Abstract
In spite of increasing public health concerns about the potential risks associated with swimming in waters contaminated with waterfowl feces, little is known about the composition of the gut microbial community of aquatic birds. To address this, a gull 16S rRNA gene clone library was developed and analyzed to determine the identities of fecal bacteria. Analysis of 282 16S rRNA gene clones demonstrated that the gull gut bacterial community is mostly composed of populations closely related to Bacilli (37%), Clostridia (17%), Gammaproteobacteria (11%), and Bacteriodetes (1%). Interestingly, a considerable number of sequences (i.e., 26%) were closely related to Catellicoccus marimammalium, a gram-positive, catalase-negative bacterium. To determine the occurrence of C. marimammalium in waterfowl, species-specific 16S rRNA gene PCR and real-time assays were developed and used to test fecal DNA extracts from different bird (n = 13) and mammal (n = 26) species. The results showed that both assays were specific to gull fecal DNA and that C. marimammalium was present in gull fecal samples collected from the five locations in North America (California, Georgia, Ohio, Wisconsin, and Toronto, Canada) tested. Additionally, 48 DNA extracts from waters collected from six sites in southern California, Great Lakes in Michigan, Lake Erie in Ohio, and Lake Ontario in Canada presumed to be impacted with gull feces were positive by the C. marimammalium assay. Due to the widespread presence of this species in gulls and environmental waters contaminated with gull feces, targeting this bacterial species might be useful for detecting gull fecal contamination in waterfowl-impacted waters.
Gulls are common shore waterfowl species, and consequently, their feces could be considered a major source of contamination in coastal and lake waters worldwide. The health risks associated with waterfowl fecal pollution are largely unknown, although they are presumed to be lower than those associated with human fecal pollution. However, because of the migratory character and feeding behavior of feral birds (3), there are increasing public health concerns regarding waterfowl fecal contamination in environmental waters due to the potential spread of microbial pathogens to humans, domesticated animals in close contact with humans, and human food sources. Indeed, several studies have shown that waterfowl feces may carry human pathogens like Campylobacter spp. (35), Salmonella spp. (2), pathogenic Escherichia coli (19, 24), microsporidia (33), and Cryptosporidium spp. (37). The role of wild birds in spreading drug-resistant genes has also been recently documented (9), further suggesting the importance of avian pollution in zoonosis. Aquatic birds are also natural reservoirs of influenza viruses and therefore are an important link in the evolution of these viruses and their spreading in the environment (18).
Most studies describing the gut microbiota of waterfowl have used culture-based methods and have focused on targeted pathogens (6, 15). As culture-based studies can provide only a limited view of natural microbial communities, recently developed molecular methods can be used to better describe the composition of waterfowl gut systems. This information is critical in order to recognize potential hazards associated with waterfowl fecal pollution and to help distinguish waterfowl fecal sources from other animal sources. Waterfowl feces has been suggested as an important source of fecal contamination in several studies (27, 34) and therefore is potentially responsible for many beach closures every year (10). Although fecal pollution in recreational waters is traced with fecal indicator bacteria, whether contamination in water is primarily associated with human, waterfowl, or other fecal sources is often undetermined. While microbial source-tracking (MST) methods that use fingerprint databases of water and fecal bacterial isolates may be able to differentiate bird feces from other fecal sources (10, 14), these methods can be laborious and time-consuming and have yet to clearly discriminate among different avian fecal sources. Alternatively, PCR-based methods that detect host-specific 16S rRNA genes and functional genes directly in water DNA extracts are becoming more popular (4, 31, 32). The latter methods have been used mainly to differentiate human and ruminant from other animal fecal sources, primarily targeting Bacteroidetes. Studies tracking sources of gull feces based on the detection of Bacteroidetes genes have had little success (12), perhaps because of the low prevalence of Bacteroidetes in bird feces and the limited knowledge of the composition of the normal microbiota of gull feces.
The aims of this study were to study microbial community composition and structure in the fecal DNA of gulls by 16S rRNA gene sequencing analysis and to develop host-specific assays for detecting gull fecal community DNA in waters.
MATERIALS AND METHODS
Bacterial strains.
DNA extracts from the following bacteria were used to test the specificity of the Catellicoccus marimammalium PCR assay: C. marimammalium DSMZ M35/04/3T (obtained from the Culture Collection of the University of Göteborg, Göteborg, Sweden), Aeromonas hydrophila ATCC 7966, Bacillus cereus ATCC 10876, B. subtilis ATCC 21332, Enterobacter aerogenes ATCC 13048, Enterococcus faecalis ATCC 29212, E. faecium ATCC 19433, Lactobacillus acidophilus ATCC 43121, and Streptococcus pyogenes ATCC 19615. With the exception of C. marimammalium, biomass from the aforementioned strains was directly harvested from agar plates and used for DNA extractions. Cells of C. marimammalium were harvested directly from lyophilized cultures.
Sample collection and DNA extraction.
