Abstract
Methane vents are of significant geochemical and ecological importance. Notable progress has been made toward understanding anaerobic methane oxidation in marine sediments; however, the diversity and distribution of aerobic methanotrophs in the water column are poorly characterized. Both environments play an essential role in regulating methane release from the oceans to the atmosphere. In this study, the diversity of particulate methane monooxygenase (pmoA) and 16S rRNA genes from two methane vent environments along the California continental margin was characterized. The pmoA phylotypes recovered from methane-rich sediments and the overlying water column differed. Sediments harbored the greatest number of unique pmoA phylotypes broadly affiliated with the Methylococcaceae family, whereas planktonic pmoA phylotypes formed three clades that were distinct from the sediment-hosted methanotrophs and distantly related to established methanotrophic clades. Water column-associated phylotypes were highly similar between field sites, suggesting that planktonic methanotroph diversity is controlled primarily by environmental factors rather than geographical proximity. Analysis of 16S rRNA genes from methane-rich waters did not readily recover known methanotrophic lineages, with only a few phylotypes demonstrating distant relatedness to Methylococcus. The development of new pmo primers increased the recovery of monooxygenase genes from the water column and led to the discovery of a highly diverged monooxygenase sequence which is phylogenetically intermediate to Amo and pMMO. This sequence potentiates insight into the amo/pmo superfamily. Together, these findings lend perspective into the diversity and segregation of aerobic methanotrophs within different methane-rich habitats in the marine environment.
Methane cycling between the ocean and the atmosphere is an important component of the global carbon budget and has been the subject of increased attention in recent years. Most methane in the oceans is concentrated along the continental margins, where the majority of ocean productivity occurs and where tectonic activity facilitates petroleum and natural gas leakage. Seafloor seeps and vents are a natural source of methane and may contribute as much as 48 Tg methane to the atmosphere each year, which is approximately 8% of the global methane budget (15, 50). The potential for methane release to the atmosphere is even greater from submarine vents and mud volcanoes, where deeply sourced geologic methane is frequently advected into the water column (3, 37, 50). Methane is a significant greenhouse gas, estimated to be at least 20 times as effective as CO2 in forcing irradiative warming (28). Thus, a fundamental understanding of the biological modulation of methane release from localized marine sources is critical for understanding the transfer of methane from the ocean to the atmosphere as it relates to the oceanic contribution of greenhouse gasses and the global carbon budget.
The initial demonstration that certain bacteria utilize methane as a carbon and energy source occurred over a century ago (58), and interest in these bacteria has increased recently, as the relevance of methane to the global carbon budget and Earth's climate has become evident (24, 29). Among the Bacteria, aerobic methane oxidizers occur within the alphaproteobacteria (type II methanotrophs), within the gammaproteobacteria (type I methanotrophs and Crenothrix spp.) (11, 59), and within acidophilic members of the Verrucomicrobiales (5, 22, 48). Uncultured members of the Archaea have been shown to participate in methane oxidation in anoxic marine sediments, a discovery which significantly expands our understanding of the diversity and evolutionary history of microorganisms capable of C1 metabolism (1, 16, 36, 45).
Aerobic bacterial methanotrophs are ubiquitous; they are present in terrestrial and marine environments under physicochemical conditions ranging from temperate to extreme (11). Dissolved methane, copper, nitrate, oxygen levels, and moisture have been shown to influence the relative distributions of type I and type II methanotrophs (8, 11). Less is known regarding the factors influencing the activities and diversity of marine aerobic methanotrophs. Studies have documented aerobic methane oxidation in deep water above areas of active seafloor venting, with methane turnover times of up to 1.5 years (62). In pelagic environments with lower concentrations of methane, turnover times have been estimated at ∼50 years (50, 51, 56, 62). The microbial assemblages in the water column responsible for methane oxidization in this habitat have not been well characterized, although a few studies have begun to identify marine methanotrophic bacteria potentially involved in this process (6, 12). Cultivation efforts have previously recovered type I methanotrophs from the marine environment (30, 39, 57). Culture-independent studies typically either probe for methanotrophic 16S rRNA by fluorescence in situ hybridization or survey the particulate methane monooxygenase (pmoA) gene (21, 55, 63).
The pmoA gene is a well-established marker for surveying the methanotrophic potential within an environment. The protein it encodes is highly conserved across the bacterial domain and is central to aerobic methanotrophy (11). The gene also displays highly conserved regions at the 5′ and 3′ ends, facilitating the development of primers with broad utility (4, 17). The pmoA gene is a satisfactory proxy for establishing general phylogenetic relationships, because 16S rRNA and pmoA phylogenies show congruence at the genus level (14, 25). Environmental pmoA surveys have begun to determine the potential for bacterial aerobic methane oxidation in marine sediments.
Environments studied to date include the Kuroshima Knoll (21), methane-rich sediments in the Gulf of Mexico (66), the Haakon Mosby mud volcano (41), and East Pacific rise and Mid-Atlantic vent fields (40). These studies revealed diverse populations of type I methanotrophs inhabiting methane-rich ocean sediments. Microorganisms related to the type II methanotrophs have also occasionally been reported (9, 34). Fewer studies of bacterial methanotrophs and pmoA diversity in the marine water column are available, with one recent study of the Eastern Pacific oxygen minimum zone (OMZ) reporting limited diversity related to type I methanotrophs (12). In the Black Sea, aerobic and anaerobic methanotrophic assemblages appear to be stratified in the euxinic water column (6, 55, 63, 64). Comparative studies documenting the phylogenetic relationship between planktonic bacterial methanotrophs inhabiting marine water columns and those associated with marine methane seep sediments are lacking. Similarly, few studies have examined both 16S rRNA and pmoA diversity from deep-sea bacterioplankton (7).
In this study, we used a combination of pmo and 16S rRNA assays to characterize the diversity of aerobic methanotrophs associated with active seafloor methane venting in both the sediment and overlying water column from the Eel River Basin (ERB) (46) and above a methane vent in the Santa Monica Basin (SMB) (61) in order to gain insight into the relationships between methane-consuming microorganisms in different habitats within marine methane vent systems. Conventional pmoA surveys were expanded to include a new forward primer targeting a conserved region in the pmoC gene, thus addressing possible primer bias introduced by the conventional pmoA forward primer and improving the level of recovery of diverse monooxygenase sequences from the marine environment.
MATERIALS AND METHODS
Site description and sample collection.
Water column and sediment samples were obtained from two methane vent environments off the coast of California on two separate research cruises, conducted during February and July of 2005 using the remotely operated vehicle (ROV) Tiburon, operated from the research vessel Western Flyer. Water column samples were collected approximately 3 m above the seafloor in active methane venting using paired 5-liter Niskin bottles mounted on the ROV. In the SMB, two methane water column samples were collected on dive T-785 (33°47.9748′N, 118°38.796′W) at a water depth of ∼763 m from low-oxygen bottom waters (∼0.12 ml/liter) in a methane bubble plume emanating from a carbonate mound. The methane concentrations of the two samples were 4,980 and 5,340 nM, respectively.
Water column samples from the ERB were collected within a methane plume from the Northern Ridge (40°48.6893′N, 124°36.6770′W) at 518-m depth, with oxygen bottom water concentrations ranging between ∼0.52 and 0.55 ml/liter at a temperature of ∼5.7°C during dive T-864. Biomass from the water column at the ERB site was collected as part of an independent study, and corresponding water column methane values for these samples were not obtained.
Water column samples were transferred from Niskin bottles immediately after collection into clean Nalgene carboys and stored at 4°C until processed (within 3 h of collection). Water column samples (0.5 to 1.7 liters) were directly filtered under vacuum set at 5 mm Hg and collected onto a 47-mm Durapore filter (0.2 μm; Millipore) using a GF/F backing filter (Whatman). Filters were stored dry at −80°C until they were further processed in the laboratory.
