the statistical tests performed are presented in Table S1 in the supplemental material. The dendrogram was constructed with Java TreeView, version 1.0.13.
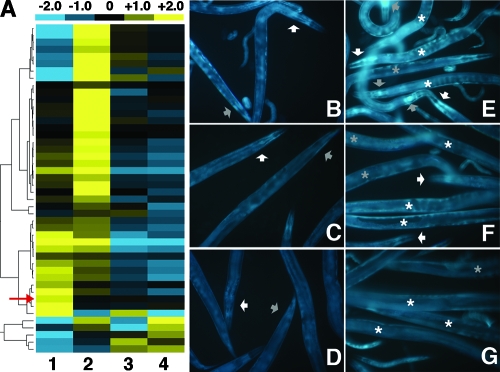
FIG. 6.
Exposure to TAN 31504 alters host gene expression. (A) Hierarchically clustered qRT-PCR gene expression sets (rows) of a panel of infection-responsive genes that are differentially expressed in adult worms after 12 h of exposure to L. chromiireducens subsp. solipictus TAN 31504 (column 1); P. aeruginosa PA14 (column 2); S. enterica serovar Typhimurium SL1344 (column 3); and the standard food source, E. coli OP50-1 (column 4). The median CT of each gene expression set has been set to zero (black). Shown above the dendrogram is a color bar illustrating the colors used to indicate the positive (yellow) and negative (blue) directions of normalized gene expression relative to the median; contrast within the color bar is illustrated for the change (fold) CT values of −2.0 through +2.0. The red arrow points to the nlp-29 expression set. The tree to the left of the dendrogram illustrates the hierarchical clustering of the gene expression sets produced by application of the Cluster 3.0 average linkage algorithm. The normalized qRT-PCR CT data represented in the dendrogram are restricted to the expression patterns of the 46 genes determined to be expressed at significantly different levels under one or more of the pathogenic conditions compared to the standard food source condition. Those 46 genes in the order presented (from top to bottom) are lys-1, lys-2, F56F10.1, Y87G2A.2, F35E12.8, crn-2, clec-3, ZK6.11, ins-18, lec-11, ins-7, tsp-1, T01D3.6, gst-38, cpr-3, K11D12.5, clc-1, tth-1, F35E12.7, skr-3, pgp-5, T25C12.3, C33H5.13, dod-19, nhr-112, Y40D12A.2, F49F1.1, lys-8, pqm-1, K05F1.10, ins-11, spp-18, T20G5.8, mpk-2, F52E1.5, asp-3, spp-1, abf-2, nlp-29, spp-12, fmo-2, C23G10.11, thn-3, cnc-2, clec-97, and ZK6.7. The median-normalized qRT-PCR CT values associated with the expression of all 91 genes assessed are shown in Table S1 in the supplemental material. (B to G) 31019 short-pass filter (GFP) fluorescence micrographs of pnlp-29::gfp reporter strain adults after 24 h of exposure to a TAN 31504 LB lawn (B and E), an OP50-1 LB lawn (C and F), and the standard food source (D and G) (×200 magnification). Images include the gravid adults of interest (large worms) and some of their progeny (small worms and eggs/developing embryos) that are not discussed here. (B to D) The white arrowheads point to the head region of one worm, and the gray arrowheads point to the tail region of another. Slight induction of the pnlp-29::gfp reporter by exposure to the OP50-1 LB lawn (C and F) over that observed on the standard food source lawn (D and G) was detected along the entire worm. A somewhat higher increase in pnlp-29::gfp reporter expression was apparent in the adults exposed to the TAN 31504 LB lawn (B and E). (E to G) White and gray asterisks placed on individual worms are positioned to the right and to the left, respectively, of the anterior portion of the worm. The white arrowheads point to the mouth and pharyngeal regions of worms in panels E and F. The gray arrowheads point to the terminal bulb of the pharynx of worms in panel E. A majority of the intestine is represented among the groups of worms displayed (E to G). The most prominent increase in GFP signal displayed by TAN 31504-exposed reporter worms was seen in the anterior intestine and in the head, especially around the terminal region of the pharynx (E). A comparison between these populations of worms further supported the idea of a TAN 31504-mediated induction of nlp-29, as suggested by the data presented in panel A.
pnlp-29::gfp reporter gene expression.
Synchronized populations of young adult reporter strain worms (approximately 44-h-old worms) were generated at 25°C on the standard food source from sterilized eggs. Approximately 100 worms were moved to each of three plates of the bacterial lawns tested. The worms were incubated at 25°C for 24 h, after which time micrographs of the gravid reporter worms were captured.
RESULTS
TAN 31504 displays a novel mode of infection on C. elegans.
L. chromiireducens subsp. solipictus strain TAN 31504 was isolated from a tainted S. enterica serovar Typhimurium SL1344 assay, in which some sterile Glp worms died more rapidly and accumulated cyst-like structures within the mid-body region. To determine if TAN 31504 caused this unusual infection, Glp adult C. elegans worms were exposed to axenic lawns of TAN 31504. Both the cyst-like structure (Fig. 1A) and the increased host mortality rate (see Fig. 3E and see Fig. S3 in the supplemental material) were recapitulated by exposure to TAN 31504 alone. On closer inspection, the cyst-like structure resulted from a TAN 31504-mediated infection of the uterus (Fig. 1B, 2A, and 3C). The detrimental effects of TAN 31504 on nematode health were not restricted to the Glp background of the host, since fertile adult C. elegans worms also developed uterine infections when exposed to TAN 31504, and in fact succumbed to infection faster than infected Glp worms (data not shown). For subsequent TAN 31504-mediated pathogenesis studies, unless specified otherwise, 1-day-old adult Glp worms were used because the progression of infection within these worms was easier to visualize in the absence of developing embryos. The Glp background also negated the confounding problem of host death due to matricide that results from progeny hatching within fertile worms.
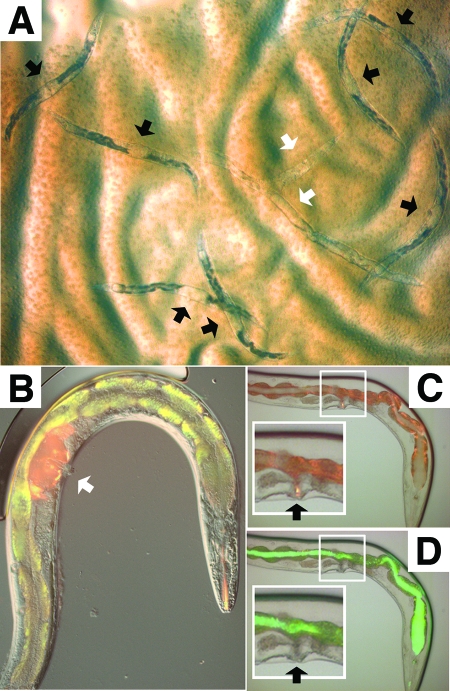
FIG. 1.
Leucobacter chromiireducens subsp. solipictus strain TAN 31504 accumulates within the uterus of C. elegans via the external vulval opening. (A) Bright-field micrograph of Glp adult worms feeding on a lawn of TAN 31504 after 5 days of exposure (×8 magnification). Black arrowheads point to the accumulated bacterial pathogen within the uterus of dead and dying worms. The white arrowheads indicate degraded worm carcasses. (B) Image of N3 filter (red) and I3 long-pass filter (green-red) fluorescence micrographs overlain on the corresponding DIC micrograph of a Glp adult worm exposed to TAN 31504 for 5 days followed by treatment with 10-kDa tetramethylrhodamine dextran (×200 magnification; images at ×400 magnification are shown in Fig. S1D to F in the supplemental material). The white arrowhead points to the accumulated red 10-kDa dye within the uterus of the infected worm. Autofluorescent granules, predominately of the worm intestine, appear as yellow in the merged image shown. The intensity of autofluorescence dramatically increases during C. elegans exposure to TAN 31504; the reason for this is unknown. (C) Image of N3 filter fluorescence micrograph overlain on the corresponding bright-field micrograph of a Glp adult worm exposed to S. enterica serovar Typhimurium SM022 for 5 days followed by treatment with 10-kDa tetramethylrhodamine dextran (×200 magnification; images at ×400 magnification are shown in Fig. S1A to C in the supplemental material). Fluorescence is predominately of the autofluorescent intestinal granules. The black arrowheads in panels C and D indicate the vulval openings. The insets, highlighted by white boxes, in panels C and D contain enlarged images of the vulva-uterus regions. The 10-kDa dye accumulated within the exterior vulval slit. (D) Corresponding I3 fluorescence micrograph overlain on the same bright-field micrograph depicted in panel C. The GFP-expressing pathogen accumulated within the worm intestinal lumen. The vulval slit was devoid of the GFP signal.