Gull fecal samples used to develop the 16S rRNA gene clone library were collected in West Virginia. Fecal DNA extracts from deposited fecal samples from the following animals were used in host specificity studies: Sus scrofa (pig), Bos taurus (bovine), Homo sapiens (human), Capra aegagrus (domestic goat), Ovis aries (sheep), Equus caballus (horse), Felis catus (cat), Canis familiaris (dog), Canis latrans (coyote), Sciurus carolinensis (gray squirrel), Odocoileus virginianus (whitetail deer), Didelphis virginiana (possum), Loragyps atratus (black vulture), Lynx rufus (bobcat), Procyon lotor (raccoon), Erinaceus sp. (hedgehog), Pongo pygmaeus (red ape), Elephas maximus (Asian elephant), Zalophus californianus (California sea lion), Callorhinus ursinus (northern fur seal), Phoca vitulina (Pacific harbor seal), Physeter macrocephalus (sperm whale), Megaptera novaeangliae (humpback whale), Phocoenoides dalli (Dall's porpoise), Phocoena phocoena (harbor porpoise), Lagenorhynchus obliquidens (Pacific white-sided dolphin), Branta canadensis (Canadian goose), Anser sp. (goose), Meleagris gallopavo (turkey), Treron sp. (pigeon), Aix sponsa (duck), Gallus gallus (chicken), Pygoscelis sp. (penguin), Psittacus sp. (parrot), Collumba livia (dove), Pelicanus sp. (pelican), Eudocimus ruber (ibis), Larus atricilla (laughing gull), and L. delawarensis (ring-billed gull). In addition, feces collected from gulls located in Georgia, Ohio, West Virginia, Florida, and Ontario (Canada) were used in host distribution studies. Feces from marine mammals were obtained directly from the animals, while samples from other animals were collected from previously deposited fecal samples. All samples were collected aseptically, placed into sterile 50-ml conical tubes with screw caps, and stored at −80°C until required. Total DNA was extracted from individual fecal samples. The Mo Bio Fecal kit (Mo Bio Laboratories, Inc., Carlsbad, CA) and the FastDNA kit (Q-Biogene, Carlsbad, CA) were used by following the protocols provided by the manufacturers. Total DNA was eluted in 50 (Mo Bio Fecal kit) or 100 μl (Fast DNA kit) of water, and DNA concentrations were measured with a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Inc., Berlin, Germany).
Water samples presumed to be impacted by gull feces were used to evaluate the potential of gull-specific assays as a source-tracking tool (see below). Freshwater samples were collected in sterile bottles from beaches in or near the Great Lakes (Grant Park Beach, Wisconsin; Lake Erie, Ohio; Lake Ontario, Canada) and from a pond adjacent to the San Juan Creek next to Doheny State Beach (Dana Point, California). Additionally, samples were collected from sites in northern Georgia and northern Ohio with no previous history of gull contamination, as well as from sites in northeastern Ohio impacted by non-waterfowl sources (i.e., chicken, cattle, swine). Water samples were transported to the laboratory in ice coolers and filtered (i.e., 100 to 300 ml) onto 47-mm polycarbonate membranes (0.2-μm pore size) as previously described (23). Membranes were then transferred into sterile conical tubes and kept at −80°C until further processing. Total community DNA was extracted from water samples with a Mo Bio Fecal kit and a FastDNA kit.
16S rRNA gene clone library analysis.
Sequence analysis of 16S rRNA gene clones was used to describe the phylogenetic affiliations of bacterial populations in gull fecal extracts. The 16S rRNA gene was amplified with general bacterial primers 27F (E. coli numbering positions 8 to 27: 5′-AGAGTTTGATCMTGGCTCAG-3′) and 785R (E. coli numbering positions 785 to 804: 5′-ACTACCRGGGTATCTAATCC-3′). DNA extracted from eight gull fecal samples from West Virginia was pooled and used as the PCR template. PCR amplifications were performed in a PTC-240 DNA Engine Tetrad 2 Cycler (MJ Research, Inc., Alameda, CA). Reaction mixtures were prepared in a 25-μl volume and subjected to the following cycling conditions: 3 min at 95°C, followed by 22 cycles of 30 s at 95°C, 30 s at 58°C, and 60 s at 72°C and a final 10-min primer extension step at 72°C. PCR products from five reactions were pooled and cloned into pCR4.1 TOPO (Invitrogen). Individual clones were sequenced by using BigDye Terminator chemistry and an Applied Biosystems PRISM 3730XL as described by Lu et al. (23).
Sequence editing and alignment were completed with Sequencher (Gene Codes Corporation, Ann Arbor, MI). The 16S rRNA gene sequences were screened for chimeras with the Check Chimera program of the Ribosome Database Project and by manual alignments of secondary structure. The Bellerophon program (http://foo.maths.uq.edu.au/∼huber/bellerophon.pl) (16) was also used to check for chimeras by comparing each sequence against the sequences from the same library. As a final check for chimeras, each sequence was split into 5′ and 3′ fragments, which were analyzed separately by BLAST searching of the GenBank database. Sequences for which either the 5′ or the 3′ fragment had significantly different closest relatives were considered probable chimeras and were removed from the data set. For 16S rRNA gene sequences, homology searches of DNA sequences in the GenBank (NR) database were done with National Center for Biotechnology Information (NCBI) BLASTn (http://www.ncbi.nlm.nih.gov/BLAST/) (1).
Conventional PCR and real-time assays.
C. marimammalium PCR primers were designed by aligning 16S rRNA gene sequences from closely related species by using Primer Designer software (version 2.01; Scientific & Educational Software, Durham, NC) and the following conditions: no hairpin, no primer dimer formation, and an annealing temperature of 64 to 65°C. Assays were optimized through temperature gradients and with various concentrations of fecal DNA templates. Primers were tested for host specificity against fecal DNA composites for each of the animal types listed above. DNA composites were generated by combining equal amounts of DNA from the individual fecal samples. Assays that showed host specificity to gull composites were further tested against DNA extracts of individual avian, human, pig, and cow fecal samples. Gull samples were also used to determine the host distribution of potential markers. Detection limits of PCR assays were determined by three different approaches: (i) 10-fold dilutions of plasmid DNA (6 to 6 × 10−7 ng) containing a targeted insert in reaction mixtures with no fecal DNA background; (ii) 10-fold dilutions of plasmid DNA (6 to 6 × 10−7 ng) containing a targeted insert in reaction mixtures spiked with bird fecal DNA (10 ng/μl) made of equal amounts of chicken, turkey, and Canadian goose fecal DNA extracts; and (iii) 10-fold dilutions of gull fecal DNA (6 to 6 × 10−7 ng). PCR assays specific to Bacteroides spp. and Clostridium coccoides were used to determine the presence of potential PCR inhibitors in DNA extracts used as templates in PCR assays (4, 25). All of the assays were performed with 1 and 10 ng μl−1 fecal DNA extracts. The presence of PCR products was visualized by 2% agarose gel electrophoresis with GelStar as the nucleic acid stain (FMC BioProducts; Rockland, ME). The cycling conditions for the PCR assays were 3 min at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 64°C, and 60 s at 72°C and a final 10-min primer extension step at 72°C. The C. marimammalium species-specific primer sequences used in this study were TGCATCGACCTAAAGTTTTGAG and GTCAAAGAGCGAGCAGTTACTA for the forward and reverse primers, respectively. We refer to this assay as Gull-2.