Sediment samples from the Northern Ridge of the ERB (518-m water depth) were collected with a push core from actively venting sediments (“bubsed” sediments) and chemosynthetic clam beds (“clamsed” sediments) (46). Sediment samples were processed shipboard immediately after recovery. Push core samples were extruded upward at 2-cm intervals, and subsamples were collected using a sterile, 5-ml disposable syringe with the tip removed as reported previously (44). Samples were immediately frozen and maintained at −80°C until they were further processed in the laboratory.
Water column methane analysis.
Methane concentration in the collected waters from the SMB was measured shipboard. For each sample, a 240-ml amber glass bottle was filled to overflowing and sealed with a polytetrafluoroethylene-silicon septum and screw cap. A 10-ml high-purity nitrogen headspace was added to the bottle and was equilibrated for 15 min using a commercial paint shaker. Methane concentration in this headspace was obtained shipboard using a Shimadzu mini-2 gas chromatograph (GC) equipped with a flame ionization detector. Methane was separated isothermally from other gases in a high-purity nitrogen carrier gas stream using a 5-ft by 1/8-in. Outer diameter stainless chromatographic column packed with 60/80-mesh Carbosieve G (Supelco, Bellefonte, PA). The oven temperature was 100°C; the detector temperature was 125°C. Gas samples were injected into the GC via a small-volume magnesium perchlorate drying trap in series with a 2-ml stainless steel sample loop. A primary methane standard (9.93 ppm in nitrogen) was run approximately every 10th sample. A 60-ml aliquot of nitrogen carrier gas was used to flush residual methane from the GC sample loop between every sample. This method has a detection limit of ∼0.5 nM.
DNA extraction.
Water column filters were cut in half using a sterile razor blade; placed into 2 ml of filter-sterilized, sucrose-based lysis buffer (0.73 mM sucrose, 40 mM EDTA, and 50 mM Tris, pH 8.3); and gently homogenized using a sterile pestle. Cells were lysed by chemical and enzymatic methods, including a lysozyme treatment (2 mg in 40 μl) with incubation at 37°C for 45 min, followed by a proteinase K treatment (1 mg in 100 μl lysis buffer) and the addition of 100 μl of 20% sodium dodecyl sulfate, and incubated at 55°C for 2 h with gentle agitation.
Samples were then extracted using a standard phenol-chloroform method (pH 8.0) (53). Extracted DNA was cleaned and purified by Centricon dialysis exchange with Tris-EDTA (Millipore, Billerica, MA). DNA from frozen sediment aliquots (0.5 g) from the upper 0- to 2-cm sediment horizon associated with active methane venting (T864 PC-55) and a chemosynthetic clam bed (T864 PC-51) from the same area was extracted using the Ultraclean soil DNA kit (Mo Bio Laboratories Inc., Carlsbad, CA).
Design of new pmo primers (pmoC617f, wcpmoA189f, and wcpmoA661r).
Available sequences from the 3′ ends of pmoC and amoC were aligned, and a new forward primer, pmoC617f (5′-ACACCTTCTGGTTCATGG-3′), was designed from this region. This primer was tested for its ability to amplify target sequences from genomic DNA from type I and type II methanotrophs, including Methylosinus trichosporium OB3b, Methylocystis parvus OBBP, Methylomicrobium album BG8, Methylosarcina lacus LW14, Methylosarcina fibrata, Methylomonas sp. strain LW13, Methylomonas methanica S1, and Methylomicrobium agile. The amo-containing nitrosifier Nitrosomonas europaea was also included in these amplifications. In combination with the primer pmoA682r, all genomic DNAs except Methylomicrobium agile served as effective templates for the primer pmoC617f. In combination with the primer mb661r, all genomic DNAs except Nitrosomonas europaea and Methylocystis parvus OBBP served as effective templates for the primer pmoC617f.
Recovered pmoA sequences from the water column that included native sequences at priming sites 189 and 661 were examined and found to contain mismatches to conventional primers (described in Results). Based on these recovered sequences, water column-specific pmoA primers (wcpmoA189f and wcpmoA661r) were designed for amplifying the amplicon from priming sites 189 to 661.
PCR.
Primers and cycling parameters used in this study are summarized in Table 1. Final reaction mixture concentrations for all amplifications include 1× AmpliTaq gold buffer, a 0.5 μM concentration of each primer, a 200 μM concentration of the deoxynucleoside triphosphates, and 1 unit AmpliTaq Gold (Applied Biosystems, Foster City, CA). Amplifications geared toward library construction were performed in triplicate (20 μl, each sample) to reduce sampling bias, and samples were pooled prior to downstream analysis. For all amplifications, a 3-minute denaturation step (94°C) was included at the beginning of the PCR. This step was followed by 25 to 32 cycles using the annealing temperatures indicated in Table 1, and a 5-minute extension step (72°C) was included at the end of the reaction. Reaction mixtures were held at 4°C until they could be further manipulated.
TABLE 1.
PCR primers and amplification conditions used in this study
| Amplification | Primer set | Primer | Primer sequence | Annealing temp (°C) (no. of cycles) | Reference(s) |
|---|---|---|---|---|---|
| pmo strategy 1 | pmoA189f | 5′-GGNGACTGGGACTTCTGG-3′ | 56 (28) | 4 | |
| mb661r | 5′-CCGGMGCAACGTCYTTACC-3′ | ||||
| pmo strategy 2 | 1 | pmoC617f | 5′-ACACCTTCTGGTTCATGG-3′ | 50 (25) | This study |
| (seminested) | pmoA682r | 5′-GAASGCNGAGAAGAASGC-3′ | 4, 18 | ||
| 2 | pmoC617f | 5′-ACACCTTCTGGTTCATGG-3′ | 50 (25) | This study | |
| pmoA661r | 5′-CCGGMGCAACGTCYTTACC-3′ | 4 | |||
| pmo strategy 3 | pmoC617f | 5′-ACACCTTCTGGTTCATGG-3′ | 48 (26) | This study | |
| pmoA682r | 5′-GAASGCNGAGAAGAASGC-3′ | 18 | |||
| Water column-specific | wcpmoA189f | 5′-GGNGACYGGGATTTCTGG-3′ | 56 (28) | This study | |
| pmoA primers | wcpmoA661r | 5′-CAGGMGCAACGTCYTTACC-3′ | |||
| 16S rRNA genes | 27f | 5′-AGAGTTTGATCCTGGCTCAG-3′ | 54 (27) | 28 | |
| 1492r | 5′-GGTTACCTTGTTACGACTT-3′ | ||||
| MethT1df | 5′-CCTTCGGGMGCYGACGAGT-3′ | 55 (32) | 69 | ||
| MethT1br | 5′-GATTCYMTGSATGTCAAGG-3′ | ||||
| pmoA, ERBWC_3B | pmoA189f | 5′-GGNGACTGGGACTTCTGG-3′ | 56 (30) | This study | |
| novelmoA634r | 5′-CTATGATGCGCAGATATTCTGG-3′ | 4 | |||
| pmoB, ERBWC_3B | 1 | novelmoA196f | 5′-GGACTTTTGGGTCGATTGGAA-3′ | 61 (30) | This study |
| (seminested) | novelmoB448r | 5′-CNGGYCCNRYDAKNGGNCC-3′ | |||
| 2 | novelmoA281f | 5′-GGCTATTTTCTATGCTCATTTTCG-3′ | 64 (28) | This study | |
| novelmoB448r | 5′-CNGGYCCNRYDAKNGGNCC-3′ |
For amplifications of pmo genes, primers pmoC617f, pmoA189f, mb661r, pmoA682r, wcpmoA189f, and wcpmoA661r were used (Table 1). To amplify pmo sequences for library construction, three different priming strategies were used in order to address potential primer bias (Table 1). Small-subunit rRNA gene sequences were amplified using either primer pair 27f-1492r (27) or the methanotroph-targeted primer pair MethT1dF-MethT1bR (65). Sequence corresponding to the diverged monooxygenase-like clone from the water column was screened directly with primers pmoA189f and novelmoA634r. Additional downstream sequence corresponding to this clone was recovered by a seminested strategy, using primers novelmoA196f and novelmoA281f sequentially with the degenerate reverse primer novelmoB448r. A 1-kilobase band from the first round was gel cleaned with Qiaquick (Qiagen, Valencia, CA) prior to the second, seminested round.
