Abstract
In the detergent industry, fungal endoglucanases have been used to release microfibrils (defibrillation) from the surface of dyed cellulosic fabrics to enhance color brightness. Although endoglucanases for laundry use must have various properties, such as a neutral or alkaline optimum pH, resistance to anionic surfactants and oxidizing agents (main components in detergents), and high defibrillation activity, all-purpose endoglucanases have not been obtained yet. As a result of screening of endoglucanases, a new family 45 endoglucanase (family 45 glycoside hydrolase), designated STCE1, was obtained and purified to apparent homogeneity from the culture supernatant of Staphylotrichum coccosporum NBRC 31817. The molecular mass of STCE1 was 49 kDa. The optimum pH for the carboxymethyl cellulase activity of STCE1 was 6.0, and the optimum temperature was 60°C. STCE1 was highly resistant to an anionic surfactant and an oxidizing agent. Furthermore, the defibrillation activities on dyed cotton and lyocell fabrics of STCE1 were higher than those of the other representative endoglucanases tested. These results indicate that STCE1 is an all-purpose enzyme for laundry use. A gene encoding STCE1, designated the stce1 gene, was cloned from S. coccosporum, and the complete sequence was determined. STCE1 consisted of three distinct domains: an N-terminal catalytic domain (family 45), a linker domain, and a C-terminal carbohydrate-binding module (family 1). The amino acid sequences of the catalytic domain of STCE1 were phylogenetically close to those of the family 45 endoglucanases EGL3, EGL4, and EGV from a Humicola sp. Hence, the stce1 gene was transferred into Humicola insolens and expressed. As a result, extremely high levels (0.90 mg protein per ml of culture supernatant, 27% of the total proteins) of the recombinant STCE1 were secreted as a mature form in the culture supernatant.
Cellulose is the major plant cell wall polysaccharide and is degraded by cellulases. This degradation is thought to be achieved by the synergistic action of three types of cellulase components: endoglucanases (EC 3.2.1.4; endo-β-d-1,4-glucanases), cellobiohydrolases (EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21) (15). The endoglucanases, which are ubiquitous enzymes that hydrolyze 1,4-β linkages adjacent to unsubstituted glucose residues (14), are produced by a broad range of organisms, including fungi (39), bacteria (4), plants (30), and insects (termites) (16).
Most fabrics consisting of cellulosic fibers, such as cotton, linen, ramie, viscose, and lyocell, have a tendency to form microfibrils. When dyed cellulosic fabrics have been washed several times in the laundry, microfibrils accumulate on the surface of the fabrics, making the fabrics look hairy and scattering incident light, thereby lessening the brightness of the original colors (17). These phenomena are known to be negative features of cellulosic fabrics. Hence, endoglucanases have been used to release the microfibrils (defibrillation) from the surface of dyed cellulosic fabrics and thus to restore the original colors (1, 5, 6, 9).
In general, detergents contain nonionic and anionic surfactants and oxidizing agents (bleaching agents). Anionic surfactants and oxidizing agents invariably denature proteins partially or completely. Therefore, endoglucanases for laundry use must have some resistance to anionic surfactants and oxidizing agents, as well as high levels of defibrillation activity (31). Furthermore, because detergent solutions have a neutral or alkaline pH, a neutral or alkaline optimum pH of endoglucanase is in demand for laundry use.
Of the various endoglucanases, the family 45 endoglucanases (family 45 glycoside hydrolase [GH45]) are mainly used for laundry use because of their neutral optimum pH (17). However, all-purpose enzymes that have all the properties mentioned above have not been obtained yet. For example, although family 45 endoglucanases from members of the Zygomycota, such as RCE1 and RCE2 from Rhizopus oryzae (23, 25), MCE1 and MCE2 from Mucor circinelloides (2), and PCE1 from Phycomyces nitens (34), had high defibrillation activities on dyed lyocell and cotton fabrics, they had little resistance to an anionic surfactant and an oxidizing agent (29, 35). By contrast, family 45 endoglucanases from members of the Ascomycota, such as EGL3 and EGL4 from Humicola grisea (38), were highly resistant to the anionic surfactant and oxidizing agent, whereas they had low defibrillation activity on the dyed lyocell fabric (35).
Several years ago, the cellulase mixtures used in the detergent industry were obtained mainly from fungi that had not been genetically engineered. However, because large quantities of these cellulase mixtures are needed to achieve the desired effect on cellulosic fabrics, fungi have been designed to overproduce endoglucanases with high defibrillation activity in order to reduce the cost of the defibrillation (5, 21). In the detergent industry, fungi are frequently used as the recombinants for enzyme production because of their high levels of enzyme secretion (5, 21). In particular, Humicola insolens has been a very suitable host for the overproduction of family 45 endoglucanases (24). However, it is extremely difficult to heterologously express family 45 endoglucanases in a mature form in H. insolens. For example, H. insolens could not produce endoglucanase RCE1 from R. oryzae because of differences in codon usage between genes derived from Rhizopus and Humicola (28). Therefore, the codon usage of the entire rce1 gene from Rhizopus oryzae was changed into the codon usage of the cellulase genes from Humicola sp. Next, four chimeric endoglucanase genes, in which each of the four regions of the egl3 gene from H. grisea was replaced by the corresponding codon usage-optimized rce1 gene, were constructed and expressed in H. insolens. Of four chimeric endoglucanases, one was not expressed at all, and the others were expressed in truncated forms (26). Thus, heterologous overexpression of a useful enzyme in a mature form in H. insolens is of great importance to the detergent industry.
