Abstract
We investigated the infection dynamics of endosymbiotic bacteria in the developmental course of the mealybugs Planococcus kraunhiae and Pseudococcus comstocki. Molecular phylogenetic analyses identified a betaproteobacterium and a gammaproteobacterium from each of the mealybug species. The former bacterium was related to the β-endosymbionts of other mealybugs, i.e., “Candidatus Tremblaya princeps,” and formed a compact clade in the Betaproteobacteria. Meanwhile, the latter bacterium was related to the γ-endosymbionts of other mealybugs but belonged to distinct clades in the Gammaproteobacteria. Whole-mount in situ hybridization confirmed the peculiar nested formation in the endosymbiotic system of the mealybugs: the β-endosymbiont cells were present in the cytoplasm of the bacteriocytes, and the γ-endosymbiont cells were located in the β-endosymbiont cells. In nymphal and female development, a large oval bacteriome consisting of a number of bacteriocytes was present in the abdomen, wherein the endosymbionts were harbored. In male development, strikingly, the bacteriome progressively degenerated in prepupae and pupae and became almost unrecognizable in adult males. In the degeneration process, the γ-endosymbionts disappeared more rapidly than the β-endosymbionts did. Quantitative PCR analyses revealed that (i) the population dynamics of the endosymbionts in female development reflected the reproductive activity of the insects, (ii) the population dynamics of the endosymbionts were strikingly different between female development and male development, (iii) the endosymbiont populations drastically decreased in male development, and (iv) the γ-endosymbiont populations decreased more rapidly than the β-endosymbiont populations in male development. Possible mechanisms underlying the uncoupled regulation of the β- and γ-endosymbiont populations are discussed in relation to the establishment and evolution of this unique prokaryote-prokaryote endosymbiotic system.
Multiple endosymbiotic bacteria are often housed in the same host organisms. Many insects harbor an obligate endosymbiotic bacterium, referred to as the “primary symbiont,” in specialized cells called bacteriocytes or mycetocytes. The primary symbiont is essential for the survival and reproduction of the host and is therefore fixed in the host population. Frequently, these insects also possess a facultative endosymbiotic bacterium, referred to as the “secondary symbiont,” often in different types of cells and tissues. The secondary symbiont is, in most cases, nonessential for the host and usually shows partial infection in the host population. The bacteriocytes harboring the primary symbiont and the bacteriocytes housing the secondary symbiont often form a huge symbiotic organ, called the bacteriome or mycetome. Endosymbiotic associations comprising an obligate primary symbiont and one or several optional secondary symbionts are commonly found in diverse insect groups (4, 9, 22).
Homopteran insects, such as aphids, scale insects, whiteflies, psyllids, planthoppers, and many others, have needle-like mouthparts and feed exclusively on plant sap throughout their life. Nutritionally, plant sap is extremely unbalanced and difficult to utilize for herbivorous animals. Although rich in carbohydrates, mostly in the form of sucrose, plant sap contains very small amounts of lipids and proteins. Most lipids can be synthesized from carbohydrates in plant sap, but proteins cannot due to deficiencies in nitrogenous precursors such as essential amino acids. Some amino acids are certainly present in plant sap, but they are mostly nonessential ones. In the homopteran insects, however, the primary endosymbiont supplements the deficient diet through provisioning of essential amino acids and other nutrients, whereby they prosper solely on the specialized food source, and some of them are rated among the most serious agricultural pests (4, 11). In aphids, for example, the primary endosymbiont Buchnera aphidicola provides the host with essential amino acids and some vitamins (10, 33), while various secondary endosymbionts confer the host with supplemental biological functions, such as tolerance to heat stress (26), resistance to parasitoid wasps (30), immunity to fungal pathogens (32), and broadening of food plant range (35).
Mealybugs (Homoptera: Coccoidea: Pseudococcidae) comprise one of the largest families of scale insects and include many agriculturally injurious species worldwide. They feed on plant sap, thereby causing damage and vectoring diseases on agricultural crops and trees (5). In the abdomen of nymphal mealybugs, a large oval bacteriome consisting of a number of bacteriocytes exists, in which numerous rod- or sausage-shaped bacteria are harbored. In the cytoplasm of the bacteriocytes, pleomorphic vesicle-like structures are present, in which the bacterial rods are located. Early researchers were puzzled by the peculiar mucoidal subcellular compartments, so-called “mucus packets,” and regarded them as specialized mucus-rich vesicles for harboring the rod-shaped endosymbiotic bacteria (6-9, 13, 23, 37). However, recent molecular phylogenetic and histochemical studies have revealed that (i) mealybugs harbor at least two distinct endosymbionts, one belonging to the Betaproteobacteria and the other being a member of the Gammaproteobacteria (21, 27); (ii) the β-endosymbiont and the γ-endosymbiont coexist in the cytoplasm of the same bacteriocytes (17); (iii) the “mucus packets” are not vesicles but actually the β-endosymbiont cells, and the γ-endosymbiont cells are harbored inside the β-endosymbiont cells (36); (iv) the β-endosymbionts are conserved among all mealybug species and exhibit cocladogenesis with the host insects, suggesting that the primary β-endosymbionts are of a single evolutionary origin in the common ancestor of extant mealybugs (2, 12); (v) the γ-endosymbionts are detected in most, but not all, mealybug species and form several distinct clades in the gammaproteobacterial phylogeny, suggesting that the secondary γ-endosymbionts are of multiple evolutionary origins in mealybugs (34); and (vi) genetic properties of the primary β-endosymbiont are different from those of the primary endosymbionts of other insects in that neither AT bias in nucleotide composition nor accelerated molecular evolution is observed (3). On account of these distinct phylogenetic and phenotypic features, the β-endosymbiont of mealybugs was given the designation “Candidatus Tremblaya princeps” (34).