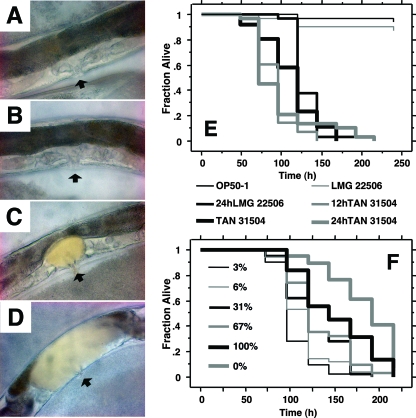
FIG. 3.
Limited exposure to TAN 31504 is sufficient to produce an increase in host mortality rate. (A to D) Bright-field micrographs of the vulva-uterus regions of Glp adult worms; the vulva is placed toward the bottom and the intestine is positioned toward the top of each worm body image, with the uterus situated directly above the vulva and below the intestine (×200 magnification). The black arrowheads point to the vulvas. (A to C) Accumulation of the food source bacteria within the nematode uterus was not found after 4 days of continuous exposure to the standard-food-source condition (A) or to L. chromiireducens subsp. chromiireducens LMG 22506 (B); however, uterine accumulation of L. chromiireducens subsp. solipictus TAN 31504 was observed (C). (D) Extensive uterine infection in a worm initially exposed to TAN 31504 for 24 h followed by 3 days of exposure to the standard food source. Similar extensive uterine infections were observed in worms exposed to mixed lawns of TAN 31504 and OP50-1 comprised of only 3 to 6% TAN 31504. (E) Survival curves of Glp adult N2 worms exposed continuously and discretely to pathogen. OP50-1, TAN 31504, and LMG 22506 indicate constant exposure to the respective pathogens. “24hLMG 22506” indicates an initial 24-h exposure to LMG 22506 followed by an extended exposure to TAN 31504. “12hTAN 31504” and “24hTAN 31504” indicate initial 12-h and 24-h exposures to TAN 31504, respectively, followed by extended exposure to standard-food-source lawns. Survival curves begin with time zero set to the time just prior to application of the infection conditions described above. The remaining worm populations on OP50-1 and LMG 22506 were censored after 10 days. Initial exposure to LMG 22506 did not appear to offer extended protection with regard to host life span extension over the TAN 31504 condition. Although from time zero the average 19.1-h extension in mean life span observed was significant (log rank P = 0.044), it was less than the 24-h period of initial LMG 22506 exposure (LTmean = 130.8 ± 3.9 h for 24 h of LMG 22506 exposure and 111.7 ± 6.5 h for TAN 31504). The brief 12-h exposure to TAN 31504 was sufficient to increase the host mortality rate (LTmean = 90.9 ± 4.2 h) significantly over that yielded by the constant exposure condition (log rank, P = 0.0041). The shortened host life span produced by the 24-h TAN 31504 condition (LTmean = 96.8 ± 7.9 h) was no different from that generated by the 12-h TAN 31504 condition (log rank, P = 0.66), nor was it significantly resolvable from that produced by the constant exposure condition (log rank, P = 0.26). (F) Survival curves of Glp adult N2 worms exposed to LB lawns consisting of varied ratios of bacteria. “100%” indicates exposure to pure lawns of TAN 31504; “0%” indicates exposure to pure lawns of OP50-1; and “3%,” “6%,” “31%,” and “67%” represent proportions of TAN 31504 to OP50-1 at the time of worm exposure. Host mortality rate was significantly enhanced by exposure to TAN 31504 (100%) compared to OP50-1 (0%) (log rank, P = 0.010; LTmean = 151.4 ± 7.7 h and 183.4 ± 5.8, respectively). Worms exposed to the 3%, 6%, and 37% lawns displayed an enhanced mortality rate compared to those exposed to the pure TAN 31504 lawn (log rank, P < 0.0001, P < 0.0001, and P = 0.0024, respectively; LTmean = 103.8 ± 3.4 h, 114.7 ± 4.0 h, and 125.7 ± 5.0 h, respectively). The mean host life span (LTmean = 132.4 ± 5.9 h) on the 67% lawn was shorter than that exhibited on the OP50-1 lawn (log rank, P < 0.0001), but was not significantly different from that displayed on the 100% lawn (log rank, P = 0.081).
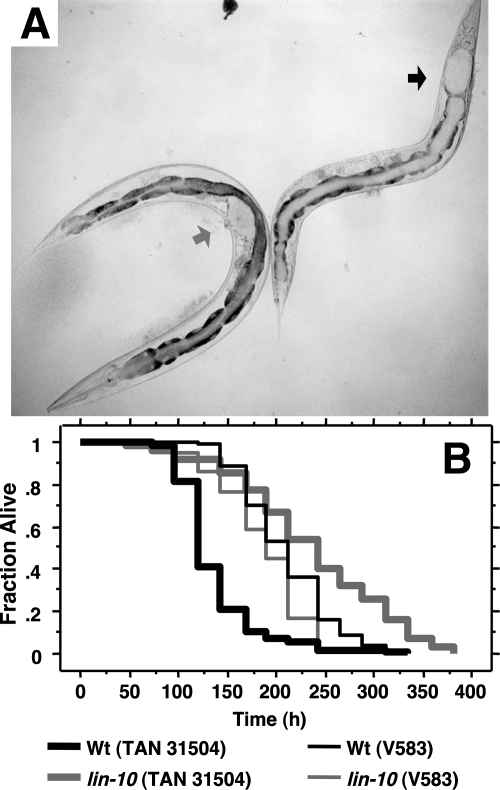
FIG. 2.
A vulvaless C. elegans strain is resistant to TAN 31504-mediated infection of the uterus and does not exhibit an enhanced host mortality rate. (A) Bright-field micrograph of TAN 31504-infected Glp adult wild-type and vulvaless lin-10(n1390) worms (left and right, respectively) taken 5 days after exposure to pathogen at 25°C (×100 magnification). The gray arrowhead points to the vulva-uterus region of the wild-type worm. The black arrowhead points to the most anterior region of the lin-10(n1390) worm intestine. Accumulation of the yellow-pigmented pathogen (represented here in off-white) within the uterine and intestinal lumens of the wild-type and lin-10(n1390) worms, respectively, was observed. Distention of the lumen of the respective tissues was seen and is indicated by the arrowheads. Magnified color images of the infected worms are shown in Fig. S2 in the supplemental material. (B) Survival curves of Glp adult wild-type (Wt; black curves) and vulvaless lin-10(n1390) (gray curves) worms exposed to TAN 31504 (thick lines) and E. faecalis V583 (thin lines) at 25°C. Curves are represented as a fraction of survivors over time. The vulvaless strain exhibited significant resistance (log rank, P < 0.0001) to TAN 31504-mediated killing compared to wild-type (LTmean = 236.4 ± 9.5 h versus 137.1 ± 4.3 h, respectively), while displaying increased sensitivity (log rank, P = 0.0003) to V583 pathogenesis (LTmean = 185.5 ± 4.8 h versus 210.3 ± 4.5 h, respectively).
The yellow color of the distended uterine lumen of infected worms was due to the accumulation of TAN 31504 cells, which produce a carotenoid that is firmly associated with the cell envelope (29). TAN 31504 could enter the uterus from the exterior through the vulval opening. Alternatively, following entry into the intestinal lumen, this pathogen could invade and transverse host tissue to gain entry into the uterus. To distinguish between these possibilities, we compared uterine cyst formation in the wild type and the vulvaless lin-10(n1390) mutant. Unlike wild-type adults, the vulvaless worms did not develop uterine infections (Fig. 2A). This result supported the idea that TAN 31504 gained entry to the nematode uterus through the external vulval opening.