Cloning.
To prepare PCR products for cloning, residual primers and primer dimers were removed from reaction mixtures using Montage size exclusion plates (Millipore, Billerica, MA) under a 10-lb/in2 vacuum for 10 minutes. DNA was resuspended in 30 μl 10 mM Tris (pH 8.0), and a 5-μl aliquot was analyzed for yield and purity by 1% agarose gel electrophoresis. Cleaned PCR products of the correct size were cloned into the pGEM T-easy vector (Promega, Madison WI). Approximately 20 ng of the PCR product was incubated with 50 ng pGEM t-easy in a total volume of 10 μl and allowed to ligate overnight according to the manufacturer's instructions. One-fifth of the ligation was transformed into high-efficiency competent JM109 cells (Promega, Madison WI) and plated on indicator LB-ampicillin agar plates. For all amplification strategies, 96 random clones were initially screened, and clones containing inserts of the correct size were selected for additional analysis. For pmo amplification strategies 1 and 2, the number of clones containing inserts of the correct size ranged from 63 to 94. For pmo amplification strategy 3, a high background resulted in fewer clones containing inserts of the correct size, and 30 clones were obtained for further analysis from each environment. For general bacterial 16S rRNA cloning, 89 (SMB water column [SMBWC]) or 91 (ERB water column [ERBWC]) clones had inserts of the correct size. For methanotroph-targeted 16S rRNA cloning, 72 (SMBWC) or 73 (ERBWC) clones had inserts of the correct size. Clones selected for analysis were digested with RsaI, and unique restriction fragment length polymorphism (RFLP) patterns were selected for further analysis.
Sequencing.
16S rRNA and pmo clones containing unique RsaI RFLP patterns were selected for sequencing. When possible, two clones of each RFLP pattern were selected to ensure that RFLP patterning corresponded well with phylotype. Prior to the sequencing, M13 forward and reverse primers were used to amplify inserts. M13-amplified products were cleaned of residual primers using Montage size exclusion plates (Millipore, Billerica, MA) under a 10-lb/in2 vacuum for 10 minutes. DNA was resuspended in 30 μl of 10 mM Tris (pH 8.0), and yield and purity were gauged using agarose gel electrophoresis. Approximately 100 ng of DNA was used in each sequencing reaction mixture. Primers M13f, M13r, pmoA189f, and mb661r were used to generate double-stranded sequences for pmo fragments. M13f and M13r primers were used to sequence 16S rRNA clones. Sequencing reactions were performed with PCR products using the GenomeLab Dye Terminator cycle sequencing quick start kit (Beckman Coulter, Fullerton, CA), and then reaction mixtures were precipitated with glycogen and sodium acetate, resuspended in 40 μl of formamide, and run on a CEQ 8800 genetic analysis system (Beckman Coulter, Fullerton, CA). Bidirectional sequences were manually verified against chromatograms in Sequencher version 4.6 (Gene Codes, Ann Arbor, MI) and, in the case of 16S rRNA genes, exported into ARB for alignment using the fast aligner function (34); they were then manually checked against the secondary structure. Related 16S rRNA sequences were imported into ARB from GenBank and aligned. The phylogenetic relationships of putative methanotroph 16S rRNA sequences to full-length reference sequences were determined in ARB using the parsimony insertion tool.
Determination of operational taxonomic units and phylogenetic analysis.
To compare sequences to the existing pmoA database, sequences were trimmed to the region from primers pmoA189f through mb661r (not inclusive; 471 base pairs). Sequences were translated in NCBI's open reading frame finder, and protein sequences were aligned in MacClade. Alignments were exported to PAUP and analyzed using neighbor-joining analysis. One thousand bootstrap analyses were performed to generate confidence levels for trees. Sequences that were greater than 90% identical at the amino acid level, as supported by bootstrap analysis, were considered to be within the same taxonomic clade. The pmoA phylogeny was generated in PAUP v4.0b10 (60) using the Kimura two-parameter model to estimate evolutionary distance. Maximum parsimony and neighbor-joining analyses were conducted with heuristic searches using 100 and 5,000 nonparametric bootstrap replicates, respectively, to assign confidence levels to nodes. Total character difference was used to estimate evolutionary distance, and a neighbor-joining analysis was conducted using 5,000 nonparametric bootstrap replicates to assign confidence levels to nodes.
Nucleotide sequence accession numbers.
Sequences identified in this study were deposited in GenBank under accession numbers EU444837 to EU444876.
RESULTS
The diversity of methanotrophic bacteria above methane vents and within vent sediments and adjacent chemosynthetic clam bed sediments was assessed using a series of previously published and newly designed pmo-targeted primers. Methanotroph-specific 16S rRNA-targeted primers (65) were also applied to a subset of samples analyzed in this study. Two active methane vent regions located approximately 1,000 km from each other off the coast of California were characterized: an organic-rich sediment-dominated methane vent and seep area within the ERB (46) and a recently discovered methane-derived authigenic carbonate mound in the SMB (13, 61). The diversity of water column methanotrophs was determined at both sites, and the diversity of sediment-associated methanotrophs was assessed at the ERB.
Diversity estimates of particulate methane monooxygenase genes in the water column are based on three independent PCR amplification strategies (Table 1). Strategy 1 employed the conventional primers pmoA189f and mb661r. Strategy 2 employed the newly designed pmoC617f primer in combination with mb661r. Strategy 3 paired pmoC617f with reverse primer pmoA682r, which is less stringent than mb661r and targets both amo and pmo gene sequences.
Benthic aerobic methanotroph diversity in the ERB methane seep system.
The diversity of sediment-associated pmoA sequences in two ecologically distinct methane seep habitats within the ERB was surveyed using amplification strategy 2 (Table 1). Surficial sediments (0 to 2 cm) from both actively venting sediments (bubsed) and bioturbated sediments from chemosynthetic clam beds (clamsed), were analyzed in this survey.
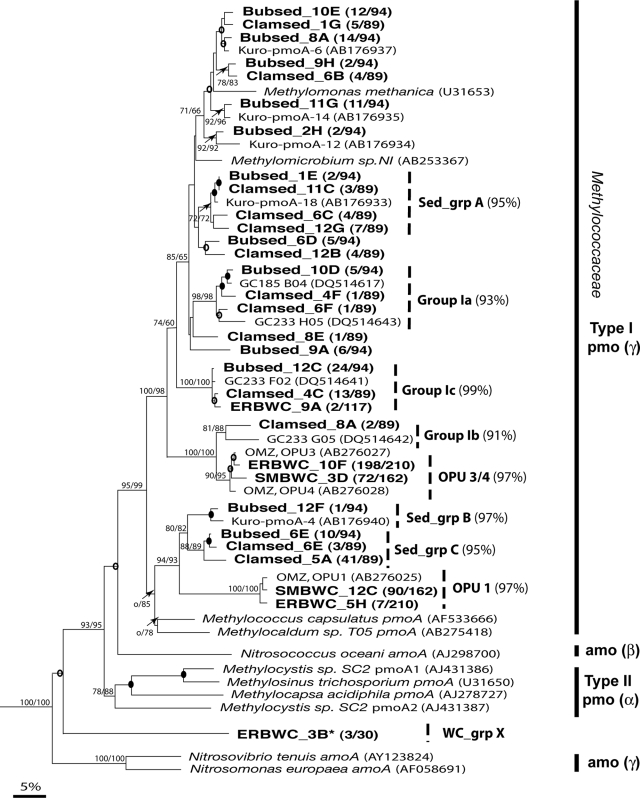
In total, 183 pmoA-containing clones were analyzed from these two sediment habitats and included 12 operational taxonomic units (OTUs) from the site of active venting and 13 OTUs from the clam bed. All recovered OTUs associated with type I methanotrophs belonging to diverse members of the Methylococcaceae and clustered with pmoA sequences recovered from other methane-rich sediment environments (21, 41, 66) (Fig. 1 and Table 2).