The construction and properties of the endosymbiotic system identified in mealybugs are unprecedented in several respects. Firstly, this system provides the only case of prokaryote-prokaryote endocellular symbiosis ever known. Among diverse endosymbiotic associations in the biological world, the host cells are always eukaryotic, whereas the symbiont cells are either prokaryotic (e.g., many nutritional endosymbioses in insects) or eukaryotic (e.g., many photosynthetic endosymbioses in cnidarians) (4, 19). Hence, the β- and γ-endosymbionts in mealybugs comprise the sole exception. Secondly, although it has been believed that endocytotic processes are restricted to eukaryotic cells (1), the endocellular location of the γ-endosymbiont cells inside the β-endosymbiont cells suggests a novel endocytosis-like mechanism operating in the β-endosymbiont cells. Thirdly, in the light of the host control over the symbiont, the nested bacterial endosymbiosis may entail unique prokaryote-prokaryote interactions that seldom occur in other systems. Fourthly, most of the primary endosymbionts of diverse insects, such as those of aphids, psyllids, whiteflies, tsetse flies, ants, weevils, etc., belong to the Gammaproteobacteria, so the primary β-endosymbiont of mealybugs is exceptional.
However, besides the above-mentioned molecular phylogenetic and histological studies, the endosymbiotic bacteria of mealybugs have been investigated poorly. The biological roles of the β- and γ-endosymbionts for the host insect are almost untouched, except for an early work describing that antibiotic and high-temperature treatments of mealybugs resulted in degeneration of the endosymbiotic system and mortality of the insects (23). Nutritional, physiological, and ecological interactions between the β-endosymbiont, the γ-endosymbiont, and the host insect are totally unknown.
In an attempt to gain insights into symbiont-symbiont and host-symbiont interactions in the nested endosymbiotic system, we investigated the population dynamics of the β- and γ-endosymbionts in the developmental course of two mealybug species.
MATERIALS AND METHODS
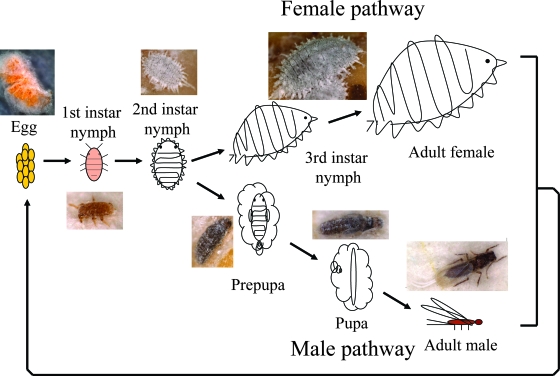
Materials.
Laboratory strains of the Japanese mealybug Planococcus kraunhiae and the comstock mealybug Pseudococcus comstocki were provided by Tamotsu Murai (Utsunomiya University, Japan). The insects were reared on germinated seeds of the broad bean, Vicia faba, at 25°C under a long-day regimen (16 h of light, 8 h of dark) in plastic containers (8 cm × 11 cm × 5 cm) as described previously (28). Figure 1 shows the life cycle of the mealybugs. In first- and second-instar nymphs, males and females are indistinguishable, and then differences between male and female become distinctly apparent. In the female pathway, unwinged adult females emerge through third-instar nymphs. In the male pathway, winged adult males develop through two immobile stages, prepupae and pupae. Note that mealybugs are hemimetabolous and thus do not have true pupae, but their immobile developmental stages prior to adult emergence in males are often designated “prepupae” and “pupae.” These insects were collected at different developmental stages and preserved in acetone (14) for molecular and histological analyses.
FIG. 1.
Life cycle of mealybugs.
Molecular biological procedures.
Each of the acetone-preserved insect samples was briefly dried in air, crushed in a 1.5-ml plastic tube, and digested in lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 0.5% sodium dodecyl sulfate, 0.8 mg/ml proteinase K) at 55°C overnight. The lysate was extracted with phenol-chloroform and subjected to ethanol precipitation, and the precipitated whole nucleic acid was dried and dissolved in TE buffer (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA). From the DNA samples, a 1.5-kb segment of the bacterial 16S rRNA gene was amplified by PCR with the primers 16SA1 (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 16SB1 (5′-TAC GGY TAC CTT GTT ACG ACT T-3′), with a temperature profile of 40 cycles of 95°C for 20 s, 55°C for 20 s, and 70°C for 2 min (16). A 0.8-kb segment of the betaproteobacterial groEL gene was amplified with the primers β-GroE-1F (5′-ATG GAC MTS AAG CGY GGB ATC GA-3′) and β-GroE-2R (5′-ACD GCR ACD CCG CCK GCC AGC TT-3′), with a temperature profile of 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. A 0.6-kb segment of the gammaproteobacterial groESL operon, ranging from a 3′ region of the groES gene to a 5′ region of the groEL gene, was amplified with the primers SymA1 (5′-AAA TTC GTC CAT TGC ATG ATC G-3′) and SymB1 (5′-GAA GTT GCA TCA AAA GCA AAC G-3′), with a temperature profile of 40 cycles of 95°C for 30 s, 43°C for 2 min, and 65°C for 3 min. A 1.2-kb segment of the insect elongation factor 1α (ef1α) gene was amplified with the primers M44-1 (5′-GCT GAG CGY GAR CGT GGT ATC AC-3′) and rcM53-2 (5′-GCA ATG TGR GCI GTG TGG CA-3′), with a temperature profile of 40 cycles of 95°C for 30 s, 50°C for 30 s, and 70°C for 2 min. The PCR products were subjected to cloning, restriction fragment length polymorphism (RFLP) genotyping, and DNA sequencing as previously described (16).