The C. marimammalium PCR assay was also used in real-time assays, with SYBR green as the detection dye. The assays were performed with a 7900 HT Fast Real-Time Sequence Detector (Applied Biosystems). Reaction mixtures (20 μl) contained 10 μl 2x SYBR Premix Ex Taq, 0.4 μl ROX reference dye (Takara Bio Inc., Shiga, Japan), 0.2 μM (final concentration) primers, and either 100 ng genomic DNA (fecal and water samples), a series of dilutions of plasmid DNA with a target insert with a log copy number of 8.8 to 1.8 (i.e., a copy number of 6.3 × 108 to about 63) per reaction mixture, or a series of dilutions of gull fecal DNA ranging from 60 to 6 × 10−7 ng per reaction mixture. All reaction mixtures were prepared in triplicate in MicroAmp Optical 96-well reaction plates with MicroAmp Optical Caps (Applied Biosystems). The amplification protocol consisted of 50°C for 2 min, followed by 95°C for 2 min and then 40 cycles of 95°C for 5 s, 64°C for 15 s, and a 72°C extension for 10 s and an additional disassociation at 60°C for 15 s. Data were initially analyzed with Sequence Detector software (version 2.2.2) with a 0.2 threshold. Gene copies in fecal samples were calculated from standard curves based on the log transformation of a known concentration versus the threshold cycle (CT). Comparison tests between gull species (laughing gull and ring-billed gull) were performed with Statistical Analysis Software (SAS Institute Inc., Cary, NC) by using PROC GLM with the contrast statement feature.
Nucleotide sequence accession numbers.
Representative 16S rRNA gene sequences from cloning experiments were deposited in GenBank with accession numbers EU181006 to EU181122.
RESULTS AND DISCUSSION
Phylogenetic analysis of 16 rRNA gene sequences.
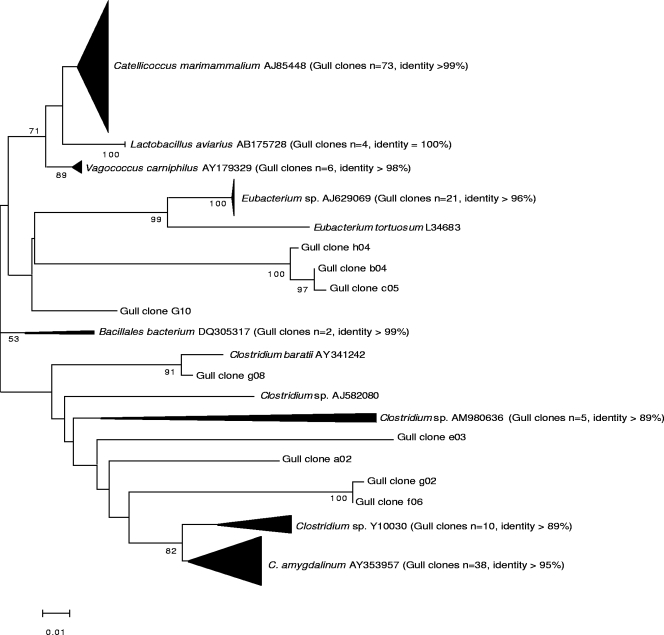
Of the 282 gull fecal 16S rRNA gene sequences analyzed, there were 85 different operational taxonomic units (i.e., 97% identical to previously deposited sequences). These sequences represent 76% coverage of the gull fecal community diversity as calculated by Lu et al. (22), indicating that the majority of the predominant populations of the fecal microbial community in gulls was represented in the clones analyzed. Excluding the sequences classified as unknowns (n = 9), 38 bacterial genera were represented in the clone library (Table 1; Fig. 1). Bacilli sequences were the most common (37%), particularly sequences closely related to the low-G+C gram-positive bacterium C. marimammalium (i.e., 99% identity), which constituted 26% of the clone library. The C. marimammalium-like sequences were nearly identical to each other, with <1% showing sequence heterogeneity among the clones recovered. Other Bacilli-like sequences were closely related to Lactobacillus aviarius and Vagococcus camiphilus. Clostridia sequences were relatively numerous (17%), specifically, sequences closely related to Clostridium sp. and Eubacterium tortuosum. Mollicute sequences (9%) pertaining to an unknown genus were also numerous. Sequences homologous to Gammaproteobacteria (identity, ≥98%) were also numerous (11%), and many were closely related to Klebsiella pneumoniae, Enterobacter sp., Pseudomonas sp., and E. coli. In contrast, sequences homologous to Bacteroides represented only 1% of the total number of clones.
TABLE 1.