FIG. 1.
Amino acid phylogeny of predicted pMMOA sequences from methane seep systems. Confidence levels above 50% are shown at nodes on trees. Open circles represent bootstrap values between 50% and 70%; filled circles represent values above 70%. Sequences from surficial sediments (bubsed and clamsed) in the ERB methane seep system and from the ERBWC and SMBWC methane seep systems are indicated. Sediment clones are broadly distributed throughout the Methylococcaceae. The majority of water column clones cluster into two distinct groups of closely related phylotypes and are phylogenetically distinct from sediment-hosted pMMOA. Clones that are less than 88% identical at the nucleotide level to previously described environmental isolates are given new designations (Table 2). *, clone ERBWC_3B is substantially diverged from all previously described sequences of pMMO and Amo.
TABLE 2.
Summary of pmoA isolates from the SMB and ERB methane seep systems
| Reference clade and/or isolated | Water column
|
Sediment
|
Representative isolate(s) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SMBWC
|
ERBWC
|
Bubsed
|
Clamsed
|
||||||||
| No. of isolates/total tested | % Identity | No. of isolates/total tested | % Identity | No. of OTUs | No. of isolates/total tested | % Identity | No. of OTUs | No. of isolates/total tested | % Identity | ||
| OPU1 | 77/140 | 96-98 | 7/210 | 96-98 | SMBWC_12C | ||||||
| OPU3/4 | 63/140 | 96-98 | 198/210 | 96-98 | ERBWC_10F | ||||||
| WC_grpXa | 3/30b | 60-62c | ERBWC_3B | ||||||||
| Group Ia | |||||||||||
| GC185_BO4 | 1 | 5/94 | 98 | 1 | 1/89 | 93 | Bubsed_10D | ||||
| GC233_HO5 | 1 | 1/89 | 89 | Clamsed_6F | |||||||
| Group Ib, GC233_G05 | 1 | 2/89 | 85 | Clamsed_8A | |||||||
| Group Ic, GC233_F02 | 2/117 | 98 | 1 | 24/94 | 96-99 | 1 | 13/89 | 98 | Clamsed_4C | ||
| Sed_grp A, Kuro_pmoA_18 | 1 | 2/94 | 96 | 3 | 14/89 | 94-96 | Clamsed_11C | ||||
| Sed_grp B, Kuro_pmoA_4 | 1 | 1/94 | 93 | Bubsed_12F | |||||||
| Sed_grp C, Kuro_pmoA_4 | 1 | 10/94 | 85 | 2 | 44/89 | 82-83 | Bubsed_6E | ||||
| Clamsed_5A | |||||||||||
| Unassigned | |||||||||||
| Kuro_pmoA_6 | 3 | 28/94 | 87-92 | 2 | 9/89 | 90 | Clamsed_6B | ||||
| Bubsed_10E | |||||||||||
| Kuro_pmoA_12 | 1 | 2/94 | 86-88 | Bubsed_2H | |||||||
| Kuro_pmoA_14 | 1 | 11/94 | 93 | Bubsed_11G | |||||||
| Kuro_pmoA_18 | 1 | 5/94 | 89 | 1 | 4/89 | 82 | Bubsed_6D | ||||
| Clamsed_12B | |||||||||||
| Methylomicrobium sp. strain NI | 1 | 6/94 | 82 | 1 | 1/89 | 80 | Bubsed_9A | ||||
| Clamsed_8E | |||||||||||
WC_grpX (isolate ERBWC_3B) could be detected in both water column environments when it was screened directly.
This sequence was recoverable only with strategy 3 (Table 1).
Percent identity relative to the best BLAST hits (uncultured organisms, GenBank accession no. AY177951 and EU193295).
Groups Ia to Ic are defined in reference 66.
Planktonic aerobic methanotroph diversity above the ERB methane seep and above the SMB methane vent.
The diversity of planktonic pmoA sequences in two vent-associated methane-rich water columns (designated ERBWC and SMBWC) was also examined, using amplification strategies 1, 2, and 3 (Table 1). For either site, the same general methanotrophic-population profiles were recovered regardless of amplification strategy used, illustrating that primer pmoC617f surveys diversity comparably to pmoA189f in these settings. In the ERBWC, a dominant type I methanotrophic phylotype (representing over 90% of ERBWC clone libraries) was consistently recovered. This phylotype is closely related (96 to 99% identity at the nucleotide level) to phylotypes OPU3 and OPU4 recovered from the Eastern Pacific OMZ (12). Less common phylotypes (representing less than 10% of ERBWC clone libraries) were also recovered from the ERB. One of these phylotypes is 96 to 99% identical at the nucleotide level to OPU1 reported from the Eastern Pacific OMZ (12). In the SMBWC, the same dominant phylotypes related to OPU1 and OPU3/4 were recovered, again regardless of amplification strategy. However, these phylotypes were present in proportions roughly equal to one another in the SMBWC libraries (Fig. 1; Table 2).
Two additional phylotypes were recovered in low abundance with pmoC617f (amplification strategies 2 and 3 in Table 1) but not with the conventional primer set (amplification strategy 1 in Table 1). These phylotypes included the recovery of a sequence (clone ERBWC_9A) highly similar to that of GC233_F02 (group Ic) recovered from Gulf of Mexico methane-rich sediments (66), as well as the recovery of a highly diverged sequence (clone ERBWC_3B) distantly related to all previously identified clades of pmo and amo genes available in GenBank (Fig. 1). This diverged sequence, which is further described below, was recovered with amplification strategy 3. Comparison of the diverse sequences recovered from amplification strategies 1, 2, and 3 suggests that the newly designed forward primer, pmoC617f, identified diversity in the water column that was missed with the conventional primer pmoA189f. Additionally, there was no obvious loss in recovered diversity through the use of the new forward primer in this environmental setting.
Isolation of a highly divergent monooxygenase sequence.
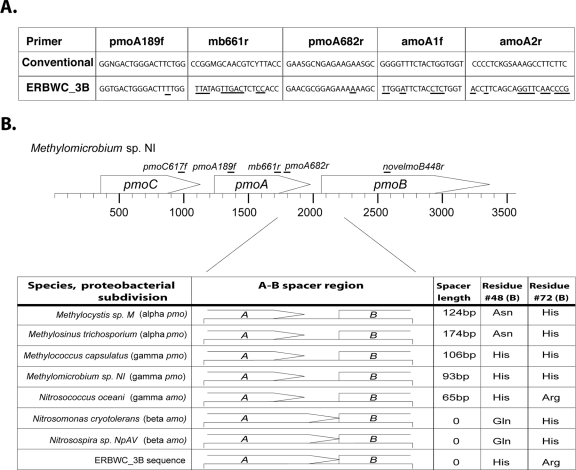
Amplification strategy 3, which amplifies sequence from the 3′ end of pmoC to the 3′ end of pmoA (pmoC617f to pmoA682r; ∼900 base pairs, depending on phylotype), led to the discovery of a highly divergent monooxygenase-like sequence phylogenetically positioned between Amo and pMMO sequences (isolate ERBWC_3B) (Fig. 1). This sequence does not have a canonical site 189 or site 682, nor does it have a recognizable site 661. Thus, it would not be easily amplified with conventional pmoA primers (2, 4). Furthermore, this sequence does not provide a good match for the traditional ammonia monooxygenase (amo) primers, amoA1f and amoA2r, indicating that it would not be identified in typical molecular screens for amoA (Fig. 2A).