Molecular phylogenetic analysis.
The nucleotide or deduced amino acid sequences were subjected to molecular phylogenetic analysis, together with sequences retrieved from the DDBJ nucleotide sequence database. Multiple alignment of the sequences was performed using the program VectorNTI (Invitrogen). Final alignments were inspected and corrected manually. Ambiguously aligned regions and sites including alignment gaps were excluded from the analysis. The program MEGA V3.1 (24) was used to construct maximum likelihood and neighbor-joining trees and to perform bootstrap tests with 500 resamplings.
Whole-mount FISH.
An oligonucleotide probe specific to the 16S rRNA sequences of the β-endosymbionts of P. kraunhiae and P. comstocki, called Cy3-fujiB-437R (Cy3-5′-ACG CCC CCC TTC TTT CCG AA-3′), and a probe specific to the 16S rRNA sequences of the γ-endosymbionts of the mealybugs, called Cy5-fujiG-440R (Cy5-5′-ATA TTG CCT TCC TCC CTA CT-3′), were used for fluorescent in situ hybridization (FISH). After being decapitated to facilitate infiltration of reagents, the acetone-preserved insects were fixed in Carnoy's solution (chloroform-ethanol-acetic acid [6:3:1]). Subsequently, the insects were incubated in ethanolic hydrogen peroxide (6% H2O2, 80% ethanol) for several days in order to quench the autofluorescence of insect tissues. After being washed thoroughly with absolute ethanol and PBSTx buffer (136.9 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8.1 mM Na2HPO4 [pH 7.4], 0.3% Triton X-100), the insects were prehybridized with hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate, 30% formamide) three times for 5 min each. The insects were then hybridized with 100 nM each of the probes Cy3-fujiB-437R and Cy5-fujiG-440R in hybridization buffer overnight. Nuclei of the host cells were counterstained with 0.5 μM of SYTOX green (Invitrogen). After being washed with PBSTx four times for 10 min each, the insects were mounted with Slowfade antifade solution (Invitrogen) and observed under an epifluorescence microscope (Axiophoto; Carl Zeiss) or a laser confocal microscope (Pascal 5; Carl Zeiss). To confirm the specificity of the hybridization signals, a series of control experiments, namely, a no-probe control, an RNase digestion control, and a competitive suppression control with excess unlabeled probes, were conducted as previously described (31).
Quantitative PCR.
Real-time PCR quantification of gene copy numbers of the β-endosymbionts, the γ-endosymbionts, and the host insects was performed using SYBR green and an Mx3000P QPCR system (Stratagene) essentially as described previously (29). DNA extraction was conducted using a NucleoSpin tissue kit (Macherey-Nagel). For preparation of DNA samples in 50 μl of TE buffer, 15 individuals were pooled and extracted for eggs and first-instar nymphs, whereas prepupae, pupae, adult males, third-instar nymphs, and adult females were extracted individually. For each of the developmental stages, about 10 DNA samples were subjected to quantitative PCR. For the DNA samples from eggs and first-instar nymphs, the quantified value was divided by 15 to yield an average value for an individual. The following primer sets were used: β-groE-AF (5′-CCG CAC AGT TGG CGA GAT GA-3′) and β-groE-AR (5′-CGA CAG TTA TGA CGC CCT CC-3′) for the groEL gene of the β-endosymbionts; γ-groE-AF (5′-AAA AAT GCT TCA CGG AGT TAA TGT TC-3′) and γ-groE-AR (5′-TGA TTT GTC CAG TAC GAC GTT G-3′) for the groEL gene of the γ-endosymbionts; and ef1α-AF (5′-CGC CAG CCA CGT AAC CTC T-3′) and ef1α-AR (5′-CCC GGA GAT AAC GTC GGT TT-3′) for the nuclear ef1α gene of the mealybugs. Each of the PCR mixtures consisted of 2.5 μl of 10× TaqGold buffer (Applied Biosystems), 1.5 μl of 25 mM MgCl2, 2.5 μl of nucleotide mixture solution (2 mM [each] of dATP, dTTP, dGTP, and dCTP), 0.3 μl of SYBR green I (1/1,000 diluted solution) (Molecular Probes), 1.5 μl of primer mixture solution (5 μM [each] of forward and reverse primers), 0.1 μl of AmpliTaqGold DNA polymerase (Applied Biosystems), 11.6 μl of distilled water, and 5 μl of DNA sample solution. The PCR temperature profile was 45 cycles of 95°C for 10 s, 65°C for 10 s, and 72°C for 20 s. Standard curves for each of the genes were drawn with standard samples that contained 101, 103, 105, and 107 copies per reaction of the target PCR product.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession numbers AB374415 to AB374424.
RESULTS
Identification of beta- and gammaproteobacterial 16S rRNA gene sequences.
A region of the bacterial 16S rRNA gene of about 1.5 kb was amplified by PCR from DNA samples of P. kraunhiae and P. comstocki. When the PCR products were cloned and subjected to RFLP genotyping, two major RFLP types were identified from each of the mealybug species (data not shown). Three or more clones for each of the RFLP types were sequenced. The sequences of the same RFLP type were identical to each other, while the sequences of different RFLP types from the same species differed remarkably. DNA database searches revealed that for both P. kraunhiae and P. comstocki, one of the sequences belongs to the Betaproteobacteria and the other belongs to the Gammaproteobacteria. The highest BLAST hits for the sequences were as follows: for the β sequence from P. kraunhiae, the β-endosymbiont “Candidatus Tremblaya princeps” of the vine mealybug Planococcus ficus (98% sequence similarity [1,464/1,486 nucleotides]; GenBank accession number AF476092); for the γ sequence from P. kraunhiae, the secondary endosymbiont of the citrus mealybug Planococcus citri (96% similarity [1,428/1,483 nucleotides]; accession no. AF476107); for the β sequence from P. comstocki, the β-endosymbiont “Candidatus Tremblaya princeps” of the mealybug Erium globosum (97% similarity [1,456/1,488 nucleotides]; accession no. AF476084); and for the γ sequence from P. comstocki, the secondary endosymbiont of the gray pineapple mealybug Dysmicoccus neobrevipes (97% similarity [1,454/1,491 nucleotides]; accession no. AF476104).