Distribution of 16S rRNA genes in the gull clone library
| Class or group (% clones of total) | Genus | No. of clones |
|---|---|---|
| Actinobacteria (6.4) | Arthrobacter | 4 |
| Corynebacterium | 8 | |
| Microbacterium | 1 | |
| Propionibacterium | 2 | |
| Unknown | 3 | |
| Bacilli (37.2) | Bacillus | 3 |
| Catellicoccus | 74 | |
| Enterococcus | 1 | |
| Lactobacillus | 9 | |
| Paenibacillus | 1 | |
| Staphylococcus | 1 | |
| Vagococcus | 9 | |
| Unknown | 7 | |
| Bacteroidetes (1.1) | Bacteroidetes | 1 |
| Unknown | 2 | |
| Clostridia (17.31) | Clostridium | 44 |
| Eubacterium | 1 | |
| Ruminococcus | 2 | |
| Unknown | 2 | |
| Fusobacteria (0.7) | Cetobacterium | 2 |
| Mollicutes (8.8) | Unknown | 25 |
| Alphaproteobacteria (6.7) | Agrobacterium | 1 |
| Bosea | 1 | |
| Devosia | 1 | |
| Fulvimarina | 1 | |
| Mesorhizobium | 1 | |
| Paracoccus | 8 | |
| Rhizobium | 1 | |
| Rhodobacter | 1 | |
| Unknown | 4 | |
| Betaproteobacteria (4.3) | Acidovorax | 6 |
| “Panaciterramonas” | 2 | |
| Polynucleobacter | 3 | |
| Zoogloea | 1 | |
| Gammaproteobacteria (11.3) | Acinetobacter | 13 |
| Enterobacter | 6 | |
| Escherichia | 6 | |
| Klebsiella | 5 | |
| Pantoea | 1 | |
| Pseudomonas | 1 | |
| Deltaproteobacteria (0.4) | Unknown | 1 |
| Epsilonproteobacteria (0.4) | Campylobacter | 1 |
| Planctomycetes (0.4) | Planctomyces | 1 |
| Spirochaetes (1.1) | Leptospira | 3 |
| Cyanobacteria (0.4) | Synechococcus | 1 |
| Archaea (0.4) | Unknown | 1 |
| Unknown class (3.2) | Unknown | 9 |
FIG. 1.
Unrooted neighbor-joining tree of 16S rRNA gene sequences of low-G+C gram-positive bacteria, including C. marimammalium-like sequences obtained from clone libraries. Sequences were aligned, and a bootstrap consensus tree was created with MEGA 3.1 (1% divergence).
Although there are very limited data on the composition of bird fecal microbial communities, some trends are emerging from 16S rRNA gene sequence analyses of chicken (22) and turkey (30) intestines and from this study. For example, the predominant bacteria in avian gut microbial communities are low-G+C gram-positive bacteria, particularly Clostridia and Bacilli in chicken and turkey systems. In contrast to other gut systems, Bacteroidetes bacteria represent a small fraction of the bacteria in avian feces (e.g., as little as ∼1% of the total community). This is an important finding to those developing markers to track avian fecal pollution in environmental waters, as it suggests that Bacteroidetes bacteria might not be practical targets for the development of avian-specific assays due to their low densities in the avian gut and considering that Bacteroidetes host-specific populations represent less that 1 to 10% of the total Bacteroidetes populations (4, 20).
Our data show that the dominant low-G+C gram-positive bacteria in gull feces are closely related to C. marimammalium. This finding is interesting, as C. marimammalium was recently described as a new group of low-G+C gram-positive bacteria isolated from a porpoise and a gray seal (21). This group is part of the Bacilli class and specifically belongs in the Enterococcaceae family, which houses the genera Enterococcus, Melissococcus, Tetragenococcus, and Vagococcus. Enterococcus and Vagococcus spp. are commonly found in animal feces, and in fact, Enterococcus spp. are often used as indicators of fecal pollution in freshwater and marine waters and as targets in source-tracking studies (8, 28). Several studies have shown that some Enterococcus populations might be host specific (5), although the ecology of this genus is still poorly understood. Since no strains of C. marimammalium have been isolated from avian systems thus far, we decided to determine if these results were unique to gulls or if this species is also present in other gut systems.
Host specificity PCR studies.
The Gull-2 PCR assay was designed to target C. marimammalium by using publicly available sequences and closely related clone sequences obtained in this study. In silico analysis showed that the primers have no mismatches to the C. marimammalium 16S rRNA gene. Only a limited number of unrelated 16S rRNA gene bacterial sequences in the NCBI database (e.g., B. vulgatus, Prochlorococcus marinus, Mycobacterium sp. strain JLS, and Jannaschia sp. strain CCS1) (as of 23 August 2007) showed similarity to either one of the primers, but these sequences contain several mismatches in the 3′ end of the sequence. Moreover, none of the primers perfectly annealed sequences from the same species, suggesting a relatively low potential for cross-hybridization with bacterial species other than C. marimammalium.
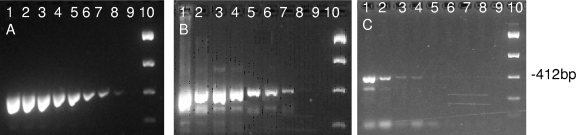
When the Gull-2 PCR assay was used to test DNA extracts from eight bacterial strains, including Firmicutes closely related to C. marimammalium (i.e., enterococci), no cross-amplification signals were obtained, further suggesting that the assay is highly species specific. DNA from the C. marimammalium type strain produced a PCR band of the expected size (i.e., approximately 412 bp). When the PCR assay was used to test composite fecal DNA extracts from a variety of animals, including marine mammals, only gull fecal DNA extracts generated a PCR product of the correct size (Table 2). As C. marimammalium was originally isolated from marine mammals, our results suggest that C. marimammalium might not be a normal or abundant inhabitant of a marine mammal's gut. Considering that this species was originally isolated from deceased animals, this organism might be an opportunistic pathogen, which will also explain the negative results. All of the fecal samples tested yielded PCR signals with Bacteroides-Prevotella 16S rRNA gene-specific primers (4), suggesting that PCR inhibition could not explain the absence of C. marimammalium-like signals in fecal samples that tested negative for the species-specific primers. The detection limit of both positive control tests (i.e., assays containing plasmid DNA in spiked and nonspiked bird fecal DNA) was 6 × 10−6 ng DNA per PCR. The PCR assay showed a detection limit of 0.006 ng of gull fecal DNA per reaction (Fig. 2).