We next assessed the penetration of a 10-kDa tetramethylrhodamine dextran dye into adult worms infected with either TAN 31504 or the GFP-tagged S. enterica serovar Typhimurium SL1344-derivative, strain SM022. When applied to worms externally, under conditions that prevented oral uptake, the dextran dye accumulated within the uteri of severely TAN 31504-infected wild-type worms but did not penetrate further than the vulval slit of SM022-infected worms (Fig. 1B to D). Furthermore, when SM022- and TAN 31504-infected worms were exposed orally to the 10-kDa dye, under conditions that permitted consumption and entry of the dye into the gut, no leakage of the small dye from the intestinal lumen into surrounding tissues was observed (data not shown). Results obtained from dye penetration assays of worms maintained on the standard food source were similar to those described for SL1344-exposed worms (data not shown). These experiments were also performed with the glp-4(bn2) strain at the restrictive temperature and yielded similar findings (data not shown). That TAN 31504, but not SM022, was able to penetrate the vulval barrier to the nematode uterus indicates that uterine accumulation is not a general mode of infection demonstrated by bacterial pathogens on compromised hosts. Together, the results suggest that TAN 31504 gains entry to the nematode uterus via an external route and further suggested that mere contact with TAN 31504 may be sufficient for adult worms to become infected.
Uterine infection contributes to increased mortality upon constant exposure to TAN 31504.
In addition to accumulation in the uterus, TAN 31504 also accumulated to a minor extent within the intestinal lumen of wild-type adults (Fig. 2A and see Fig. S2A and S2C in the supplemental material). On the other hand, large amounts of TAN 31504 accumulated in the intestine of lin-10(n1390) adults (Fig. 2A and see Fig. S2B in the supplemental material), causing severe distention of the intestinal lumen. Similar expansion of the intestinal lumen was not observed in wild-type worms, although some distention did occur. To determine whether infection of the uterus or the intestine was the primary mode of TAN 31504-mediated pathogenesis, we compared survival of wild-type and lin-10(n1390) adults upon constant exposure to TAN 31504. The vulvaless strain did not develop a uterine infection (Fig. 2A) and survived 1.7-fold longer than the wild-type strain (Fig. 2B). Resistance of the lin-10(n1390) strain to TAN 31504 pathogenesis was not due to a general increase in resistance to pathogenic challenge, as lin-10(n1390) worms were more susceptible than the wild type to a gram-positive intestinal pathogen, Enterococcus faecalis V583 (Fig. 2B). Neither did lin-10(n1390) worms exhibit an increased life span relative to wild-type worms on the standard food source [log rank, P = 0.89; mean lethal time (LTmean) = 203.0 ± 3.4 h, lin-10(n1390); 207.0 ± 8.9 h, N2]. Another vulvaless strain, lin-2(e1309), also survived longer on TAN 31504 and, like lin-10(n1390), did not develop a uterine infection (data not shown). These findings suggest that the absence of a vulval opening and uterine infection in the vulvaless mutants was the underlying basis for their increased life span compared to the wild type upon constant exposure to TAN 31504. Furthermore, the data indicated that the virulence exerted by TAN 31504 on or from within the uterus was the primary contributor to the wild-type worms' demise.
The above conclusion was further supported by a strong positive correlation between the development of a TAN 31504-mediated uterine infection and an enhanced host mortality rate in experiments involving different growth media and temperatures (see Fig. S3A to D in the supplemental material). Ninety-five percent of adult wild-type worms exposed to TAN 31504 lawns generated on LB medium developed visible uterine infections prior to death. In contrast, fewer than 10% of worms fed on NGM lawns of TAN 31504 developed uterine infections and their mean life span was marginally longer (1.14-fold) than those fed the standard food source (see Fig. S3B in the supplemental material).
We investigated the temperature dependence of TAN 31504 pathogenicity on C. elegans. Uterine infection was observed at 25°C, 20°C, and 15°C (data not shown). An increased host mortality rate also accompanied the development of a uterine infection at the lower temperatures (see Fig. S3A, C, and D in the supplemental material). The ability to infect the nematode uterus and reduce the host life span was not a general characteristic of L. chromiireducens. L. chromiireducens subsp. chromiireducens strain LMG 22506, the closest phylogenetic relative and only subspecies partner of TAN 31504 described to date (28, 29), had a negligible affect on host life span (Fig. 3E, and see Fig. S3A to D in the supplemental material) and did not cause uterine infections in the wild-type worms assessed (Fig. 3B). These results further substantiated the correlation of increased host mortality with uterine infection. These findings also indicated that TAN 31504 possesses a relatively robust virulence potential for C. elegans.
Limited exposure of C. elegans to TAN 31504 allows for considerable pathogenicity.
We next asked if TAN 31504 was able to infect and kill worms after a limited exposure by introducing adult worms to TAN 31504 for brief time periods and then shifting them to Escherichia coli OP50-1 grown on NGM agar, the standard growth condition for maintaining C. elegans; for the rest of the article, this condition is referred to as the “standard food source.” We then monitored the development of uterine infection and compared the life spans of these worms to those feeding continuously on TAN 31504 (Fig. 3A to F and see Fig. S2C and D in the supplemental material). Worms feeding on the standard food source and on the relatively benign strain LMG 22506 served as controls (Fig. 3A, B, and E). Neither OP50-1 (Fig. 3A) nor LMG 22506 (Fig. 3B) accumulated in the uterine lumen of worms. In contrast, TAN 31504 was able to establish a persistent uterine infection in worms exposed to pathogen continuously (Fig. 3C and see Fig. S2C in the supplemental material), for 12 to 24 h (Fig. 3D and see Fig. S2D in the supplemental material), and for as few as 4 h (data not shown). The worms exposed for 12 to 24 h died more quickly than worms that were continuously exposed to TAN 31504 (Fig. 3E). The notable and unexpected increase in susceptibility of the briefly exposed hosts is addressed in the Discussion section.
In a second approach, host survival on mixed lawns containing various ratios of TAN 31504 and OP50-1 was compared to that on the pure TAN 31504 and pure OP50-1 lawns (Fig. 3F). The lower the proportion of TAN 31504 in the mixed lawn producing the same effect on worm life span as the pure TAN 31504 lawn, the greater the virulence potential of this pathogen. A lawn composed of as little as 3% TAN 31504 was sufficient to cause an extensive uterine infection and resulted in a markedly shorter host life span compared to either the pure TAN 31504 or pure OP50-1 lawns (Fig. 3F). This result was also unexpected and is addressed in the Discussion section. Both sets of experiments highlight the ability of TAN 31504 to readily colonize and infect C. elegans.
We also tested the idea that preexposure of the host to LMG 22506, a comparatively avirulent conspecific, could prime host resistance against TAN 31504 pathogenesis. Exposure of C. elegans to LMG 22506 for a 24-h period prior to the introduction of TAN 31504 did not prevent or delay the development of a uterine infection once exposure to TAN 31504 commenced (data not shown). Nearly all of the preexposed worms developed a visual uterine infection prior to death. Similarly, initial exposure to LMG 22506 did not provide significant survival benefit. The mean time to death of worms preexposed to LMG 22506 was not much different from that of worms exposed solely to TAN 31504 (Fig. 3E).
Naïve C. elegans L1 larvae prefer LMG 22506 or OP50-1 over TAN 31504.
C. elegans prefers food with higher nutritional value. When given a choice between bacterial lawns of different food qualities, naïve C. elegans larvae predominately seek out and remain within the food source lawn that is of higher quality and better supports growth (39). To gain insight into the interactions of young worms with TAN 31504, we performed unbiased- and biased-food-choice tests on L1 larvae that had no prior experience with food (Fig. 4A to F and H) and assessed their developmental growth rate in the presence of the pathogen generated on LB medium supplemented with cholesterol (Fig. 5A). In the unbiased-food-choice experiments, naïve L1 larvae were placed equidistant from the two food source lawns (Fig. 4G, top). In the biased-food-choice tests, the naïve L1 larvae were placed in a location that guaranteed experience with one of the food sources prior to the other (Fig. 4G, bottom). Unbiased-food-choice assessments were expected to reveal any chemoattraction C. elegans displayed for TAN 31504 and whether or not the young worms would choose to feed on the pathogen. As implied above, C. elegans does not exhibit a strong leaving behavior when experiencing a high-quality food source. The leaving behavior displayed during unbiased- and biased-food-choice tests was anticipated to reveal the degree of preference C. elegans had for the food sources assessed, as well as indicate the relative nutritional value of TAN 31504 in comparison to the other food sources tested. The biased-food-choice assessments were expected to further reveal the preference, if any, the worms exhibited for TAN 31504.