Identification of beta- and gammaproteobacterial groEL gene sequences.
A region of the betaproteobacterial groEL gene of about 0.8 kb was amplified by PCR and cloned from DNA samples of P. kraunhiae and P. comstocki. For both species, RFLP genotyping and sequencing of the clones identified a single sequence type. DNA database searches retrieved the groEL gene sequence from the β-endosymbiont “Candidatus Tremblaya princeps” of the pineapple mealybug Dysmicoccus brevipes (95% similarity [749/788 nucleotides]; GenBank accession no. AF481102) as the highest BLAST hit. A region of the gammaproteobacterial groESL gene of about 0.6 kb was also amplified by PCR and cloned from DNA samples of P. kraunhiae and P. comstocki. For both species, RFLP genotyping and sequencing of the clones identified a single sequence type. BLAST searches revealed that the groEL sequence region from P. kraunhiae was related to the groEL gene sequence from the facultative γ-endosymbiont Sodalis glossinidius of the tsetse fly Glossina morsitans (85% similarity [216/253 nucleotides]; accession no. AP008232), and the sequence region from P. comstocki was similar to the groEL gene sequence from the obligate γ-endosymbiont Buchnera aphidicola of aphids (84% similarity [211/250 nucleotides]; accession no. U77380).
Molecular phylogenetic analysis of β- and γ-endosymbionts.
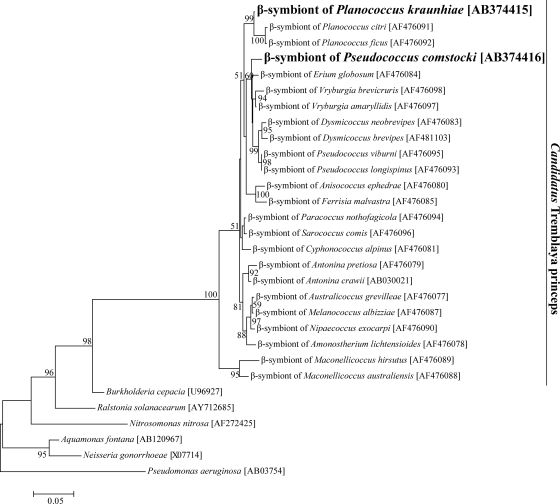
Figure 2 shows the phylogenetic relationships of the β sequences from P. kraunhiae and P. comstocki to betaproteobacterial representatives on the basis of 16S rRNA gene sequences. Both sequences were placed in the monophyletic group of the primary endosymbiont of mealybugs, “Candidatus Tremblaya princeps,” with high statistical support. The β sequence from P. kraunhiae formed a highly supported clade with those from Planococcus species, reflecting the host insect systematics. Meanwhile, the β sequence from P. comstocki did not cluster with those from Pseudococcus species.
FIG. 2.
Molecular phylogenetic analysis of the β-endosymbionts of mealybugs and allied betaproteobacteria on the basis of 16S rRNA gene sequences. A maximum likelihood tree inferred from 1,250 unambiguously aligned nucleotide sites is shown, whereas a neighbor-joining tree exhibited essentially the same topology (data not shown). Bootstrap values of >50% are shown at the nodes. Accession numbers are shown in brackets.
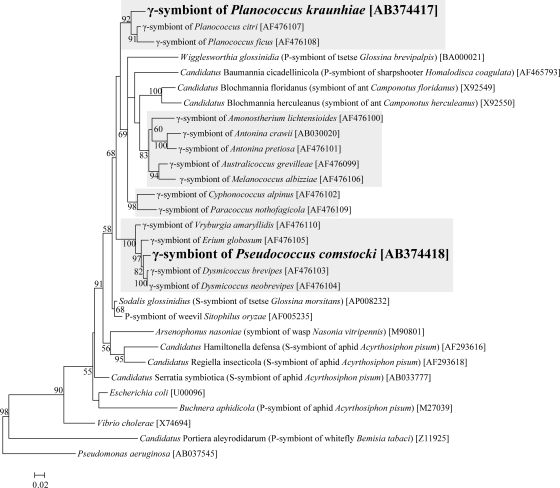
Figure 3 shows the phylogenetic relationships of the γ sequences from P. kraunhiae and P. comstocki to gammaproteobacterial representatives on the basis of 16S rRNA gene sequences. As previously reported (34), γ-endosymbionts of mealybugs were divided into several clusters in the gammaproteobacterial phylogeny. The γ sequence from P. kraunhiae formed a well-supported clade with those from Planococcus species. Meanwhile, the γ sequence from P. comstocki formed a distinct clade with those from D. brevipes, D. neobrevipes, E. globosum, and Vryburgia amaryllidis.
FIG. 3.
Molecular phylogenetic analysis of the γ-endosymbionts of mealybugs and allied gammaproteobacteria on the basis of 16S rRNA gene sequences. A maximum likelihood tree inferred from 1,293 unambiguously aligned nucleotide sites is shown, whereas a neighbor-joining tree exhibited essentially the same topology (data not shown). Bootstrap values of >50% are shown at the nodes. P-symbiont and S-symbiont indicate the primary symbiont and secondary symbiont, respectively. Accession numbers are shown in brackets. Shading indicates the clades of γ-endosymbionts of mealybugs.