FIG. 2.
Sequence information from diverged monooxygenase clone ERBWC_3B. Numbering corresponds to the M. capsulatus protein sequence. (A) Base pair mismatches to conventional primers in the diverged monooxygenase sequence, illustrating that the sequence would not easily be detected with conventional priming strategies. (B) Predicted open reading frames reveal the lack of a noncoding spacer region between the presumptive A and B genes in this operon. This arrangement has been documented for betaproteobacterial amo operons but not for gammaproteobacterial amo operons or for pmo operons. Two amino acid residues (residues 48 and 72 in the presumptive B protein) are implicated in mononuclear copper coordination and correlate with enzyme function in previously reported sequences from Amo and pMMO.
Additional sequence information for this genetic unit was obtained to gain potential insight into its subdivision affiliation and function. A nested amplification with forward primers specific to the diverged water column sequence (novelmoA196f and novelmoA281f), in combination with a degenerate reverse primer targeting a conserved region within the B gene (novelmoB448r), was used to recover sequence data from the subunit B gene. Portions of this sequence are summarized in Fig. 2B.
Phylogenetic analysis of the recovered sequence reveals comparable levels of similarity to both the Amo sequence and the pMMO sequence (50% identity and 70 to 71% similarity at the amino acid level to either Amo or pMMO). The operon structure between the A and B ammonia and particulate methane monooxygenase genes, including the length of the A gene as well as the arrangement of the stop codon for the A gene relative to the start codon for the B gene, may be informative (43). The A protein from the diverged sequence is predicted to be ∼20 amino acids longer than those of previously described pMMOA representatives and of a length that is consistent with betaproteobacterial AmoA. Likewise, the recovered sequence predicts that the A and B genes overlap by 1 nucleotide, which mirrors the arrangement of the corresponding codons in Betaproteobacteria rather than in Gamma- or Alphaproteobacteria (43). However, an examination of several predicted subunit B residues does not suggest a typical putative Amo mononuclear copper coordination site (Fig. 2B) (31). The diverged monooxygenase sequence does not conform to any one previously described Amo or pMMO clade, using these metrics.
We designed a reverse primer specific to this unusual sequence (novelmoA634r) (Table 1) and screened for the sequence directly in both water column environments as well as in the ERB methane vent sediments. The diverged sequence was recovered from both SMB and ERB benthic water columns, but not from sediment samples, with 99% sequence identity observed between the two water column environments (data not shown). The successful detection of this sequence in two geographically distant locations suggests that this sequence may be broadly distributed within water columns from regions of active methane venting. However, a survey of available environmental metagenomic data recovered from pelagic marine environments (52) did not reveal sequences related to this novel gene, and it is possible that this monooxygenase sequence derives from an organism inhabiting deep-water habitats associated with methane vents. Direct sequence comparisons to pmoA genes from Verrucomicrobia (5, 48), Crenothrix (59), and the recently isolated methylotrophic marine Betaproteobacteria strain HTCC2181 (S. Giovannoni, personal communication) also did not produce any significant matches.
Improved molecular assay for the detection of water column methanotrophs.
Analysis of pmoA sequences recovered with amplification strategies 2 and 3 reveals that methane vent-associated aerobic methanotrophs encode mismatches to primers pmoA189f and mb661r. These primers were designed from terrestrial pmoA and amoA sequences and may not be optimized for marine settings (17). Analysis of the pmoC617f-mb661r and pmoC617f-A682r sequence fragments recovered in this study indicate that 100% of water column clones and 90% of sediment clones have a mismatch to primer pmoA189f, encoding 5′-GGNGACTGGGATTTCTGG-3′ (where underlining indicates the mismatch). Furthermore, 40% of water column clones (but not sediment clones) have a second mismatch to pmoA189f, encoding 5′-GGNGACCGGGATTTCTGG-3′. Additionally, 90% of water column clones have a mismatch to primer mb661r, encoding 5-′CAGGMGCAACGTCYTTACC-3′. Primers wcpmoA189f and wcpmoA661r were designed with these modifications, and the amplification efficiency of the modified primers was tested in parallel with those of the conventional primers on DNA from the SMBWC and ERBWC. The modified primers provide a substantial enhancement (more than twofold) in product yield (data not shown). Although the conventional primers are capable of amplifying pmoA phylotypes from the water columns, the modifications reported here may provide more-sensitive assays for pmoA screens in marine environments.
16S rRNA gene analysis of water column methanotrophs.
rRNA gene amplifications with the general bacterial 16S rRNA primers 27f and 1492r (27) were conducted in order to further describe potential methanotrophic lineages; however, these rRNA-based surveys from ERBWC and SMBWC failed to recover sequences affiliated with known methanotrophs. Bacterial sequences recovered included those of Colwellia spp., Alcanivorax spp., members of the Bacteroidetes, Erythrobacter spp., Pseudomonas spp., Pseudoalteromonas spp., Moritella spp., members of the Chloroflexi, Nitrospina spp., members of the Verrucomicrobia, and Shewanella spp. (data not shown).
Subsequent amplifications using primers MethTIdf and MethTIbr (65), which specifically target type I methanotrophic 16S rRNA genes, also primarily identified nonmethanotrophic sequences, including species of the genera Ectothiorhodospira, Alcanivorax, Halomonas, and Marinobacter, although in this screen, some sequences that are moderately related to Methylococcaceae were also recovered. Nine percent of the recovered sequences, representing 4 OTUs, were ∼90 to ∼91% identical to Methylococcus capsulatus, and one recovered sequence, representing 3% of the clone library, demonstrated 96% relatedness to an uncultured putative methanotroph from profundal sediments in Lake Constance (49). This degree of relatedness is consistent with that seen in the pmo phylogeny (Fig. 1), and these 4 OTUs may represent methanotrophic 16S rRNA sequences in marine water columns. However, the slight similarity to previously described cultured methanotrophs precludes specific taxonomic assignment.
DISCUSSION
Identification of the organisms involved in aerobic methanotrophy within and above methane vents and seeps is a fundamental requirement for a complete understanding of the methane cycle in these environments and for evaluating the potential effectiveness of the biological methane filter in the ocean (15). Within the past decade, significant emphasis has been placed on determining the diversity and activity of microorganisms involved in the anaerobic oxidation of methane, estimated to consume up to 80% of the total methane flux from marine sediments (15, 50). Yet, comparably little is known about the diversity of microorganisms catalyzing aerobic methane oxidation in water columns. On areas of the seafloor where the methane flux through the sediments exceeds the capacity of the anaerobic biological filter, for example, in continuously venting systems, such as mud volcanoes (37, 41) or geothermal gas vents (18, 26, 35), these aerobic methanotrophs may serve as a significant sink for methane, precluding its release to the atmosphere (33). In an earlier study focused on quantifying the activity and magnitude of water column aerobic methane oxidation in the ERB region, elevated water column methane concentrations (maximum, ∼200 nM) and significant rates of methanotrophy were reported in the deep-water (∼370-m) column (50).
The SMB and the ERB methane vent and seep systems are similar in many respects. Both lie along the North American continental margin between 500 and 700 m of water depth and are sites of active methane venting into the water column. Benthic bottom waters are dysoxic (dissolved oxygen, <0.6 ml/liter) and are at stable, low temperatures of approximately 6°C. The SMB carbonate mound is covered with a thin sediment drape and bacterial mats and houses at least two active methane bubble vents (42, 61). In contrast, the ERB site receives significant terrigenous input from organic-material-rich sediments from the Eel River. Although there may be contrasting amounts of sediment cover on the seafloor and differences in seafloor morphology at the study sites, the diversity of the water column aerobic-bacterium methanotroph clades appears not to be sensitive to these gross differences.