TABLE 2.
Host specificity results of Gull2 assay against feces from various animals
| Animal | Location of sample | No. of samples/no. of composites testeda | Gull2
|
|
|---|---|---|---|---|
| PCR | Real-time CT | |||
| Pig | Delaware | 10/2 | − | BDLb |
| Cow | West Virginia | 17/3 | − | BDL |
| Cow | Delaware | 11/1 | − | BDL |
| Human | West Virginia | 16/3 | − | BDL |
| Goat | Delaware | 10/2 | − | BDL |
| Sheep | Delaware | 11/3 | − | BDL |
| Horse | West Virginia | 5/1 | − | BDL |
| House cat | West Virginia | 11/1 | − | BDL |
| Domestic dog | West Virginia | 13/1 | − | BDL |
| Coyote | Texas | 10/1 | − | BDL |
| Gray squirrel | Texas | 4/1 | − | BDL |
| Deer | West Virginia | 6/1 | − | BDL |
| Possum | Texas | 2/1 | − | BDL |
| Black vulture | Texas | 1/1 | − | BDL |
| Raccoon | Texas | 1/1 | − | BDL |
| Hedgehog | West Virginia | 1/1 | − | BDL |
| Bobcat | Texas | 1/1 | − | BDL |
| Red ape | Ohio | 1/1 | − | BDL |
| Asian elephant | Ohio | 1/1 | − | BDL |
| California sea lion | California | 10/10 | − | NDc |
| Northern elephant seal | California | 8/8 | − | ND |
| Pacific harbor seal | California | 6/6 | − | ND |
| Sperm whale | California | 2/2 | − | ND |
| Humpback whale | California | 1/1 | − | ND |
| Dall's porpoise | California | 1/1 | − | ND |
| Harbor porpoise | California | 2/2 | − | ND |
| Pacific white-sided dolphin | California | 1/1 | − | ND |
| Canadian goose | West Virginia | 20/20 | − | BDL |
| Canadian goose | New Jersey | 5/5 | − | BDL |
| Canadian goose | Georgia | 16/16 | − | BDL |
| Canadian goose | Oregon | 4/4 | − | BDL |
| Canadian goose | Ohio | 4/4 | − | BDL |
| Geese | Ohio | 13/13 | − | BDL |
| Turkey | Delaware | 11/1 | − | BDL |
| Turkey | Ohio | 8/8 | − | BDL |
| Pigeon | West Virginia | 2/1 | − | BDL |
| Pigeon | Ohio | 3/3 | − | BDL |
| Duck | Georgia | 21/21 | − | BDL |
| Duck | Ohio | 4/4 | − | BDL |
| Chicken | West Virginia | 14/1 | − | BDL |
| Penguin | Ohio | 3/3 | − | BDL |
| Parrot | Ohio | 4/4 | − | BDL |
| Dove | Ohio | 2/2 | − | BDL |
| Pelican | Ohio | 1/1 | − | BDL |
| Ibis | Ohio | 1/1 | − | BDL |
| Seagull | West Virginia | 8/1 | + | 23.41 |
The number of samples tested and the number of composites provided for each sample type are shown.
BDL, below detection limit.
ND, not determined.
FIG. 2.
Detection limits of the Gull-2 PCR assay. (A) Plasmid DNA (10-fold dilutions of plasmid DNA ranging from 6 to 6 × 10−7 ng) containing a targeted insert in reaction mixtures with no fecal DNA background from lane 1 to lane 8 (negative control in lane 9). (B) Tenfold dilutions of plasmid DNA (6 to 6 × 10−7ng) containing a targeted insert in reaction mixtures spiked with bird fecal DNA (10 ng/μl) made of equal amounts of chicken, turkey, and Canadian goose fecal DNA extracts from lane1 to lane 8 (only bird DNA in lane 9). (C) Tenfold dilutions of seagull fecal DNA ranging from 6 to 6 × 10−7 ng/PCR mixture from lane1 to lane 8 (negative control in lane 9).
The geographic and host distribution of C. marimammalium was determined by PCR assays against individual gull fecal samples (n = 58) collected from Florida (Larus atricilla), West Virginia (L. delawarensis), Ohio (L. delawarensis), Georgia (L. atricilla and L. delawarensis), and Ontario, Canada (L. delawarensis) (Table 3). Positive signals in the Gull-2 PCR assay were obtained with approximately 71% of the gull fecal specimens tested in this study. Wide distribution and high prevalence of the gull marker were obtained for the gull species tested, regardless of the locations at which the samples were collected. Larus atricilla and L. delawarensis are among the most common gulls in North America, and therefore, the C. marimammalium-specific PCR assay results suggested that this bacterial species is ubiquitous in the gull gastrointestinal tract as it was detected in gull feces and gull-impacted water samples from different geographic locations. The results also suggested that this species has restricted host specificity, as it was detected only in gulls and not in any other animal fecal samples, including several avian species. It should be noted that C. marimammalium was originally isolated from the mesentery, kidney, pericardial fluid, peritoneum, and small intestine of a dead harbor porpoise exhibiting severe enteritis and peritonitis. This is interesting from the standpoint that it suggests that some C. marimammalium strains are potentially pathogenic and that gull fecal contamination could be implicated in the transmission of aquatic mammal pathogens. As both C. marimammalium strains were isolated in mammals swimming in coastal waters of Scotland, their global distribution also merits future attention.
TABLE 3.