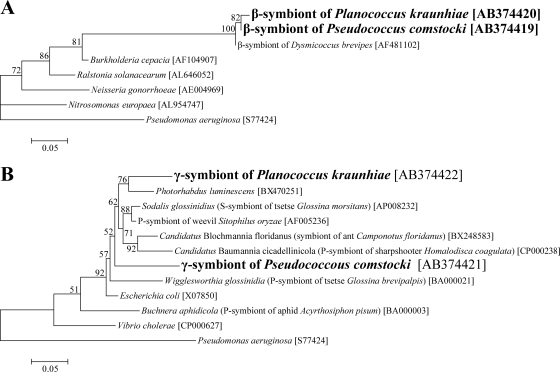
Figure 4 shows the phylogenetic placement of the GroEL protein sequences from P. kraunhiae and P. comstocki. The β sequences were placed in the Betaproteobacteria, forming a clade with the β-endosymbiont of the mealybug D. brevipes (Fig. 4A). The γ sequences were certainly placed in the Gammaproteobacteria, but they did not form a cluster (Fig. 4B).
FIG. 4.
Molecular phylogenetic analysis of β-endosymbionts and γ-endosymbionts of mealybugs on the basis of deduced groESL protein sequences. (A) Phylogenetic placement in the Betaproteobacteria. A maximum likelihood tree inferred from 264 unambiguously aligned amino acid sites is shown. (B) Phylogenetic placement in the Gammaproteobacteria. A maximum likelihood tree inferred from 187 unambiguously aligned amino acid sites is shown. Neighbor-joining trees exhibited essentially the same topologies (data not shown). Bootstrap values of >50% are shown at the nodes. Accession numbers are shown in brackets.
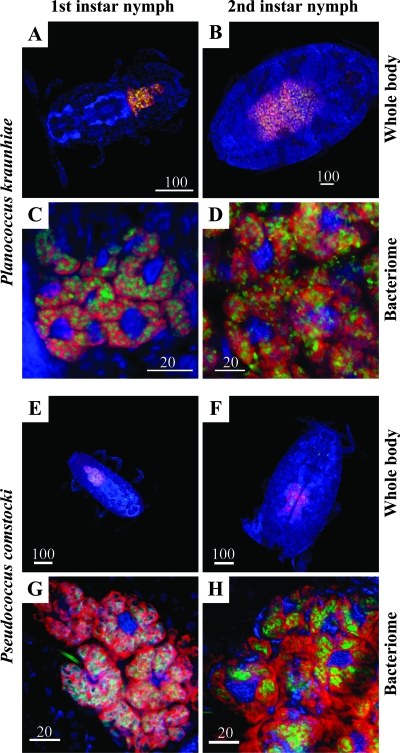
Localization of β- and γ-endosymbionts in nymphs and females.
Localization of the beta- and gammaproteobacterial 16S rRNA sequences was analyzed by FISH. In nymphs of P. kraunhiae, both the β and γ sequences were specifically located in a large oval bacteriome in the abdomen (Fig. 5A and B). The bacteriome consisted of a number of bacteriocytes, and signals of the β and γ sequences were restricted to the cytoplasm of the bacteriocytes. In the cytoplasm, the signals of the β sequence were observed as large pleomorphic formations surrounding the nucleus, reflecting the β-endosymbiont cells (Fig. 5C and D). The signals of the γ sequence were identified as a number of sausage-shaped structures, indicating the γ-endosymbiont cells, in the area of the β-endosymbiont cells (Fig. 5C and D). In mature adult females, signals of the β and γ sequences were also associated with oocytes in the ovary, indicating vertical transmission of the endosymbionts (data not shown).
FIG. 5.
Localization of β- and γ-endosymbionts in nymphs of mealybugs. Red, signals of the β-endosymbiont; green, signals of the γ-endosymbiont; blue, signals of the host nuclei. (A to D) P. kraunhiae. (E to H) P. comstocki. (A, C, E, and G) First-instar nymphs. (B, D, F, and H) Second-instar nymphs. (A, B, E, and F) Whole insects. (C, D, G, and H) Bacteriomes.
In nymphs and adult females of P. comstocki, the same localization patterns of the β- and γ-endosymbionts were observed (Fig. 5E to H).
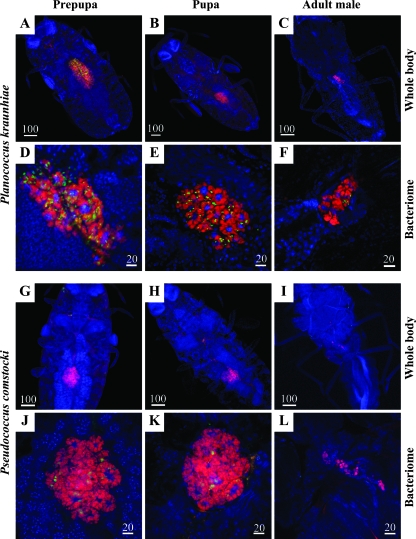
Degeneration of endosymbiotic system in males.
In the developmental pathway toward males, the endosymbiont infection dynamics exhibited strikingly different patterns. In P. kraunhiae, the bacteriome progressively degenerated in prepupae and pupae and became almost unrecognizable in adult males (Fig. 6A to C). In prepupae and pupae, the γ-endosymbionts significantly decreased in number, while the β-endosymbionts still remained (Fig. 6D to F).