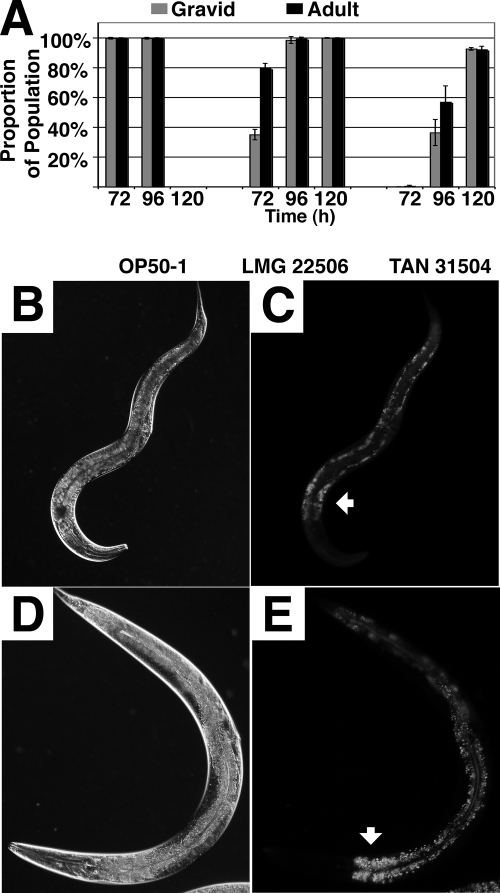
FIG. 5.
C. elegans worms develop at a reduced rate, achieve a thinner body size, and accumulate decreased intestinal fat deposits on TAN 31504 food source lawns. (A) Developmental progress of worms raised from eggs at 25°C on lawns of E. coli OP50-1, L. chromiireducens subsp. chromiireducens LMG 22506, and L. chromiireducens subsp. solipictus TAN 31504 generated on LB medium supplemented with cholesterol. Each growth condition (food source) assessed is listed on the x axis below the corresponding set of data. The percentage of worms that reached adulthood relative to the total number of worms assessed for the particular condition (black) and the percentage of gravid adults among that population (gray) are shown on the y axis for each time point. A developmental delay was observed for worms reared on LMG 22506 and TAN 31504 compared to OP50-1, with the most extensive delay in development occurring on TAN 31504. Only on OP50-1 did a majority (97%) of the worm population reach adulthood by 48 h (not shown). (B and D) DIC micrographs of age-matched young adult worms reared from eggs at 25°C in the presence of cholesterol on Nile red-containing TAN 31504 (B) and Nile red-containing standard food source lawns (D) (×200 magnification). The head of the worm is positioned down, and the vulva is located on the right exterior side of the mid-body region. The images are representative of the overall thinner body size TAN 31504-reared worms displayed compared to the average body size achieved by young adult worms raised on the standard food source. Magnified images of these worms are found in Fig. S4 in the supplemental material. (C and E) Corresponding N3 filter fluorescence micrographs of images shown in panels B and D, respectively. The white arrowhead points to the anterior region of the intestine. The fluorescence shows the Nile red-stained fat deposits of the worm intestinal cells. The images are representative of the general decrease in intestinal fat stores observed in worms raised on TAN 31504 compared to those accumulated by worms reared on the standard food source.
Naïve L1 C. elegans worms did not preferentially seek out TAN 31504 or LMG 22506 lawns over OP50-1 lawns in unbiased-food-choice assays (Fig. 4E and F). In these assays, worms left the Leucobacter lawns in greater numbers and for longer periods than they did from OP50-1 lawns (Fig. 4A to F). These behaviors suggested that TAN 31504 and LMG 22506 did not match the higher nutrient value of the OP50-1 lawn.
A strong preference for OP50-1 over the Leucobacter food sources was further supported from the behavior of worms in biased-food-choice assays. C. elegans larvae left their original TAN 31504 or LMG 22506 food sources at high frequency to seek out the available OP50-1 lawn (Fig. 4H). In both the unbiased- and biased-food-choice assessments, young worms rarely moved off of OP50-1 lawns once contact with this food source was established (Fig. 4A, E, F, and H). In contrast, a more pronounced leaving behavior was exhibited by the young worms after initial experience with the TAN 31504 and LMG 22506 lawns (Fig. 4B to F and H). Young worms did, however, display a slight preference for LMG 22506 over TAN 31504, suggesting that LMG 22506 was more nutritious for developing worms (Fig. 4D and H).
C. elegans hatchlings can utilize TAN 31504 as the sole food source.
Although young worms did not prefer TAN 31504 or LMG 22506 over OP50-1 lawns generated on LB medium supplemented with cholesterol, they could develop into adults on both Leucobacter strains, albeit at a slower rate than those reared on OP50-1 (Fig. 5A). Within 48 h after hatching, 97% of worms feeding on OP50-1 reached adulthood. In contrast, most worms feeding on LMG 22506 or TAN 31504 required 72 h and 96 h, respectively, to develop into adults (Fig. 5A). Adults developed on TAN 31504 lawns were thinner and accumulated markedly decreased amounts of intestinal fat stores compared to worms grown on the standard food source (Fig. 5B to E). These findings further substantiate the poorer food quality of the Leucobacter lawns suggested by the food choice assays described above (Fig. 4). The data also supported the idea that TAN 31504 provides less nutrition to developing worms than LMG 22506. Alternatively, bacterial virulence may contribute to the further delay in nematode development exhibited on TAN 31504 compared to LMG 22506.
LMG 22506-reared worms also developed into comparatively thin adults (data not shown). However, worms could persist for generations on the LMG 22506 food source, whereas they could not on TAN 31504. On average, 40% and 80% of adults reared on TAN 31504 and LMG 22506 lawns, respectively, yielded progeny (approximately 50 individual worms were assessed for each food source). Over the course of three generations, adults developed on TAN 31504 and LMG 22506 produced an average of 0.41 ± 0.16 and 2.61 ± 1.00 viable progeny, respectively (approximately 130 worms of multiple populations were assessed for each food source). Worms reared on the standard food source produced more than 160 progeny per gravid adult. Although progeny production on LMG 22506 was poor, it was sufficient for a sustainable population, whereas this was not the case on TAN 31504.
The cumulative results suggested that LMG 22506 exerted slight virulence on either larval or adult-stage C. elegans, whereas TAN 31504 was mostly pathogenic to adult C. elegans. This finding was in line with the idea that TAN 31504 infection of the uterus via the vulva was the primary contributor to the death of the host. The morphogenesis of the vulval tissue is such that the mature vulval opening to the uterus is not exposed to the external environment until adulthood (see Fig. 8B to I in reference 1a and see reference 22a). Besides the decreased virulence of LMG 22506 compared to TAN 31504, other differences between the conspecifics were observed. Unlike TAN 31504, LMG 22506 lacked expression of the light-inducible carotenoid pigment (28, 29). Carotenoids are known to protect invading microbes from reactive oxygen species commonly elicited during host defense responses. For example, Staphylococcus aureus expresses a series of carotenoids, including 4,4′-diaponeurosporene, that protect S. aureus from neutrophil-mediated killing (23, 25). The same pigment when heterologously expressed in Streptococcus pyogenes conferred increased virulence and resistance to singlet oxygen (23). We investigated the contribution of the yellow carotenoid produced by TAN 31504 in pathogenicity on C. elegans. Under predominately dark conditions (light was required periodically to monitor survival rates), dark-generated pigment-deficient TAN 31504 lawns actually enhanced the rate of host mortality by 13% to 15% relative to that observed on light-generated lawns (data not shown). Furthermore, carotenogenesis was not induced in TAN 31504 cells by the presence of 0.2 to 2.0 mM paraquat under dark growth conditions, although a reduction in growth rate was observed for bacteria exposed to the free radical producer (data not shown). These data indicate that the light-inducible carotenoid of TAN 31504 does not account for the pathogen's ability to inflict disease on C. elegans and also imply that TAN 31504 is more virulent to worms under dark conditions.
TAN 31504 forms a biofilm.
LMG 22506 and TAN 31504 lawns were notably different in both their appearance and biofilm-like characteristics. One-day-old TAN 31504 lawns were extremely hydrophobic (see Fig. S5A and C in the supplemental material), sticky, and exceedingly resistant to shear force stress. In contrast, 1-day-old LMG 22506 lawns were not particularly hydrophobic (see Fig. S5A and B in the supplemental material) and could be easily and quickly dissociated in water (see Fig. S5D in the supplemental material). At 25°C, TAN 31504 lawns formed a hardened, gel-like film within 12 to 24 h of inoculation and became progressively more rigid over time. This behavior appeared to reflect a response to the increased desiccation of the medium plate, as lawns on plates protected from rapid drying remained pliable for a longer period. TAN 31504 lawns would eventually fragment underwater (see Fig. S5E and F in the supplemental material). However, the lawn fragments retained a film-like quality for an extended period, and could be lifted away as flakes of various sizes from the medium substrate by moderate agitation (see Fig. S5F in the supplemental material). In contrast, LMG 22506 lawns did not produce stable flakes or aggregates when submerged (data not shown) and the bacteria within the submerged lawns were dispersed easily by gentle mixing (see Fig. S5D in the supplemental material). The differences in lawn quality between TAN 31504 and LMG 22506 could not be attributed to a faster growth rate and correspondingly more advanced biofilm formation for the TAN 31504 lawns, since LMG 22506 grows more quickly than TAN 31504 on LB agar (29).