Host distribution and estimated average copy number of gull-specific marker
| Target | Sampling location | No. of fecal samples tested | Gull2 assay
|
||
|---|---|---|---|---|---|
| No. of PCR-positive samples | Quantitative PCR
|
||||
| No. of samples detected | Avg copy no./ng DNA ± SD | ||||
| Larus domesticus | Georgia | 13 | 10 | 10 | 6,117 ± 12,428 |
| Larus atricilla | Georgia | 20 | 10 | 12 | 905 ± 1,040 |
| Larus atricilla | Ohio | 3 | 3 | 3 | 414 ± 496 |
| Larus domesticus | Ohio | 3 | 2 | 3 | 52 ± 73 |
| Larus domesticus | West Virginia | 8 | 7 | 6 | 896 ± 932 |
| Larus atricilla | Florida | 7 | 5 | 5 | 216 ± 171 |
| Larus domesticus | Ontario, Canada | 4 | 4 | 4 | 93,044 ± 71,792 |
Real-time PCR assays.
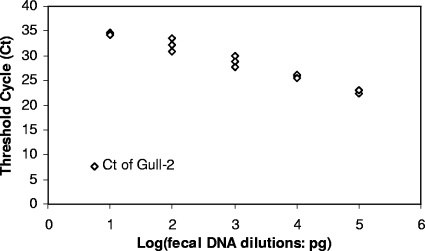
When the gull primer sets were used in real-time PCR assays, only the composite gull samples tested positive, while the CT values for other fecal DNA extracts were below the detection limit. Standard curves for real-time PCR assays were constructed with plasmid inserts with an equivalent of 6.3 × 108 to 63 plasmid copies. There was a linear relationship between CT values and copy numbers of the targeted DNA fragment according to plasmid standard curves for a wide range of targets (r2 = 0.98; data not shown). CT values were linear from 60 ng to 6 pg for the primer when using fecal DNA dilutions (Fig. 3), suggesting that neither PCR inhibitors nor the presence of large amounts of heterologous DNA inhibited amplifications (7). The detection limit of the real-time PCR assay was similar to that of the conventional PCR assay with the original composite gull DNA extract (i.e., 6 pg fecal DNA; Fig. 2). These results allowed us to estimate the copy numbers of fecal DNA signals from several locations (West Virginia, Georgia, Ohio, Florida, and Ontario, Canada) and in different gull species.
FIG. 3.
Performance of C. marimammalium 16S rRNA gene PCR assay against 10-fold dilutions of seagull fecal DNA starting with 60 ng (indicated as 5 log units on the x axis) to 6 pg (indicated as 1 log unit on the x axis).
The copy number of the targeted sequence varied considerably among individual gull fecal samples from the same species collected in the same location or at different locations and between different gull species (Table 3). Overall, the range was 2 to 166,701 DNA copies per ng of fecal DNA, indicating that the densities of C. marimammalium could greatly fluctuate in the gull gut system. The copy number in the ring-billed gulls was, on average, 10 times higher than in laughing gulls from Georgia (P = 0.0094, n = 25). Factors influencing C. marimammalium fecal densities are yet to be determined, but it is important to know whether physiological changes in the gull due to age, diet, or the surrounding environment affect the densities of C. marimammalium in the gull intestine. These fluctuations are relevant to source-tracking studies for several reasons. For example, average signals obtained in one locality might not be applicable to another study site due to inter- and intraspecies fluctuations. This is critical to presence/absence assays from the standpoint of assay sensitivity, as it suggests that detection limits in environmental waters might vary among different sites. When using quantitative PCR assays, it will also be necessary to understand the level of variability between hosts in order to better estimate fecal loads for a given source, information that is needed for regulatory activities and for microbial quantitative risk analysis (29). Fluctuations in E. coli and Enterococcus densities in gull feces have previously been documented (11). However, this is the first study showing variations in gull fecal bacterial populations other than fecal bacterial indicators.
Gull PCR-based signals in water samples.
C. marimammalium was found to be ubiquitous and specific to gull feces, suggesting that assays targeting this bacterial species might be used as an indicator of gull fecal pollution in MST studies. Gull contamination is prevalent in many coastal areas, as well as in recreational waters in the Great Lakes. However, thus far, no assays have been published that can determine the presence of gull fecal contamination in surface waters. To address the value of the C. marimammalium assay in detecting gull feces in environmental waters, we tested DNA extracts from waters presumed to have a history of gull contamination against the Gull-2 assay (Table 4). All samples suspected to have been impacted with gull feces showed strong PCR signals in the Gull-2 assay, suggesting that this assay can be used to track sources in different geographic locations. Additionally, we used the assay to test waters that are impacted by other waterfowl fecal sources (i.e., Canadian geese). Water samples collected from the Toledo Botanical Garden pond (Toledo, OH), which is known to be impacted by Canadian geese (i.e., as the only waterfowl species), showed an average of 26,900 fecal coliform CFU/100 ml (W. Von Sigler, personal communication) and were negative by the Gull2 assay. Samples taken from environmental waters known to be impacted by non-waterfowl sources (i.e., swine, cattle, chickens) were also negative. In contrast, all three samples taken from a site near the Toledo Botanical Garden pond where gulls are often seen (near Lake Erie) were positive. Similar results were obtained with freshwater samples collected from Wisconsin, California, and Ontario beaches known to be frequented by gulls. More importantly, the Gull-2 assay was positive for water samples collected over beach seasons at three different Lake Ontario beaches known to be highly contaminated by gull droppings (10). A library-dependent MST study at Bayfront Park Beach on Lake Ontario demonstrated that most of the E. coli contamination at this beach was from birds rather than municipal wastewater or pets (10). Our results are relevant to the latter study, as the previous assays used could not discriminate between the importance of gull droppings and that of Canada goose droppings at these sites. Overall, our results showed that the Gull-2 assay can be used to detect the presence of gull fecal impacts at different geographic locations and that it can be used across multiple seasons (i.e., it is temporally stable).