FIG. 6.
Localization of β- and γ-endosymbionts in prepupae, pupae, and adult males of mealybugs. Red, signals of the β-endosymbiont; green, signals of the γ-endosymbiont; blue, signals of the host nuclei. (A to F) P. kraunhiae. (G to L) P. comstocki. (A, D, G, and J) Prepupae. (B, E, H, and K) Pupae. (C, F, I, and L) Adult males. (A to C and G to I) Whole insects. (D to F and J to L) Bacteriomes.
In prepupae, pupae, and adult males of P. comstocki, similar infection dynamics of the β- and γ-endosymbionts were observed (Fig. 6G to L).
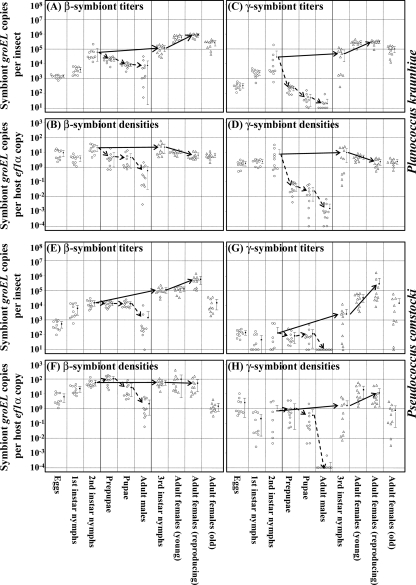
Population dynamics of β- and γ-endosymbionts during host development.
Infection titers and densities of the β- and γ-endosymbionts throughout the developmental course of the host insects were examined by quantitative PCR in terms of numbers of groEL gene copies of the endosymbionts. In P. kraunhiae, the titers and densities of the β-endosymbiont increased from eggs to second-instar nymphs. In the developmental course toward females, the β-endosymbiont titers increased from third-instar nymphs to mature adult females, attained a peak in ovipositing females, and dropped in old females at postoviposition (Fig. 7A). Meanwhile, the β-endosymbiont densities were largely constant from third-instar nymphs to mature adult females and declined in old females (Fig. 7B). In the developmental course toward males, in contrast, the β-endosymbiont titers and densities significantly decreased in prepupae and pupae, resulting in very low values for adult males (Fig. 7A and B). Infection dynamics of the γ-endosymbiont were generally similar to those of the β-endosymbiont in P. kraunhiae, except that the bacterial titers and densities decreased more rapidly in prepupae and pupae, leading to a substantial absence of the γ-endosymbiont in adult males (Fig. 7C and D).
FIG. 7.
Infection titers and densities of β- and γ-endosymbionts in the developmental course of mealybugs. Circles, eggs, first-instar nymphs, and second-instar nymphs without sexing; triangles, third-instar nymphs and adult females; diamonds, prepupae, pupae, and adult males. The developmental pathway toward females and the developmental pathway toward males are highlighted by solid arrows and dotted arrows, respectively. Means and standard deviations are indicated by dots and bars. (A to D) P. kraunhiae. (E to H) P. comstocki. (A, B, E, and F) β-Endosymbionts. (C, D, G, and H) γ-Endosymbionts. (A, C, E, and G) Infection titers in terms of groEL gene copies per insect. (B, D, F, and H) Infection densities in terms of groEL gene copies per ef1α gene copy.
In P. comstocki, the infection dynamics of the β- and γ-endosymbionts were largely similar to those in P. kraunhiae, except that in the developmental course of males, the bacterial titers and densities declined more slowly than in P. kraunhiae (Fig. 7E to H).
DISCUSSION
In agreement with previous studies on other mealybug species (17, 36), our test species, P. kraunhiae and P. comstocki, harbored both the β- and γ-endosymbionts in a huge bacteriome in the abdomen (Fig. 2 to 4). FISH analyses confirmed their characteristic localization as previously described (36): the β-endosymbionts are harbored in the cytoplasm of the bacteriocytes, and the γ-endosymbionts are present in the cells of the β-endosymbionts (Fig. 5). Such prokaryote-prokaryote endocellular symbiosis has been found nowhere else in nature. Phylogenetic analyses suggested that the mealybug β-endosymbionts are monophyletic, whereas the mealybug γ-endosymbionts were acquired independently multiple times (Fig. 2 and 3). In the bacteriocytes of the mealybugs, therefore, the closely related betaproteobacteria harbor the phylogenetically distinct gammaproteobacteria endocellularly, providing a unique opportunity to investigate the infection dynamics of the different bacterial lineages coexisting in the nested endosymbiotic system.
In both P. kraunhiae and P. comstocki, the β-endosymbiont titers increased during nymphal and female development, attained a peak in actively reproducing females, and declined in old females that had ceased reproduction (Fig. 7A and E). Meanwhile, the β-endosymbiont densities were almost constant from third-instar nymphs to mature adult females (Fig. 7B and F), wherein proliferation of the endosymbionts is probably cancelled out by rapid growth of the insects. The infection dynamics plausibly reflects important biological roles of the primary β-endosymbionts for the nutritional and reproductive physiology of the host mealybugs. It was reported that ingestion of antibiotics by mealybugs or exposure to high temperatures resulted in degeneration of the endosymbiotic system within a few days, and the insects died soon after (23). The host-symbiont cocladogenesis over evolutionary time (2, 12) also favors the idea that the primary β-endosymbionts are essential for the host mealybugs. In the pea aphid Acyrthosiphon pisum, similarly, infection titers of the essential endocellular symbiont Buchnera aphidicola attain a peak in actively reproducing females in the course of development (22, 31).