Exposure to TAN 31504 elicits a change in expression of a subset of C. elegans innate immunity-related genes.
TAN 31504 colonizes worms and establishes a persistent infection within 12 h (see Fig. S2D in the supplemental material). We wondered if C. elegans responds to the initial stages of TAN 31504 infection with a measurable defense response. A panel of infection-responsive genes known to be associated with the C. elegans innate immune response to bacteria and fungal infections and to various other environmental stresses was assembled and used to assess the transcriptional profile of TAN 31504-exposed worms by qRT-PCR (Fig. 6A) (S. S. Slutz, E. A. Evans, and M.-W. Tan, unpublished data). Expression of the majority of the immunity genes included within the panel has been shown to change significantly during exposure to P. aeruginosa strain PA14 (37, 44).
We compared the transcript levels of 91 genes from age-matched fertile adult worms after 12 h of exposure to TAN 31504, PA14, SL1344, or the standard food source. Expression levels of roughly half of the genes assessed were significantly different during host exposure to one or more of the pathogenic conditions compared to those for worms maintained on the standard food source (Fig. 6A and see Table S1 in the supplemental material). Interestingly, only three genes responded in a significant fashion during host exposure to SL1344 (see Table S1 in the supplemental material). This is in contrast to the 34 and 26 significant alterations in C. elegans gene expression exhibited during challenge with PA14 and TAN 31504, respectively (see Table S1 in the supplemental material). Analysis of the transcription profiles using the Cluster 3.0 average linkage algorithm revealed that the expression profile obtained from TAN 31504-exposed worms was most similar to that of PA14-challenged worms. Half of the TAN 31504-responsive genes shared similar patterns of gene expression in the PA14-generated profile. The remaining 13 gene expression patterns distinct to the TAN 31504-exposed worms included seven genes that were induced and six genes that were downregulated. Details pertaining to the distinct TAN 31504-responsive genes are presented in the Discussion section.
Interestingly, the upregulation of nlp-29, an infection-regulated gene associated with the C. elegans response to the fungal pathogen D. coniospora, was included within the group of genes exhibiting increased transcript levels distinctly during host exposure to TAN 31504 (Fig. 6A) (9, 46). D. coniospora interacts with the nematode vulva, and strong expression of nlp-29 in perivulval cells of C. elegans responding to early events of D. coniospora pathogenesis has been shown (9, 19). The nlp-29 gene encodes a neuropeptide-like protein that is expressed in the nematode hypodermis and intestine and is thought to encode an antimicrobial peptide that is induced in response to both fungal infection and physical wounding of worm hypodermal tissue (30, 34, 46).
The nlp-29 transcripts reached a fourfold-higher level in TAN 31504-exposed worms compared to those in worms fed the standard food source (Fig. 6A). The transcript abundance of nlp-29 was also slightly elevated, though not significantly (see Table S1 in the supplemental material), in SL1344-challenged hosts, achieving a 1.3-fold-higher level than that of worms maintained on the standard food source (Fig. 6A). Osmotic stress can induce nlp-29 expression (46). For that reason, we considered if the increased expression of nlp-29 in TAN 31504- and SL1344-exposed hosts could be attributed to the higher osmotic strength of the LB agar, on which these hosts were exposed, relative to the NGM agar of the standard food source lawns. However, the SL1344 lawns were prepared under slightly higher osmotic conditions than the TAN 31504 lawns (see Materials and Methods), and yet the nlp-29 transcript level of SL1344-exposed hosts was significantly lower than that of TAN 31504-exposed hosts (see Table S1 in the supplemental material). It is therefore likely that factors other than the osmotic strength of the media contributed to the induction of nlp-29 expression observed during C. elegans exposure to TAN 31504.
To further investigate the TAN 31504-mediated increase in nlp-29 expression, we employed a pnlp-29::gfp reporter strain. Expression of the reporter gene was observed to occur in adult worms maintained on the standard food source (Fig. 6D and G), a result anticipated from previous findings (9, 30). A slight increase in expression of the pnlp-29::gfp reporter was evident in adult reporter worms subjected to OP50-1 LB conditions (Fig. 6C and F) over that observed in the reporter worms exposed solely to standard-food-source conditions (Fig. 6D and G), presumably due to the increase in osmotic strength of the LB agar. However, the data do not exclude the possibility of the induced pnlp-29::gfp expression resulting from the increased virulence characteristic of OP50-1 on LB agar at 25°C (see Fig. S3A in the supplemental material). In line with the pattern of nlp-29 gene expression described above, increased expression of the pnlp-29::gfp reporter was seen to occur throughout the hypodermal and intestinal tissues of adult TAN 31504-infected worms (Fig. 6B and E) compared to worms exposed solely to the standard food source (Fig. 6D and G). The increase in reporter gene expression could not be attributed simply to the change in osmotic conditions accompanying the movement of the adult reporter worms from NGM agar to LB agar, as TAN 31504-exposed worms accumulated higher levels of the GFP reporter than did worms exposed to OP50-1 LB lawns for the same 24-h period.
DISCUSSION
We have presented evidence for a pathogenic interaction between L. chromiireducens subsp. solipictus TAN 31504 and C. elegans. TAN 31504 colonizes the nematode uterus through the vulval opening (Fig. 1 to 3 and see Fig. S1 in the supplemental material), a mode of infection that has not been described for other bacterial pathogens of C. elegans. TAN 31504-mediated infection of the uteri of adult worms is the primary cause of early mortality, as adult vulvaless lin-10(n1390) worms did not develop uterine infections and die at a slower rate during TAN 31504 exposure (Fig. 2 and see Fig. S2 in the supplemental material).
TAN 31504 also exerts a low level of virulence on the nematode intestine. Infected worms exhibited moderate distention of the intestinal lumen that was associated with the presence of the yellow-pigmented pathogen (Fig. 2 and 3 and see Fig. S2 in the supplemental material). Intact bacteria were present within the intestinal lumen after 3 days of exposure to the pathogen (data not shown). Interestingly, vulvaless lin-10(n1390) adults, despite showing a greater intestinal distension and accumulation of TAN 31504 after 5 days of exposure, continued to survive for an average of an additional 5 days (Fig. 2). By comparison, nearly half of the wild-type adults were dead after 5 days of exposure to TAN 31504. The remaining population typically exhibited a severe uterine infection with only mild to moderate intestinal distention (see Fig. S2 in the supplemental material). A majority of these survivors were dead the following day (Fig. 2). Together, these data suggested that the intestinal infection with TAN 31504 made only a marginal contribution to the host life span. We hypothesized that the increased intestinal distress observed for the TAN 31504-infected lin-10(n1390) worms may have resulted from the loss of a Lin-10-dependent immune function and was not a general characteristic of vulvaless strains, as lin-2(e1309) worms did not exhibit this rapid intestinal accumulation of TAN 31504 (data not shown). Consistent with this idea, adult lin-10(n1390) worms were significantly more susceptible than the wild-type strain to intestinal pathogens, such as Enterococcus faecalis V583 (Fig. 2), P. aeruginosa PA14, S. enterica serovar Typhimurium SL1344, and S. aureus 8325-4 (data not shown).
An unanticipated finding was that worms feeding on mixtures of OP50-1 and TAN 31504 or worms exposed as adults to TAN 31504 for brief periods displayed more rapid mortality rates and more severe uterine infections than worms exposed to pure lawns of TAN 31504 (Fig. 3 and see Fig. S2 in the supplemental material). Since OP50-1 is pathogenic to C. elegans when grown on LB agar, as opposed to being a benign food source when generated on NGM (see Fig. S3 in the supplemental material), the decrease in host life span might be a combinatorial effect of both pathogens. We also speculated that the extremely large uterine infection observed in these worms might contain the more virulent, LB medium-generated OP50-1 in addition to TAN 31504. However, these hypotheses could not account for the results of the brief exposure survival assays, since in these experiments worms exposed to TAN 31504 for 12 h and 24 h were maintained on OP50-1 NGM lawns (the benign standard food source) after pathogen challenge. Under this condition, TAN 31504-infected worms developed uterine infections that were as extensive as those produced in worms feeding on the LB medium-generated mixed lawns and exhibited similarly reduced life spans (Fig. 3).