This study has contributed additional molecular assays to assess environmental methanotroph diversity. Previous pmoA screens have routinely relied on the forward primer pmoA189f designed from, and applied to, terrestrial and freshwater environments (4, 14, 19). To address the concern that primer pmoA189f may impose constraints on the recovery of diverse organisms in the marine environment, we designed and evaluated a new forward primer (pmoC617f) that lies at the 3′ end of pmoC. The use of pmoC617f revealed that marine methanotrophs encode mismatches to primer pmoA189f at priming site 189, which may impact amplification efficiency. However, despite these mismatches, the predominant patterns in diversity and clone library abundance were similar for both water column sites regardless of the forward primer used.
In this study, the functional metabolic gene for aerobic methanotrophy, pmoA, in methane-rich marine environments was probed using conventional and newly designed PCR priming strategies. Our results demonstrate that although the methane vents examined here are over 1,000 km apart, the water columns overlying these habitats host similar methanotrophic-bacterial species that fall into only a few taxonomic groups. The major groups identified in the water columns cluster with organisms of phylotypes OPU1 and -3 recovered from the Eastern Pacific OMZ (12). Phylogenetic comparison of the closest cultured relatives of the planktonic pmoA genes suggests that they are affiliated with the family Methylococcaceae. The recovery of similar planktonic pmoA phylotypes from both methane seep/vent water columns and the OMZ, all with oxygen levels below 0.6 ml/liter but highly varied methane concentrations from 10 nM to 5 μM, suggests a widespread distribution of these as-yet-uncultured methanotrophs in the water column and their adaptation to a range of methane concentrations. It remains to be determined if similar methanotrophic phylotypes are also associated with regions of higher oxygen tension or if dysoxia is a defining feature of the pmoA phylotypes recovered from these deep-water environments.
In contrast to the limited pmoA diversity recovered from methane-rich waters, substantial pmoA diversity was observed in the surficial sediment from the ERB. Furthermore, sediment-associated sequences were more closely related to previously described methanotrophic genera than to those found in the water column. Both clam bed and vent sediment habitats in the ERB displayed the same general pattern of broad diversity, differing primarily in the relative abundances of specific phylotypes (Table 2). These differences in microbial population profile between sediment sites may be a direct result of the variance in gross geochemical features, such as oxygen tension and methane partial pressures, or may be influenced by sulfide levels which may be toxic to bacterial methanotrophs. This physicochemical heterogeneity within the sediment matrix and corresponding microenvironments likely contributes to the increased diversity in the methanotrophic pmoA phylotypes recovered.
The findings of this study demonstrate that there is very little overlap between the diverse methanotrophs recovered from sediment and those of the water column in the ERB. Samples were collected concurrently; thus, this discrepancy was not due to temporal changes in community structure. It instead indicates that sediment-affiliated aerobic methanotrophs and planktonic methanotrophs from the overlying water column, in this case separated by only 3 m of distance, host distinct populations, as indicated by pmoA diversity. This illustrates that methane oxidation in the water column is mediated by a group of bacteria distinct from those that facilitate aerobic methanotrophy in the underlying sediment. This segregation in diversity is a strong indication that methanotroph sequences recovered from the deep-water column are not simply the result of sediment entrainment from vigorous methane advection and that sediment methanotrophs are not the product of the settling of planktonic methanotrophs.
Analysis of 16S rRNA genes in the water column reveals potential methanotroph sequences that are significantly diverged from previously described type I methanotrophs, with identities to methanotrophic 16S rRNA sequences ranging from 89 to 96%. The assessment of methanotrophic 16S rRNA genes in the water column was less conclusive than our pmoA gene surveys, despite the use of a specific, methanotroph-targeted 16S rRNA primer set. However, the moderate 16S rRNA similarity to Methylococcus and Methylomonas, both of which are within the family Methylococcaceae, is congruent with the diversity observed in pmoA phylotypes from the water column, and it is possible that these sequences are associated with previously unrecognized planktonic methanotrophs. Follow-up studies employing methane incubations, stable isotope probing, and/or microfluidic digital PCR (47) may provide a more direct link between these diverged vent-associated pmoA genes and their corresponding 16S rRNA ribotypes.
New priming strategies used in this study facilitated the identification of a highly diverged monooxygenase-like sequence from the water column. This putative monooxygenase encodes a gene order of C-A-B, suggesting membership in the bacterial domain and not in the ubiquitous, ammonia-oxidizing planktonic and soil Crenarchaeota, which have an amo operon structure of B-C-A (10, 54). Phylogenetic analysis of this sequence (Fig. 1) demonstrates that it is not typical of an established pmo or amo bacterial clade, and thus no firm conclusions can be made regarding subdivision affiliation or function. Potential insight into lineage and function may be suggested through analysis of signature features in the sequence, however.
Two predicted amino acids in the diverged sequence (B48, histidine, and B72, arginine) are characteristic of gammaproteobacterial nitrosifying bacteria (Fig. 2B). Residues B48 and B72 are highly conserved among Amo and pMMO sequences and may potentially participate in copper coordination (31), and their identities correlate with function and subdivision affiliation. Only the gammaproteobacterial Amo from Nitrosococcus oceani contains a histidine (H) at position 48 and an arginine (R) at position 72 (Fig. 2B), mirroring the identities found in the diverged sequence and suggesting possible membership among the Gammaproteobacteria and a potential ammonia-oxidizing role for this diverged sequence. Alternatively, the gross operon structure of the diverged sequence may suggest affiliation with the betaproteobacterial subdivision. Specifically, among proteobacterial subdivisions harboring pmo or amo, there is currently an absolute correspondence between the existence of a noncoding spacer between the A and B genes and affiliation with the alpha- or gammaproteobacterial subdivisions of proteobacteria (43). This noncoding spacer is found in all pmo operons (members of the Alpha- and Gammaproteobacteria) as well as in the gammaproteobacterial amo operon from Nitrosococcus oceani. In contrast, betaproteobacterial-subdivision ammonia oxidizers do not have a spacer between the A and B genes; instead, the stop codon for the A gene overlaps the start codon for the B gene (Fig. 2B) (43). The diverged sequence from the water column exhibits the lack of noncoding spacer and overlapping codon arrangement typical of the betaproteobacterial subdivision (Fig. 2B). However, the highly diverged nature of this sequence suggests that it is not a typical betaproteobacterial Amo protein. The possibility that the sequence may represent a betaproteobacterial methane monooxygenase sequence is worthy of some consideration. Although specific pmo operons have not yet been identified from these groups, recent indications that Betaproteobacteria are capable of methylotrophy and are possibly involved in methane oxidation may suggest a role for these organisms in methane cycling (20, 23, 32, 38).
Conclusions.
The diversity of pmoA genes in the water column and sediment revealed that discrete methanotrophic populations are present within the sediments and overlying water column from dysoxic, methane-rich vent and seep systems. pmo diversity in the water column appears restricted to a small number of moderately diverged clades within the Methylococcaceae, whereas sediment methanotrophs are less diverged and span numerous genera within the Methylococcaceae. The greater diversity recovered with the newly designed molecular assays in this study highlights our limited understanding of the breadth of the diversity of aerobic bacterial methanotrophs found within these deep-sea habitats. A highly divergent member of the amo/pmo superfamily was identified in both methane vent water columns examined but was not recovered from seep sediments, indicating the potential for future insight into methane and ammonia oxidation and insight into the evolution of monooxygenase genes, with further study of these poorly understood environments.
Acknowledgments
We are grateful to M. Lidstrom and M. Kalyuzhnaya (University of Washington) and D. Arp and N. Hommes (Oregon State University) for providing reference strains and S. Goffredi, S. Ussler, and A. Bures for technical assistance with various aspects of this work. We also thank S. Goffredi, D. Fike, C. Gammon, and A. Dekas for critical reading of the manuscript and constructive insight. We are indebted to the pilots of the ROV Tiburon and the crew and shipboard research party of the research vessel Western Flyer on a cruise sponsored by NOAA-NURP in 2005. We also thank three anonymous reviewers for their comments.