The γ-endosymbiont titers and densities exhibited similar dynamics to those of the β-endosymbionts in P. kraunhiae and P. comstocki (Fig. 7C, D, G, and H). The infection dynamics might be relevant to some biological roles of the γ-endosymbionts either for the host insects or for the β-endosymbionts. Meanwhile, considering that the γ-endosymbionts of the mealybugs have polyphyletic evolutionary origins (34) and some mealybug species are suggested to lack the γ-endosymbiont (9, 34), the secondary γ-endosymbionts might be less important than the primary β-endosymbionts in mealybug biology and evolution. If so, it appears likely that the infection dynamics of the γ-endosymbionts simply reflect those of the β-endosymbionts wherein they are harbored.
In the developmental pathway of males, we found that, interestingly, the endosymbiotic system degenerated progressively, and the β- and γ-endosymbionts were almost lost in adult males (Fig. 6). Why the endosymbiotic system collapses in a male-specific manner is currently unknown. It should be noted that in the male pathway, the insects neither feed nor grow but just molt and metamorphose (Fig. 1). Considering the putative nutritional roles of the endosymbionts for the sap-feeding host insects (4, 11), the endosymbiotic system is plausibly not necessary for males of the mealybugs, which might be relevant to the male-specific degeneration. Furthermore, the metamorphosis in males requires much energy and resources, and the large nymphal bacteriome might be consumed for that purpose. Similar male-specific absence and/or degeneration of the endosymbiotic system has been reported for aphids, scale insects, and lice (9, 15, 18).
In the developmental pathway of nymphs and females, the population dynamics of the β-endosymbiont was synchronous with that of the γ-endosymbiont (Fig. 7), which appears to reflect, at a glance, the intimate endocellular symbiotic association between the different bacteria. However, the uncoupled population dynamics of the β- and γ-endosymbionts in males (Fig. 6 and 7) revealed a previously unknown aspect of the endosymbiotic system, namely, proliferation and/or degeneration of the endosymbionts coexisting in a nested manner can be regulated independently. Although the mechanism underlying the preferential disappearance of the γ-endosymbiont in males is unknown, considering the spatial topology of the γ-endosymbiont cells inside the β-endosymbiont cells, we suggest two possible mechanisms that may involve the γ-endosymbiont-specific disappearance. One possibility is that the β-endosymbiont cells consume the γ-endosymbiont cells. Another possibility is that β-endosymbiont cells release the γ-endosymbiont cells into the cytoplasm of the bacteriocyte, and the γ-endosymbiont cells are digested there. In either of these scenarios, the β-endosymbiont must play a pivotal role in the process. It is also notable that although the γ-endosymbionts of P. kraunhiae and P. comstocki are phylogenetically distinct from each other (Fig. 3), the preferential disappearance of the γ-endosymbionts in the male pathway was observed in both mealybug species (Fig. 6 and 7), which favors the idea that the β-endosymbiont rather than the γ-endosymbiont mainly governs the process. Of course, these aspects of cellular interactions between the β-endosymbiont, the γ-endosymbiont, and the host bacteriocyte should be examined thoroughly in future studies by making use of electron microscopy.
The endosymbiotic theory for the origin of eukaryotic cells hypothesizes that an ancestral prokaryotic cell engulfed other prokaryotic cells, giving rise to endocellular membranous structures and compartments, such as the nucleus, mitochondria, and chloroplasts (25). As for mitochondria and chloroplasts, their bacterial endosymbiont origins have been established unequivocally (20). There is no doubt that an endocytotic process must have been crucial in the initial step toward the evolution of eukaryotes. Thus far, however, endocytosis has been unknown among prokaryotes. It is believed that endocytosis and exocytosis are cellular processes characteristic of eukaryotes, enabling a variety of eukaryotic cellular functions, such as phagocytosis, pinocytosis, cellular secretion, synaptic transmission, membrane trafficking, etc. (1). Remarkably, for establishment and control of the nested endosymbiotic system in mealybugs, it is suspected that a novel endocytosis-like mechanism may operate in the β-endosymbiont cells. By investigating the peculiar endosymbiotic system in mealybugs, we would be able to gain unique insights into the origin of endocytosis and also into the evolutionary process of the eukaryotic cells.
Acknowledgments
We thank Tamotsu Murai for providing the mealybug strains.
Footnotes
Published ahead of print on 9 May 2008.
REFERENCES
- 1.Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. Molecular biology of the cell, 4th ed. Garland Science, New York, NY.
- 2.Baumann, L., and P. Baumann. 2005. Cospeciation between the primary endosymbionts of mealybugs and their hosts. Curr. Microbiol. 50:84-87. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, L., M. L. Thao, J. M. Hess, M. W. Johnson, and P. Baumann. 2002. The genetic properties of the primary endosymbionts of mealybugs differ from those of other endosymbionts of plant sap-sucking insects. Appl. Environ. Microbiol. 68:3198-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann, P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155-189. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Dov, Y. 1994. A systematic catalogue of the mealybugs of the world. Intercept Limited, Andover, United Kingdom.
- 6.Buchner, P. 1955. Endosymbiosestudien an Schildläusen. III. Macrocerococcus und Puto, zwei primitive Pseudococcinen. Z. Morphol. Ökol. Tieraertz 43:523-577. [Google Scholar]
- 7.Buchner, P. 1957. Endosymbiosestudien an Schildläusen. IV. Hippococcus, eine myrmekophile Pseudococcine. Z. Morphol. Ökol. Tieraertz 45:379-410. [Google Scholar]
- 8.Buchner, P. 1957. Endosymbiosestudien an Schildläusen. V. Die Gattung Rastrococcus Ferris (Ceroputo Sulc). Z. Morphol. Ökol. Tieraertz 46:111-148. [Google Scholar]
- 9.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience, New York, NY.