Our results did not support the idea that TAN 31504 was more virulent in the presence of OP50-1, since worms exposed to increasing amounts of TAN 31504 in mixed lawns displayed progressively longer survival times (Fig. 3). In addition, TAN 31504 did not grow better on NGM harboring OP50-1 than on NGM alone (data not shown). This suggested that the increased virulence of TAN 31504 in the brief-exposure experiments did not result from direct utilization of OP50-1 as an additional food source by the pathogen. On the other hand, the rapid progression of TAN 31504 infections in the host could be an indirect effect of better host nutrition. In line with this idea, worms exposed to TAN 31504 for 12 h and 24 h that were then moved to a standard food source exhibited notably better intestinal health than worms maintained on the pathogen (Fig. 3 and see Fig. S2 in the supplemental material), as did worms feeding on the mixed lawns of 3% and 6% TAN 31504 (data not shown). Worms with uncompromised intestines appeared to be more susceptible to TAN 31504-mediated infection of the uterus (Fig. 3 and see Fig. S2 in the supplemental material). We hypothesized that the TAN 31504 bacteria accumulating in the uterus of otherwise healthy worms feeding on OP50-1 might have access to an abundant supply of nutrients provided by the worm tissues. In contrast, adult worms exposed to pure lawns of TAN 31504 for extended periods were feeding on lower-quality food and displayed signs of mild to moderate intestinal distress, which included slight distention of the intestinal lumen and decreased pigmentation of the intestinal cells, reflecting the reduced abundance of intestinal fat stores (Fig. 3 and 5 and see Fig. S2 and S4 in the supplemental material) (26). It seems likely that these hosts provided less nutrition to the TAN 31504 bacteria infecting the nematode uterus. Such a scenario could reasonably account for the relative decrease in pathogen virulence observed during continuous exposure of C. elegans to pure TAN 31504 lawns over the limited-exposure conditions. This appears to be an unusual example of a case in which the increased health of the host is actually detrimental during infection.
As mentioned previously, Glp worms were used in a number of the infection and survival analyses reported to allow clear visualization of the TAN 31504-mediated infection of the worm and to eliminate the complication of matricide common to fertile worms (Fig. 1 to 3 and see Fig. S1 to S3 in the supplemental material). Despite the fact that Glp [cdc-25.1(RNAi)] worms possess an abnormal uterine environment and survive twice as long as fertile worms on the standard food source, a somewhat enhanced rate of death for and the development of similar uterine infections in fertile worms exposed to TAN 31504 were observed (2, 38; data not shown). It remains possible that the infection processes occurring in fertile gravid worms may differ from those occurring in Glp adults. However, from our observations it appears that significant aspects of the TAN 31504-mediated infection of C. elegans are likely common to both fertile and Glp backgrounds.
The ability of TAN 31504 to efficiently infect and kill adult worms at low temperatures, within brief periods of host exposure, and at low titer indicated a significant virulence potential of TAN 31504 for adult C. elegans (Fig. 2 and 3 and see Fig. S3 in the supplemental material). The data obtained from food choice tests, developmental growth rate assessments, and fat accumulation of young worms feeding on TAN 31504 showed that the pathogen could adversely affect the behavior and physiology of immature worms (Fig. 4 and 5 and see Fig. S4 in the supplemental material). However, the data also indicated that TAN 31504 could suffice as a food source for the development of young worms into adults. Although TAN 31504 seemed to exert the most detrimental effects on adult hosts, TAN 31504 was observed to be harmful to young worms as well. A small percentage of hatchlings fed TAN 31504 as the sole food source did not develop beyond the first or second larval stages (data not shown). In addition, a number of developing worms reared on TAN 31504 exhibited a slight Dar phenotype that was lost by the time the worms reached adulthood (data not shown). Interestingly, these phenomena resemble the effects the related nematode-specific pathogen “M. nematophilum” exerts on other Caenorhabditis species, with the weak Dar phenotype of surviving worms specifically occurring in Caenorhabditis briggsae strain AF16 (1). TAN 31504 virulence toward young worms, though not as robust as that wielded on adult hosts, was not inconsequential.
Worms infected with TAN 31504 produced drastically fewer progeny. Although C. elegans could reproduce to some extent on lawns of TAN 31504, the number of individuals in consecutive generations declined by more than 50%. Adults developed on TAN 31504 laid a small percentage of unfertilized oocytes, and, on average, 30 times fewer embryos than worms grown on the standard food source. Embryos from TAN 31504-reared adults produced nonviable progeny that arrested between the two- and threefold stage of embryogenesis (data not shown). The uterine infection could contribute to the decreased numbers of viable progeny. Most TAN 31504-infected worms succumbed to infection early in adulthood and were afforded a relatively small window of progeny production. The low quality of food provided by TAN 31504 lawns could be another contributor to reduced progeny production. The precise events contributing to the arrest of the developing embryos remain to be determined.
The ability of TAN 31504 to form a stable biofilm may contribute to its virulence on the nematode host. We have found that 12 h and as little as 4 h of exposure to TAN 31504 is sufficient for worms to develop a lethal uterine infection and that colonization of the exterior vulval cuticle precedes the vulva-uterus infection of these briefly exposed worms (Fig. 3) (R. E. Muir and M.-W. Tan, unpublished data). It is speculated that the nonmotile pathogen ultimately gains access to the worm uterus through expansive growth within a biofilm, which nucleates from the external site of initial colonization. The pathogenicity exhibited by TAN 31504 was found not to be a general trait of the L. chromiireducens species. LMG 22506 did not form a robust biofilm on a solid LB medium substrate (see Fig. S5 in the supplemental material) and exerted relatively mild virulence on C. elegans (Fig. 3 and 5 and see Fig. S3 in the supplemental material). Furthermore, LMG 22506 accumulation was not detected on the vulval cuticle of exposed worms (R. E. Muir and M.-W. Tan, unpublished data). Preliminary findings showed that the extracellular polysaccharide (EPS)-associated material that was precipitated from TAN 31504 lawns contained more nucleic acid and protein than LMG 22506 lawns (data not shown). Nucleic acid and protein together with EPS and other organic material are all well-characterized components of the general bacterial biofilm matrix (6). TAN 31504 produced copious extracellular material both in liquid culture and on solid media, though the identity of the material as EPS has not been determined (29). The capacity to form biofilms may account for the skewed virulence potential between the two L. chromiireducens subspecies partners.
The results obtained from the qRT-PCR assessments of pathogen-induced gene expression were taken primarily as an indication that C. elegans exposure to TAN 31504 results in a host innate immune response (Fig. 6). A shared role in the C. elegans innate immune response elicited by both TAN 31504 and PA14 is suggested for a number of the infection-responsive genes in the panel assessed (Fig. 6 and see Table S1 in the supplemental material). Of the 46 C. elegans genes found to respond in a significantly different fashion during exposure to one or more of the pathogens relative to the standard growth condition, the expression patterns of 13 genes were distinct from the profile generated from TAN 31504-exposed worms (Fig. 6). Of these genes, the immunity-regulated expression of fmo-2 (flavin-containing monoxygenase), C23G10.1 (gene of unknown function), spp-12 (member of the saposin-like protein family), and clec-97 (C-type lectin) had not been described in association with the C. elegans innate immune response to either bacteria or fungal pathogens previously (gene descriptions derived from WormBase release WS188; http://www.wormbase.org). As well, the TAN 31504-evoked downregulation of lys-1 and lys-2 differed from the induced expression of the two lysozyme-encoding genes seen during host exposure to PA14 (Fig. 6) (37, 44). Reduced transcript levels were produced from 6 of the 13 TAN 31504-specific genes. Interestingly, five of these genes are included among the group of new/newly regulated immunity genes mentioned. The remaining gene that exhibited a significant reduction in transcript accumulation during C. elegans exposure to TAN 31504 was cnc-2, which encodes a caenacin peptide (9). The reduced expression of cnc-2 was also observed during the C. elegans response to at least two other bacterial pathogens (46).