TABLE 4.
Detection of gull feces in environmental samples by the Gull2 assay
| Sampling location(s) | Sample type | Collection time | No. of water samples | No. of samples Gull2 assay positive | Presumed gull contaminationa |
|---|---|---|---|---|---|
| Grant Park Beach, Milwaukee, Wisconsin (Lake Michigan) | Freshwater | September-October 2007 | 8 | 8 | Yes |
| Maumee Bay, Oregon; Lake Erie, Ohio | Freshwater | October 2007 | 3 | 3 | Yes |
| Toledo Botanical Garden pond, Toledo, Ohio | Freshwater | October 2007 | 2 | 0 | No |
| Northeastern Ohio | Chicken pit | May 2007 | 9 | 0 | No |
| Northeastern Ohio | Swine pit | February 2008 | 3 | 0 | No |
| Northeastern Ohio | Cow manure lagoon | February 2008 | 1 | 0 | No |
| Northern Georgia | Freshwater | May 2006 | 9 | 0 | No |
| Bayfront Park Beach, Toronto (Lake Ontario, Canada) | Freshwater | May-August 2007 | 10 | 10 | Yes |
| Bluffers Park Beach, Toronto (Lake Ontario, Canada) | Freshwater | May-August 2007 | 10 | 10 | Yes |
| Sunnyside Beach (Lake Ontario, Canada) | Freshwater | May-August 2007 | 10 | 10 | Yes |
| Doheny State Beach pond (Dana Point, California) | Freshwater | June-July 2007 | 7 | 7 | Yes |
For all sites presumed positive, there is historical knowledge that gulls are present during a significant part of the year, particularly during warm months. At the Canadian sites, the number of gulls present on collection dates ranged from 2 to 220. No numbers were available for the other sites.
Nonpoint fecal pollution sources are increasingly being recognized as important contributors to elevated levels of E. coli and Enterococcus indicator bacteria in recreational waters. As waterfowl are an important source of pollution in beach areas (17, 26) and can serve as potential reservoirs of human infections (13, 36), the assays described herein would be useful in health risk-based analyses (i.e., epidemiological studies and quantitative microbial risk studies) of nonhuman fecal pollution in recreational waters. Additionally, having gull-specific markers in the fecal source-tracking toolbox will help beach managers better assess potential causes of beach postings beyond familiar fecal pollution sources such as municipal wastewater and therefore implement remediation practices that target the most relevant sources of pollution.
Acknowledgments
This research was funded in part by a New Start Award from the EPA National Center for Computational Toxicology to J.W.S.D. and in part by a National Research Council fellowship to J.L.
We are grateful to Stacy Pfaller for access to her laboratory; to Don Stoeckel, Margie Lee, Ali Boehm, William Von Sigler, Jody Harwood, John Griffith, Julie Kinzelman, and Michael Yabsley for providing fecal and water samples; to Frances Gulland and Liz Wheeler (The Marine Mammalian Center) for providing fecal samples; and to Noreen Adcock, Dennis Lye, and Randy Revetta for providing bacterial strains.
Any opinions expressed in this paper are ours and do not necessarily reflect the official positions and policies of the United States Environmental Protection Agency. Any mention of products or trade names does not constitute a recommendation for use.
Footnotes
Published ahead of print on 9 May 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudart, J., K. Lemarchand, A. Brisabois, and P. Lebaron. 2000. Diversity of Salmonella strains isolated from the aquatic environment as determined by serotyping and amplification of the ribosomal DNA spacer regions. Appl. Environ. Microbiol. 66:1544-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, R. W., and A. W. Anderson. 1972. Salmonellae and Edwardsiella tarda in gull feces: a source of contamination in fish processing plants. Appl. Microbiol. 24:501-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Call, D. R., D. M. Satterwhite, and M. Soule. 2007. Using DNA suspension arrays to identify library-independent markers for bacterial source tracking. Water Res. 41:3740-3746. [DOI] [PubMed] [Google Scholar]
- 6.Cragg, J., and Y. M. Clayton. 1971. Bacterial and fungal flora of seagull droppings in Jersey. J. Clin. Pathol. 24:317-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dick, L. K., and K. G. Field. 2004. Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes. Appl. Environ. Microbiol. 70:5695-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickerson, J. W., Jr., J. B. Crozier, C. Hagedorn, and A. Hassall. 2007. Assessment of the 16S-23S rDNA intergenic spacer region in Enterococcus spp. for microbial source tracking. J. Environ. Qual. 36:1661-1669. [DOI] [PubMed] [Google Scholar]
- 9.Dolejska, M., A. Cizek, and I. Literak. 2007. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J. Appl. Microbiol. 103:11-19. [DOI] [PubMed] [Google Scholar]
- 10.Edge, T. A., and S. Hill. 2007. Multiple lines of evidence to identify the sources of fecal pollution at a freshwater beach in Hamilton Harbour, Lake Ontario. Water Res. 41:3585-3594. [DOI] [PubMed] [Google Scholar]
- 11.Fogarty, L. R., S. K. Haack, M. J. Wolcott, and R. L. Whitman. 2003. Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces. J. Appl. Microbiol. 94:865-878. [DOI] [PubMed] [Google Scholar]