- 10.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 11.Douglas, A. E. 2003. The nutritional physiology of aphids. Adv. Insect Physiol. 31:73-140. [Google Scholar]
- 12.Downie, D. A., and P. J. Gullan. 2005. Phylogenetic congruence of mealybugs and their primary endosymbionts. J. Evol. Biol. 18:315-324. [DOI] [PubMed] [Google Scholar]
- 13.Fink, R. 1952. Morphologische und physiologische Untersuchungen an den intrazellularen Symbionten von Pseudococcus citri Risso. Z. Morphol. Ökol. Tieraertz 41:78-146. [Google Scholar]
- 14.Fukatsu, T. 1999. Acetone preservation: a practical technique for molecular analysis. Mol. Ecol. 8:1935-1945. [DOI] [PubMed] [Google Scholar]
- 15.Fukatsu, T., and H. Ishikawa. 1992. Soldier and male of an eusocial aphid Colophina arma lack endosymbiont: implications for physiological and evolutionary interaction between host and symbiont. J. Insect Physiol. 38:1033-1042. [Google Scholar]
- 16.Fukatsu, T., and N. Nikoh. 1998. Two intracellular symbiotic bacteria of the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl. Environ. Microbiol. 64:3599-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukatsu, T., and N. Nikoh. 2000. Endosymbiotic microbiota of the bamboo pseudococcid Antonina crawii (Insecta, Homoptera). Appl. Environ. Microbiol. 66:643-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukatsu, T., R. Koga, W. A. Smith, K. Tanaka, N. Nikoh, K. Sasaki-Fukatsu, K. Yoshizawa, C. Dale, and D. H. Clayton. 2007. Bacterial endosymbiont of the slender pigeon louse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Appl. Environ. Microbiol. 73:6660-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodson, M. S., L. F. Whitehead, and A. E. Douglas. 2001. Symbiotic dinoflagellates in marine Cnidaria: diversity and function. Hydrobiologia 461:79-82. [Google Scholar]
- 20.Hirt, R. P., and D. S. Horner. 2004. Organelles, genomes, and eukaryote phylogeny: an evolutionary synthesis in the age of genomics. Routledge, Oxon, United Kingdom.
- 21.Kantheti, P., K. S. Jayarama, and H. S. Chandra. 1996. Developmental analysis of a female-specific 16S rRNA gene from mycetome-associated endosymbionts of a mealybug, Planococcus lilacinus. Insect Biochem. Mol. Biol. 26:997-1009. [DOI] [PubMed] [Google Scholar]
- 22.Koga, R., T. Tsuchida, and T. Fukatsu. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. R. Soc. Lond. B 270:2543-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köhler, M., and W. Schwartz. 1962. Untersuchungen über die Symbiose von Tieren mit Pilzen und Bakterien. IX. Über die Beziehungen zwischen Symbionten und Wirtsorganismus bei Pseudococcus citri, Ps. maritimus und Orthezia insignis. Z. Allg. Mikrobiol. 2:190-208. [Google Scholar]
- 24.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 25.Margulis, L. 1981. Symbiosis in cell evolution. W. H. Freeman, San Francisco, CA.
- 26.Montllor, C. B., A. Maxmen, and A. H. Purcell. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27:189-195. [Google Scholar]
- 27.Munson, M. A., P. Baumann, and N. A. Moran. 1992. Phylogenetic relationships of the endosymbionts of mealybugs (Homoptera: Pseudococcidae) based on 16S rDNA sequences. Mol. Phylogenet. Evol. 1:26-30. [DOI] [PubMed] [Google Scholar]
- 28.Narai, Y., and T. Murai. 2002. Individual rearing of the Japanese mealybug, Planococcus kraunhiae (Kuwana) (Homoptera: Pseudococcidae) on germinated broad bean seeds. Appl. Entomol. Zool. 37:295-298. [Google Scholar]
- 29.Narita, S., D. Kageyama, M. Nomura, and T. Fukatsu. 2007. Unexpected mechanism of symbiont-induced reversal of insect sex: feminizing Wolbachia continuously acts on the butterfly Eurema hecabe during larval development. Appl. Environ. Microbiol. 73:4332-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver, K. M., J. A. Russell, N. A. Moran, and M. S. Hunter. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. USA 100:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakurai, M., R. Koga, T. Tsuchida, X.-Y. Meng, and T. Fukatsu. 2005. Rickettsia symbiont of the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on the host fitness, and interaction with the essential symbiont Buchnera. Appl. Environ. Microbiol. 71:4069-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scarborough, C. L., J. Ferrari, and H. C. J. Godfray. 2005. Aphid protected from pathogen by endosymbiont. Science 310:1781. [DOI] [PubMed] [Google Scholar]
- 33.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 34.Thao, M. Y., P. J. Gullan, and P. Baumann. 2002. Secondary (γ-proteobacteria) endosymbionts infect the primary (β-proteobacteria) endosymbionts of mealybugs multiple times and coevolve with their hosts. Appl. Environ. Microbiol. 68:3190-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuchida, T., R. Koga, and T. Fukatsu. 2004. Host plant specialization governed by facultative symbiont. Science 303:1989. [DOI] [PubMed] [Google Scholar]
- 36.von Dohlen, C. D., S. Kohler, S. T. Alsop, and W. R. McManus. 2001. Mealybug β-proteobacterial endosymbionts contain γ-proteobacterial symbionts. Nature 412:433-436. [DOI] [PubMed] [Google Scholar]
- 37.Walczuch, A. 1932. Studien an Coccidensymbionten. Z. Morphol. Ökol. Tieraertz 25:623-729. [Google Scholar]