A majority of the TAN 31504-induced genes have also been shown to respond positively to a number of other microbial infections, including gram-positive and -negative bacteria and fungal pathogens (9, 24, 32, 37, 44, 46). In addition to fmo-2, the only other predominately intestinally expressed genes upregulated specifically during TAN 31504 exposure were K05F1.10, which encodes a putative secreted TIL domain protease inhibitor (WormBase release WS188; http://www.wormbase.org), and spp-1, which encodes a member of the saposin-like protein superfamily (4). Although the induction of K05F1.10 was not seen here for PA14-challenged worms, increased expression of the gene in response to PA14 infection has been reported (37, 44). The remaining C. elegans genes up-regulated distinctly during TAN 31504 exposure were immunity genes associated with nonintestinal tissues, including the hypodermis, excretory gland, and some neuronal tissues. These genes included a putative as well as a demonstrated antimicrobial peptide (nlp-29 [9] and abf-2 [21], respectively), a secreted surface protein (T20G5.8), and a mitogen-activated protein kinase (mpk-2) (WormBase release WS188; http://www.wormbase.org).
The transcription profile generated from TAN 31504-infected worms indicated that exposure to the pathogen can alter gene expression throughout the entire body of the host (Fig. 6). The broad alteration in host gene expression induced by TAN 31504 exposure likely reflects a systemic innate immune response. It is expected that a more in-depth analysis of the C. elegans whole-genome gene expression profile generated from TAN 31504-challenged worms will provide insight into novel aspects of C. elegans innate immunity. Such studies are hoped to uncover not only nematode defense responses that are specifically directed against the TAN 31504 infection, but also more general responses that may be applicable to other microbial pathogens.
The putative antimicrobial gene nlp-29 was one of the seven C. elegans innate immunity genes found to be upregulated distinctly in TAN 31504-infected worms (Fig. 6). Whereas wild-type and lin-10(n1390) worms exposed for 18 h to the relatively benign conspecific LMG 22506 expressed nlp-29 at comparable levels (2.1 and 4.1 average arbitrary transcript values, respectively; see Table S2 in the supplemental material), TAN 31504-exposed wild-type worms induced eightfold-higher nlp-29 expression than LMG 22506-exposed wild-type worms (average arbitrary transcript values, 16.4 versus 2.1; see Table S2 in the supplemental material). In contrast, vulvaless lin-10(n1390) worms exhibited only 1.3-fold nlp-29 induction (average arbitrary transcript values, 5.4 versus 4.1; see Table S2 in the supplemental material). The lower induction of nlp-29 was not specific to the lin-10(n1390) mutation, since a similar 1.5-fold increase in nlp-29 expression was observed in vulvaless lin-2(e1309) worms (data not shown). These findings, together with the data presented in the dendrogram (Fig. 6), support the idea that wild-type worms are responding specifically to infection by TAN 31504 during the first 12 to 18 h of exposure and that the defense response is not elicited to the same degree in vulvaless worms. It is tempting to speculate that the higher induction of nlp-29 is a component of the C. elegans innate immune response directed against TAN 31504 infection of the nematode vulva and uterus.
The induction of nlp-29 and other neuropeptide-like protein-encoding genes was first observed during the C. elegans response to the fungal pathogen D. coniospora (9, 46). More recently, upregulation of nlp-29 was demonstrated in response to wounding of the C. elegans major hypodermal syncytium (34). The use of a pnlp-29::gfp reporter strain revealed that the infection-responsive gene was expressed strongly in the perivulval cells of worms on which spores of D. coniospora had attached, as well as being highly induced in the hypodermal tissues of infected worms (9). Broad and rapid induction of pnlp-29::gfp was also seen in hypodermally wounded worms after insult with a femotosecond exposure to a laser or by needle prick (34). Using the same pnlp-29::gfp reporter strain, we found increased expression of the pnlp-29::gfp reporter to be evident throughout the intestine and hypodermis of TAN 31504-infected worms after the first 24 h of exposure (Fig. 6). High levels of the GFP reporter were observed in the anterior region of the intestine and around the head of TAN 31504-exposed worms, with many worms exhibiting an intense GFP signal near the terminal region of the pharynx (Fig. 6). These data again hint at a role for nlp-29 in the C. elegans innate immune response to bacteria as well as fungal pathogens, in addition to its function induced by physical injury.
We have shown that TAN 31504 exhibits significant virulence on C. elegans at temperatures ranging from 15 to 25°C (see Fig S3 in the supplemental material). TAN 31504 infection of the nematode uterus occurs via an external non-oral route (Fig. 1 and 2 and see Fig. S1 and S2 in the supplemental material). TAN 31504 can form a biofilm that is expected to function in adhesion of the bacteria to each other and to accessible substrates as well as in protection from dehydration (see Fig. S5 in the supplemental material). Naïve L1 C. elegans worms will initially seek out a TAN 31504 food source (Fig. 4). TAN 31504 is a sufficient food source for larval development through adulthood (Fig. 5). C. elegans exposure to LMG 22506, the closest relative and only known subspecies partner of TAN 31504, prior to encounter with TAN 31504 does not appear to prime host resistance, as the worms remain susceptible to TAN 31504-mediated infection (Fig. 3). Lastly, worms exposed for limited periods to TAN 31504 and to bacterial food sources consisting of smaller amounts of TAN 31504 inevitably develop fatal uterine infections and exhibit a faster mortality rate than control populations (Fig. 3 and see Fig. S2 in the supplemental material). These are ideal characteristics of a bacterium that benefits from an association with a soil-dwelling nematode, such as C. elegans.
Although the natural habitat of TAN 31504 remains to be determined, the chemotaxonomic and biochemical characteristics of the strain indicate that it is likely a saprophytic soil bacterium that grows best in moist, low-light environments at temperatures of 4°C to <25°C (29). It is therefore possible that TAN 31504 may be found in soils similar to those harboring C. elegans and other nematodes. The Microbacteriaceae family, of which TAN 31504 is a member, includes numerous species known to associate with soil, plants, and nematodes (12). Several members within this family have been identified as secondary inhabitants of plant galls induced by nematodes. Some phytopathogenic species are known to use nematodes as vectors in order to gain entry into their respective plant hosts. A relevant feature of some nematode-associated species is their role in providing a food source for larval development within the plant gall. Furthermore, these bacteria have been suggested to play a part in protecting the developing nematodes from other harmful microbes. When considering the natural soil environments of C. elegans and potentially of TAN 31504, it is intriguing to consider that host-pathogen interactions between these two organisms may be displayed outside of the laboratory.
Is TAN 31504 a broad-host-range pathogen with the potential to be used as a biological control agent against plant-pathogenic nematodes? Is TAN 31504 an opportunistic plant pathogen that aids in the life cycle of infective nematodes? It will be interesting in future work to investigate the potential interactions of TAN 31504 with other agriculturally relevant nematodes to answer such questions. Furthermore, with the use of the tractable nematode host C. elegans, we can begin to investigate characteristics of the host-pathogen interaction shared between the nematode and TAN 31504 that include those ranging from the role of biofilm formation in TAN 31504-mediated pathogenesis to the functions served by novel aspects of the C. elegans innate immune response.
Supplementary Material
Acknowledgments
This work was supported by grant R01 GM66269 from the National Institutes of Health. R.E.M. was supported, in part, by Stanford Genome Training Program grant T32 HG00044 from the National Human Genome Research Institute and by Microbiology and Immunology Postdoctoral Training Program grant NRSA 5T32 A107328 from the National Institutes of Health.
We thank Jonathan J. Ewbank for providing us with the pnlp-29::gfp reporter strain. We are grateful to Marianne C. Campbell for performing some replicate qRT-PCR experiments, Sandra S. Slutz and Eric A. Evans for the construction of the infection-responsive panel of qRT-PCR primers, and Eric A. Evans for assistance with statistical analyses. We also thank Jeffrey L. Moseley for critical reading of the manuscript.
Footnotes
Published ahead of print on 16 May 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akimkina, T., K. Yook, S. Curnock, and J. Hodgkin. 2006. Genome characterization, analysis of virulence and transformation of Microbacterium nematophilum, a coryneform pathogen of the nematode Caenorhabditis elegans. FEMS Microbiol. Lett. 264:145-151. [DOI] [PubMed] [Google Scholar]
- 1a.Altun, Z. F., and D. H. Hall. 2005, posting date. Introduction to C. elegans anatomy. In Z. F. Altun and D. H. Hall (ed.), Handbook of C. elegans anatomy. WormAtlas. http://www.wormatlas.org/handbook/contents.htm.