This work was made possible by grants from NASA (NNG06GB34G), NOAA (UAF 05-0132), the National Science Foundation (MCB-0348492), and the Gordon and Betty Moore Foundation (V.J.O.) and by the support provided to MBARI by the David and Lucile Packard Foundation (W.U.).
Footnotes
Published ahead of print on 16 May 2008.
REFERENCES
- 1.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 2.Bourne, D. G., I. R. McDonald, and J. C. Murrell. 2001. Comparison of pmoA PCR primer sets as tools for investigating methanotroph diversity in three Danish soils. Appl. Environ. Microbiol. 67:3802-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark, J. F., L. Washburn, J. S. Hornafius, and B. P. Luyendyk. 2000. Dissolved hydrocarbon flux from natural marine seeps to the southern California Bight. J. Geophys. Res. Oceans 105:11509-11522. [Google Scholar]
- 4.Costello, A. M., and M. E. Lidstrom. 1999. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunfield, P. F., A. Yuryev, P. Senin, A. V. Smirnova, M. B. Stott, S. Hou, B. Ly, J. H. Saw, Z. Zhou, Y. Ren, J. Wang, B. W. Mountain, M. A. Crowe, T. M. Weatherby, P. L. Bodelier, W. Liesack, L. Feng, L. Wang, and M. Alam. 2007. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450:879-882. [DOI] [PubMed] [Google Scholar]
- 6.Durisch-Kaiser, E., L. Klauser, B. Wehrli, and C. Schubert. 2005. Evidence of intense archaeal and bacterial methanotrophic activity in the Black Sea water column. Appl. Environ. Microbiol. 71:8099-8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elsaied, H. E., T. Hayashi, and T. Naganuma. 2004. Molecular analysis of deep-sea hydrothermal vent aerobic methanotrophs by targeting genes of 16S rRNA and particulate methane monooxygenase. Mar. Biotechnol. (New York) 6:503-509. [DOI] [PubMed] [Google Scholar]
- 8.Graham, D. W., J. A. Chaudhary, R. S. Hanson, and R. G. Arnold. 1993. Factors affecting competition between type-I and type-II methanotrophs in 2-organism, continuous-flow reactors. Microb. Ecol. 25:1-17. [DOI] [PubMed] [Google Scholar]
- 9.Guezennec, J., and A. Fiala-Medioni. 1996. Bacterial abundance and diversity in the Barbado Trench determined by phospholipid analysis. FEMS Microbiol. Ecol. 19:83-93. [Google Scholar]
- 10.Hallam, S. J., T. J. Mincer, C. Schleper, C. M. Preston, K. Roberts, P. M. Richardson, and E. F. DeLong. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi, T., H. Obata, T. Gamo, Y. Sano, and T. Naganuma. 2007. Distribution and phylogenetic characteristics of the genes encoding enzymes relevant to methane oxidation in oxygen minimum zones of the eastern Pacific Ocean. Res. J. Environ. Sci. 1:275-284. [Google Scholar]
- 13.Hein, J. R., W. R. Normark, B. R. McIntyre, T. D. Lorenson, and C. L. Powell. 2006. Methanogenic calcite, C-13-depleted bivalve shells, and gas hydrate from a mud volcano offshore southern California. Geology 34:109-112. [Google Scholar]
- 14.Heyer, J., V. F. Galchenko, and P. F. Dunfield. 2002. Molecular phylogeny of type II methane-oxidizing bacteria isolated from various environments. Microbiology 148:2831-2846. [DOI] [PubMed] [Google Scholar]
- 15.Hinrichs, K.-U., and A. Boetius. 2002. The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistry, p. 457-477. In G. Wefer, D. Billett, D. Hebbeln, B. B. Jørgensen, M. Schlüter, and T. Van Weering (ed.), Ocean margin systems. Springer-Verlag, Heidelberg, Germany.
- 16.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 17.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 18.Hornafius, J. S., D. Quigley, and B. P. Luyendyk. 1999. The world's most spectacular marine hydrocarbon seeps (Coal Oil Point, Santa Barbara Channel, California): quantification of emissions. J. Geol. Res. Oceans 104:20703-20711. [Google Scholar]
- 19.Horz, H. P., M. T. Yimga, and W. Liesack. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchens, E., S. Radajewski, M. G. Dumont, I. R. McDonald, and J. C. Murrell. 2004. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 6:111-120. [DOI] [PubMed] [Google Scholar]
- 21.Inagaki, F., U. Tsunogai, M. Suzuki, A. Kosaka, H. Machiyama, K. Takai, T. Nunoura, K. H. Nealson, and K. Horikoshi. 2004. Characterization of C1-metabolizing prokaryotic communities in methane seep habitats at the Kuroshima Knoll, southern Ryukyu Arc, by analyzing pmoA, mmoX, mxaF, mcrA, and 16S rRNA genes. Appl. Environ. Microbiol. 70:7445-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Islam, T., S. Jensen, L. Reigstad, O. Larsen, and N. Birkeland. 2008. Methane oxidation at 55 degrees C and pH 2 by a thermoacidiphilic bacterium belonging to the Verrucomicrobium phylum. Proc. Natl. Acad. Sci. USA 105:300-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalyuzhnaya, M., P. De Marco, S. Bowerman, C. Pacheco, J. Lara, M. Lidstrom, and L. Chistoserdova. 2006. Methyloversatilis universalis gen. nov., sp. nov., a novel taxon within the Betaproteobacteria represented by three methylotrophic isolates. Int. J. Syst. Evol. Microbiol. 56:2517-2522. [DOI] [PubMed] [Google Scholar]
- 24.Kennett, J. P., K. G. Cannariato, I. L. Hendy, and R. J. Behl. 2000. Carbon isotopic evidence for methane hydrate instability during quaternary interstadials. Science 288:128-133. [DOI] [PubMed] [Google Scholar]
- 25.Kolb, S., C. Knief, S. Stubner, and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kvenvolden, K. A., and B. W. Rogers. 2005. Gaia's breath—global methane exhalations. Mar. Petrol. Geol. 22:579-590. [Google Scholar]
- 27.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY.