- 12.Fogarty, L. R., and M. A. Voytek. 2005. Comparison of Bacteroides-Prevotella 16S rRNA genetic markers for fecal samples from different animal species. Appl. Environ. Microbiol. 71:5999-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, J. G., N. S. Taylor, S. Howe, M. Tidd, S. Xu, B. J. Paster, and F. E. Dewhirst. 2006. Helicobacter anseris sp. nov. and Helicobacter brantae sp. nov., isolated from feces of resident Canada geese in the greater Boston area. Appl. Environ. Microbiol. 72:4633-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genthner, F. J., J. B. James, D. F. Yates, and S. D. Friedman. 2005. Use of composite data sets for source-tracking enterococci in the water column and shoreline interstitial waters on Pensacola Beach, Florida. Mar. Pollut. Bull. 50:724-732. [DOI] [PubMed] [Google Scholar]
- 15.Holländer, R. 1982. The aerobic bacterial intestinal flora of various wintering geese species. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 252:394-400. [PubMed] [Google Scholar]
- 16.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 17.Kirschner, A. K., T. C. Zechmeister, G. G. Kavka, C. Beiwl, A. Herzig, R. L. Mach, and A. H. Farnleitner. 2004. Integral strategy for evaluation of fecal indicator performance in bird-influenced saline inland waters. Appl. Environ. Microbiol. 70:7396-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krauss, S., C. A. Obert, J. Franks, D. Walker, K. Jones, P. Seiler, L. Niles, S. P. Pryor, J. C. Obenauer, C. W. Naeve, L. Widjaja, R. J. Webby, and R. G. Webster. 2007. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog. 3:e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kullas, H., M. Coles, J. Rhyan, and L. Clark. 2002. Prevalence of Escherichia coli serogroups and human virulence factors in faeces of urban Canada geese (Branta canadensis). Int. J. Environ. Health Res. 12:153-162. [DOI] [PubMed] [Google Scholar]
- 20.Lamendella, R., J. W. Domingo, D. B. Oerther, J. R. Vogel, and D. M. Stoeckel. 2007. Assessment of fecal pollution sources in a small northern-plains watershed using PCR and phylogenetic analyses of Bacteroidetes 16S rRNA gene. FEMS Microbiol. Ecol. 59:651-660. [DOI] [PubMed] [Google Scholar]
- 21.Lawson, P. A., M. D. Collins, E. Falsen, and G. Foster. 2006. Catellicoccus marimammalium gen. nov., sp. nov., a novel gram-positive, catalase-negative, coccus-shaped bacterium from porpoise and grey seal. Int. J. Syst. Evol. Microbiol. 56:429-432. [DOI] [PubMed] [Google Scholar]
- 22.Lu, J., U. Idris, B. Harmon, C. Hofacre, J. J. Maurer, and M. D. Lee. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, J., J. Santo Domingo, and O. C. Shanks. 2007. Identification of chicken-specific fecal microbial sequences using a metagenomic approach. Water Res. 41:3561-3574. [DOI] [PubMed] [Google Scholar]
- 24.Makino, S., H. Kobori, H. Asakura, M. Watarai, T. Shirahata, T. Ikeda, K. Takeshi, and T. Tsukamoto. 2000. Detection and characterization of Shiga toxin-producing Escherichia coli from seagulls. Epidemiol. Infect. 125:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Miyamoto, T. Takada, K. Matsumoto, H. Oyaizu, and R. Tanaka. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLellan, S. L. 2004. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 70:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLellan, S. L., and A. K. Salmore. 2003. Evidence for localized bacterial loading as the cause of chronic beach closings in a freshwater marina. Water Res. 37:2700-2708. [DOI] [PubMed] [Google Scholar]
- 28.McQuaig, S. M., T. M. Scott, V. J. Harwood, S. R. Farrah, and J. O. Lukasik. 2006. Detection of human-derived fecal pollution in environmental waters by use of a PCR-based human polyomavirus assay. Appl. Environ. Microbiol. 72:7567-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santo Domingo, J. W., D. G. Bambic, T. A. Edge, and S. Wuertz. 2007. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Water Res. 41:3539-3552. [DOI] [PubMed] [Google Scholar]
- 30.Scupham, A. J. 2007. Examination of the microbial ecology of the avian intestine in vivo using bromodeoxyuridine. Environ. Microbiol. 9:1801-1809. [DOI] [PubMed] [Google Scholar]
- 31.Shanks, O. C., J. W. Domingo, J. Lu, C. A. Kelty, and J. E. Graham. 2007. Identification of bacterial DNA markers for the detection of human fecal pollution in water. Appl. Environ. Microbiol. 73:2416-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shanks, O. C., J. W. Santo Domingo, R. Lamendella, C. A. Kelty, and J. E. Graham. 2006. Competitive metagenomic DNA hybridization identifies host-specific microbial genetic markers in cow fecal samples. Appl. Environ. Microbiol. 72:4054-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slodkowicz-Kowalska, A., T. K. Graczyk, L. Tamang, S. Jedrzejewski, A. Nowosad, P. Zduniak, P. Solarczyk, A. S. Girouard, and A. C. Majewska. 2006. Microsporidian species known to infect humans are present in aquatic birds: implications for transmission via water? Appl. Environ. Microbiol. 72:4540-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Standridge, J. H., J. J. Delfino, L. B. Kleppe, and R. Butler. 1979. Effect of waterfowl (Anas platyrhynchos) on indicator bacteria populations in a recreational lake Madison, Wisconsin. Appl. Environ. Microbiol. 38:547-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waldenström, J., T. Broman, I. Carlsson, D. Hasselquist, R. P. Achterberg, J. A. Wagenaar, and B. Olsen. 2002. Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldenström, J., S. L. On, R. Ottvall, D. Hasselquist, and B. Olsen. 2007. Species diversity of campylobacteria in a wild bird community in Sweden. J. Appl. Microbiol. 102:424-432. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, L., H. Kassa, M. L. Tischler, and L. Xiao. 2004. Host-adapted Cryptosporidium spp. in Canada geese (Branta canadensis). Appl. Environ. Microbiol. 70:4211-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]