- 2.Ashcroft, N., and A. Golden. 2002. CDC-25.1 regulates germline proliferation in Caenorhabditis elegans. Genesis 33:1-7. [DOI] [PubMed] [Google Scholar]
- 3.Ashrafi, K., F. Y. Chang, J. L. Watts, A. G. Fraser, R. S. Kamath, J. Ahringer, and G. Ruvkun. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421:268-272. [DOI] [PubMed] [Google Scholar]
- 4.Banyai, L., and L. Patthy. 1998. Amoebapore homologs of Caenorhabditis elegans. Biochim. Biophys. Acta 1429:259-264. [DOI] [PubMed] [Google Scholar]
- 5.Beanan, M. J., and S. Strome. 1992. Characterization of a germ-line proliferation mutation in C. elegans. Development 116:755-766. [DOI] [PubMed] [Google Scholar]
- 6.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 7.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cipollo, J. F., A. M. Awad, C. E. Costello, and C. B. Hirschberg. 2004. srf-3, a mutant of Caenorhabditis elegans, resistant to bacterial infection and to biofilm binding, is deficient in glycoconjugates. J. Biol. Chem. 279:52893-52903. [DOI] [PubMed] [Google Scholar]
- 9.Couillault, C., N. Pujol, J. Reboul, L. Sabatier, J. F. Guichou, Y. Kohara, and J. J. Ewbank. 2004. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5:488-494. [DOI] [PubMed] [Google Scholar]
- 10.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 11.Entchev, E. V., and T. V. Kurzchalia. 2005. Requirement of sterols in the life cycle of the nematode Caenorhabditis elegans. Semin. Cell Dev. Biol. 16:175-182. [DOI] [PubMed] [Google Scholar]
- 12.Evtushenko, L. I., and M. Taekeuchi. 2006. The family Microbacteriaceae, p. 1020-1098. In S. F. Martin Dworkin, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 3. Springer, Berlin, Germany. [Google Scholar]
- 13.Gravato-Nobre, M. J., and J. Hodgkin. 2005. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell. Microbiol. 7:741-751. [DOI] [PubMed] [Google Scholar]
- 14.Gravato-Nobre, M. J., H. R. Nicholas, R. Nijland, D. O'Rourke, D. E. Whittington, K. J. Yook, and J. Hodgkin. 2005. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 171:1033-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodgkin, J., P. E. Kuwabara, and B. Corneliussen. 2000. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr. Biol. 10:1615-1618. [DOI] [PubMed] [Google Scholar]
- 16.Hoflich, J., P. Berninsone, C. Gobel, M. J. Gravato-Nobre, B. J. Libby, C. Darby, S. M. Politz, J. Hodgkin, C. B. Hirschberg, and R. Baumeister. 2004. Loss of srf-3-encoded nucleotide sugar transporter activity in Caenorhabditis elegans alters surface antigenicity and prevents bacterial adherence. J. Biol. Chem. 279:30440-30448. [DOI] [PubMed] [Google Scholar]
- 17.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 18.Horvitz, H. R., and J. E. Sulston. 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96:435-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansson, H.-B. 1994. Adhesion of conidia of Drechmeria coniospora to Caenorhabditis elegans wild type and mutants. J. Nematol. 26:430-435. [PMC free article] [PubMed] [Google Scholar]
- 20.Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, D. P. Welchman, P. Zipperlen, and J. Ahringer. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231-237. [DOI] [PubMed] [Google Scholar]
- 21.Kato, Y., T. Aizawa, H. Hoshino, K. Kawano, K. Nitta, and H. Zhang. 2002. abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem. J. 361:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, S. K., and H. R. Horvitz. 1990. The Caenorhabditis elegans gene lin-10 is broadly expressed while required specifically for the determination of vulval cell fates. Genes Dev. 4:357-371. [DOI] [PubMed] [Google Scholar]
- 22a.Lints, R., and D. H. Hall. 2005, posting date. Reproductive system. IVa. Uterus, vulva. In Z. F. Altun and D. H. Hall (ed.), Handbook of C. elegans anatomy. WormAtlas. http://www.wormatlas.org/handbook/contents.htm.
- 23.Liu, G. Y., A. Essex, J. T. Buchanan, V. Datta, H. M. Hoffman, J. F. Bastian, J. Fierer, and V. Nizet. 2005. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallo, G. V., C. L. Kurz, C. Couillault, N. Pujol, S. Granjeaud, Y. Kohara, and J. J. Ewbank. 2002. Inducible antibacterial defense system in C. elegans. Curr. Biol. 12:1209-1214. [DOI] [PubMed] [Google Scholar]
- 25.Marshall, J. H., and G. J. Wilmoth. 1981. Pigments of Staphylococcus aureus, a series of triterpenoid carotenoids. J. Bacteriol. 147:900-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKay, R. M., J. P. McKay, L. Avery, and J. M. Graff. 2003. C. elegans: a model for exploring the genetics of fat storage. Dev. Cell 4:131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendoza De Gives, P. M., K. G. Davies, S. J. Clark, and J. M. Behnke. 1999. Predatory behaviour of trapping fungi against srf mutants of Caenorhabditis elegans and different plant and animal parasitic nematodes. Parasitology 119:95-104. [DOI] [PubMed] [Google Scholar]
- 28.Morais, P. V., R. Francisco, R. Branco, A. P. Chung, and M. S. da Costa. 2004. Leucobacter chromiireducens sp. nov, and Leucobacter aridicollis sp. nov., two new species isolated from a chromium contaminated environment. Syst. Appl. Microbiol. 27:646-652. [DOI] [PubMed] [Google Scholar]
- 29.Muir, R. E., and M.-W. Tan. 2007. Leucobacter chromiireducens subsp. solipictus subsp. nov., a pigmented bacterium isolated from the nematode Caenorhabditis elegans, and emended description of L. chromiireducens. Int. J. Syst. Evol. Microbiol. 57:2770-2776. [DOI] [PubMed] [Google Scholar]
- 30.Nathoo, A. N., R. A. Moeller, B. A. Westlund, and A. C. Hart. 2001. Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc. Natl. Acad. Sci. USA 98:14000-14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholas, H. R., and J. Hodgkin. 2004. The ERK MAP kinase cascade mediates tail swelling and a protective response to rectal infection in C. elegans. Curr. Biol. 14:1256-1261. [DOI] [PubMed] [Google Scholar]
- 32.O'Rourke, D., D. Baban, M. Demidova, R. Mott, and J. Hodgkin. 2006. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 16:1005-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, J. O., K. A. El-Tarabily, E. L. Ghisalberti, and K. Sivasithamparam. 2002. Pathogenesis of Streptoverticillium albireticuli on Caenorhabditis elegans and its antagonism to soil-borne fungal pathogens. Lett. Appl. Microbiol. 35:361-365. [DOI] [PubMed] [Google Scholar]
- 34.Pujol, N., S. Cypowyj, K. Ziegler, A. Millet, A. Astrain, A. Goncharov, Y. Jin, A. D. Chisholm, and J. J. Ewbank. 2008. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr. Biol. 18:481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 36.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shapira, M., B. J. Hamlin, J. Rong, K. Chen, M. Ronen, and M.-W. Tan. 2006. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. USA 103:14086-14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapira, M., and M.-W. Tan. 2008. Genetic analysis of Caenorhabditis elegans innate immunity. Methods Mol. Biol. 415:429-442. [DOI] [PubMed] [Google Scholar]
- 39.Shtonda, B. B., and L. Avery. 2006. Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol. 209:89-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somvanshi, V. S., E. Lang, P. Schumann, R. Pukall, R. M. Kroppenstedt, S. Ganguly, and E. Stackebrandt. 2007. Leucobacter iarius sp. nov., in the family Microbacteriaceae. Int. J. Syst. Evol. Microbiol. 57:682-686. [DOI] [PubMed] [Google Scholar]
- 41.Styer, K. L., G. W. Hopkins, S. S. Bartra, G. V. Plano, R. Frothingham, and A. Aballay. 2005. Yersinia pestis kills Caenorhabditis elegans by a biofilm-independent process that involves novel virulence factors. EMBO Rep. 6:992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan, M.-W., S. Mahajan-Miklos, and F. M. Ausubel. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Timmons, L., and A. Fire. 1998. Specific interference by ingested dsRNA. Nature 395:854. [DOI] [PubMed] [Google Scholar]
- 44.Troemel, E. R., S. W. Chu, V. Reinke, S. S. Lee, F. M. Ausubel, and D. H. Kim. 2006. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vazquez-Torres, A., J. Jones-Carson, A. J. Baumler, S. Falkow, R. Valdivia, W. Brown, M. Le, R. Berggren, W. T. Parks, and F. C. Fang. 1999. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401:804-808. [DOI] [PubMed] [Google Scholar]
- 46.Wong, D., D. Bazopoulou, N. Pujol, N. Tavernarakis, and J. J. Ewbank. 2007. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 8:R194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yook, K., and J. Hodgkin. 2007. Mos1 mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 175:681-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.