- 28.Lelieveld, J., P. J. Crutzen, and C. Bruhl. 1993. Climate effects of atmospheric methane. Chemosphere 26:739-768. [Google Scholar]
- 29.Lelieveld, J., P. J. Crutzen, and F. J. Dentener. 1998. Changing concentration, lifetime and climate forcing of atmospheric methane. Tellus Ser. B 50:128-150. [Google Scholar]
- 30.Lidstrom, M. E. 1988. Isolation and characterization of marine methanotrophs. Antonie van Leeuwenhoek 54:189-199. [DOI] [PubMed] [Google Scholar]
- 31.Lieberman, R. L., and A. C. Rosenzweig. 2005. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature 434:177-182. [DOI] [PubMed] [Google Scholar]
- 32.Lin, X., S. G. Wakeham, I. F. Putnam, Y. M. Astor, M. I. Scranton, A. Y. Chistoserdov, and G. T. Taylor. 2006. Comparison of vertical distributions of prokaryotic assemblages in the anoxic Cariaco Basin and Black Sea by use of fluorescence in situ hybridization. Appl. Environ. Microbiol. 72:2679-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mau, S., D. L. Valentine, J. F. Clark, J. Reed, R. Camilli, and L. Washburn. 2007. Dissolved methane distributions and air-sea flux in the plume of a massive seep field, Coal Oil Point, California. Geophys. Res. Lett. 34:L22603. doi: 10.1029/2007GL031344. [DOI] [Google Scholar]
- 34.McDonald, I. R., K. Smith, and M. E. Lidstrom. 2005. Methanotrophic populations in estuarine sediment from Newport Bay, California. FEMS Microbiol. Lett. 250:287-293. [DOI] [PubMed] [Google Scholar]
- 35.McDonald, I. R., K. L. Warner, C. McAnulla, C. A. Woodall, R. S. Oremland, and J. C. Murrell. 2002. A review of bacterial methyl halide degradation: biochemistry, genetics and molecular ecology. Environ. Microbiol. 4:193-203. [DOI] [PubMed] [Google Scholar]
- 36.Michaelis, W., R. Seifert, K. Nauhaus, T. Treude, V. Thiel, M. Blumenberg, K. Knittel, A. Gieseke, K. Peterknecht, T. Pape, A. Boetius, R. Amann, B. B. Jorgensen, F. Widdel, J. Peckmann, N. V. Pimenov, and M. B. Gulin. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013-1015. [DOI] [PubMed] [Google Scholar]
- 37.Milkov, A. V., R. Sassen, T. V. Apanasovich, and F. G. Dadashev. 2003. Global gas flux from mud volcanoes: a significant source of fossil methane in the atmosphere and the ocean. Geophys. Res. Lett. 30:1037. doi: 10.1029/2002GL016358. [DOI] [Google Scholar]
- 38.Morris, S. A., S. Radajewski, T. W. Willison, and J. C. Murrell. 2002. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura, T., T. Hoaki, S. Hanada, A. Maruyama, Y. Kamagata, and H. Fuse. 2007. Soluble and particulate methane monooxygenase gene clusters in the marine methanotroph Methylomicrobium sp. strain NI. FEMS Microbiol. Lett. 277:157-164. [DOI] [PubMed] [Google Scholar]
- 40.Nercessian, O., N. Bienvenu, D. Moreira, D. Prieur, and C. Jeanthon. 2005. Diversity of functional genes of methanogens, methanotrophs and sulfate reducers in deep-sea hydrothermal environments. Environ. Microbiol. 7:118-132. [DOI] [PubMed] [Google Scholar]
- 41.Niemann, H., T. Losekann, D. de Beer, M. Elvert, T. Nadalig, K. Knittel, R. Amann, E. J. Sauter, M. Schluter, M. Klages, J. P. Foucher, and A. Boetius. 2006. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443:854-858. [DOI] [PubMed] [Google Scholar]
- 42.Normark, W. R., J. R. Hein, C. L. Powell II, T. D. Lorenson, H. J. Lee, and B. D. Edwards. 2003. Methane hydrate recovered from a mud volcano in Santa Monica Basin, offshore southern California, abstr. OS51B-0855. EOS Trans. Am. Geophys. Union 84 (Fall Meet. Suppl.)
- 43.Norton, J. M., J. J. Alzerreca, Y. Suwa, and M. G. Klotz. 2002. Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch. Microbiol. 177:139-149. [DOI] [PubMed] [Google Scholar]
- 44.Orphan, V. J., K. U. Hinrichs, W. Ussler III, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. Delong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 46.Orphan, V. J., W. Ussler III, T. H. Naehr, C. H. House, K. U. Hinrichs, and C. K. Paull. 2004. Geological, geochemical, and microbiological heterogeneity of the seafloor around methane vents in the Eel River Basin, offshore California. Chem. Geol. 205:265-289. [Google Scholar]
- 47.Ottesen, E. A., J. W. Hong, S. R. Quake, and J. R. Leadbetter. 2006. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science 314:1464-1467. [DOI] [PubMed] [Google Scholar]
- 48.Pol, A., K. Heijmans, H. R. Harhangi, D. Tedesco, M. S. Jetten, and H. J. Op den Camp. 2007. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450:874-878. [DOI] [PubMed] [Google Scholar]
- 49.Rahalkar, M., and B. Schink. 2007. Comparison of aerobic methanotrophic communities in littoral and profundal sediments of Lake Constance by a molecular approach. Appl. Environ. Microbiol. 73:4389-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reeburgh, W. S. 2007. Oceanic methane biogeochemistry. Chem. Rev. 107:486-513. [DOI] [PubMed] [Google Scholar]
- 51.Rehder, G., R. S. Keir, E. Suess, and M. Rhein. 1999. Methane in the northern Atlantic controlled by microbial oxidation and atmospheric history. Geophys. Res. Lett. 26:587-590. [Google Scholar]
- 52.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrews-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y. H. Rogers, L. I. Falcon, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingham, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 54.Schleper, C. 2006. Diversity and ecology of archaea: perspectives from microbial ecology and metagenomics. In H. P. Klenk and R. Garrett (ed.), Archaea: evolution, physiology & molecular biology. Blackwell Publishing, Williston, VT.
- 55.Schubert, C. J., M. J. L. Coolen, L. N. Neretin, A. Schippers, B. Abbas, E. Durisch-Kaiser, B. Wehrli, E. C. Hopmans, J. S. S. Damste, S. Wakeham, and M. M. M. Kuypers. 2006. Aerobic and anaerobic methanotrophs in the Black Sea water column. Environ. Microbiol. 8:1844-1856. [DOI] [PubMed] [Google Scholar]
- 56.Scranton, M. I., and P. G. Brewer. 1978. Consumption of dissolved methane in deep ocean. Limnol. Oceanogr. 23:1207-1213. [Google Scholar]
- 57.Sieburth, J. M., P. W. Johnson, M. A. Eberhardt, M. E. Sieracki, M. Lidstrom, and D. Laux. 1987. The 1st methane-oxidizing bacterium from the upper mixing layer of the deep ocean—Methylomonas-Pelagica sp.-nov. Curr. Microbiol. 14:285-293. [Google Scholar]
- 58.Soehngen, N. L. 1906. Ober Bakterien, welche Methan als Kohlenstoffnahrung und Energiequelle gebranehen. Zentralbl. Bakteriol. II Abt. 15:513-517. [Google Scholar]
- 59.Stoecker, K., B. Bendinger, B. Schoning, P. H. Nielsen, J. L. Nielsen, C. Baranyi, E. R. Toenshoff, H. Daims, and M. Wagner. 2006. Cohn's Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc. Natl. Acad. Sci. USA 103:2363-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swofford, D. 1998. PAUP*, version 4. Phylogenetic analysis using parsimony (and other methods). Sinauer Associates, Sunderland, MA.
- 61.Ussler, W., III, C. K. Paull, and W. Normark. 2006. Methane gas emanation from an active carbonate mound in Santa Monica Basin, offshore Southern California, abstr. 05223. Geophys. Res. Abstr. 8.
- 62.Valentine, D. L., D. C. Blanton, W. S. Reeburgh, and M. Kastner. 2001. Water column methane oxidation adjacent to an area of active hydrate dissociation, Eel River Basin. Geochim. Cosmochim. Acta 65:2633-2640. [Google Scholar]
- 63.Vetriani, C., H. V. Tran, and L. J. Kerkhof. 2003. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl. Environ. Microbiol. 69:6481-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wakeham, S. G., E. C. Hopmans, S. Schouten, and J. S. S. Damste. 2004. Archaeal lipids and anaerobic oxidation of methane in euxinic water columns: a comparative study of the Black Sea and Cariaco Basin. Chem. Geol. 205:427-442. [Google Scholar]
- 65.Wise, M. G., J. V. McArthur, and L. J. Shimkets. 1999. Methanotroph diversity in landfill soil: isolation of novel type I and type II methanotrophs whose presence was suggested by culture-independent 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 65:4887-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan, T., Q. Ye, J. Zhou, and C. L. Zhang. 2006. Diversity of functional genes for methanotrophs in sediments associated with gas hydrates and hydrocarbon seeps in the Gulf of Mexico. FEMS Microbiol. Ecol. 57:251-259. [DOI] [PubMed] [Google Scholar]