In an effort to obtain new endoglucanases having a neutral or alkaline optimum pH, high defibrillation activity, and high resistance to anionic surfactants and oxidizing agents, we screened a large number of fungal culture supernatants. As a result, we found a new family 45 endoglucanase, designated STCE1, in the culture supernatant of Staphylotrichum coccosporum. In the present study, we purified and characterized STCE1. Furthermore, we isolated the encoding gene, designated the stce1 gene, and overexpressed this gene in H. insolens.
MATERIALS AND METHODS
Bacterial strains and cultural conditions.
S. coccosporum NBRC 31817 was obtained from the NITE Biological Resource Center, Department of Biotechnology, National Institute of Technology and Evaluation, Chiba, Japan. S. coccosporum NBRC 31817 was used as the source of chromosomal DNA. For chromosomal DNA preparation, S. coccosporum NBRC 31817 was cultured aerobically at 28°C for 72 h in a medium containing 2.0% Avicel, 2.0% corn steep liquor, 2.0% yeast extract, 1.0% glucose, and 0.2% potassium phosphate (ACYG medium).
Enzyme purification. (i) Step 1.
S. coccosporum NBRC 31817 was shake cultured at 28°C in ACYG medium. After incubation for 72 h, the culture was centrifuged at 27,000 × g for 30 min at 4°C. The supernatant was collected and used in the following steps.
(ii) Step 2.
After ammonium sulfate was added to the culture supernatant to a final concentration of 1.5 M, the mixture was loaded on a HiTrap phenyl HP column (18 by 50 mm; Amersham Pharmacia Biotech) equilibrated with 1.5 M ammonium sulfate for hydrophobic chromatography. After it was washed with 4 bed volumes of the same solution, the column was eluted with a stepwise ammonium sulfate gradient (1.5, 0.9, 0.75, 0.6, 0.15, and 0 M), followed by distilled water, at a flow rate of 2 ml/min. High defibrillation activity was found in a fraction that eluted with 0 M ammonium sulfate.
(iii) Step 3.
The fraction obtained in step 2 was diluted 10-fold with 50 mM acetate buffer (pH 4.0) and was then separated by cation-exchange chromatography on a Mono S HR 5/5 column (5 by 50 mm; Amersham Pharmacia Biotech) equilibrated with the same buffer. After it was washed with 3 bed volumes of the same buffer, the column was eluted with a continuous linear gradient from 50 mM acetate buffer (pH 4.0) to 1 M NaCl in 50 mM acetate buffer (pH 5.0) at a flow rate of 1 ml/min. The active fraction eluted at 50 mM NaCl.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (19) using a ready-made 10% polyacrylamide gel (Tefco). After electrophoresis, the gel was stained using 2D-silver stain II “Daiichi” (Daiichi Pure Chemicals). SDS-PAGE low-molecular-weight standards were used as the molecular mass markers (Bio-Rad).
Protein sequencing.
For determination of the amino acid sequences of peptide fragments derived from the internal regions of STCE1, purified STCE1 was lyophilized and dissolved in 50 mM Tris-HCl (pH 8.0). The enzyme was digested with lysylendopeptidase added at a molar ratio of STCE1 to lysylendopeptidase of 100:1. The reaction mixture was incubated at 37°C for 48 h, and the peptides generated were separated by using a model 172 micropreparative high-performance liquid chromatography (HPLC) system (Applied Biosystems). N-terminal amino acid sequencing of the purified STCE1 and determination of the amino acid sequences of peptide fragments derived from internal regions of STCE1 were performed using an ABI model 491 protein sequencer (Applied Biosystems).
Protein and enzyme assays.
For the assay of carboxymethyl cellulase (CMCase) activity, reaction mixtures containing 0.32 μg of STCE1 and 10 mg of carboxymethyl cellulose in 1.0 ml of 50 mM sodium phosphate buffer (pH 6.0) were incubated for 15 min at 60°C. The liberated reducing sugars were then measured as d-glucose equivalents using the dinitrosalicylic acid assay (22). One unit of activity was defined as the amount of enzyme that released 1 μmol of reducing sugar per min.
The CMCase activity of STCE1 was measured under several different pH conditions using 50 mM sodium citrate (pH 3 to 4), sodium acetate (pH 4 to 6), sodium phosphate (pH 6 to 8), and glycine-NaOH (pH 8 to 10) buffers. The effect of temperature was examined under standard conditions by varying the temperature (20 to 70°C). Protein concentrations were determined by Bradford's method using a protein assay kit with bovine serum albumin as the standard (8).
Estimation of defibrillation activity of endoglucanase.
Dyed cotton or lyocell knit fabric (brown knit fabric [L value, 28.47], 6 by 8 cm, 2.0 g) and four rubber balls were added to various amounts of endoglucanase in 40 ml of 10 mM acetate buffer (pH 4.0 for BCE1, EGI, and EGII) and 2 mM sodium phosphate buffer (pH 6.0 for STCE1, EGL3, EGL4, EGVI, RCE1, RCE2, MCE1, MCE2, and PCE1), and then the reaction mixtures were incubated for 60 min at 40°C in the 500-ml stainless steel pots of an L-20 Launder Meter (Daiei Kagaku Seiki MFG) rotating at 40 revolutions per min. After incubation, the defibrillation activity was evaluated by determining the amount of microfibrils released by measuring the turbidity of each reaction mixture, expressed as the optical density at 595 nm (OD595), as described previously (12). One unit of activity was defined as the amount of enzyme that released microfibrils that increased the OD595 to 0.3 in 60 min. The defibrillation activity of endoglucanase was expressed as the number of units per mg of enzyme.
Measurement of resistance to an anionic surfactant and an oxidizing agent of endoglucanase.
The concentration of linear alkyl benzene sulfonate (LAS) (an anionic surfactant) or sodium hypochlorite (an oxidizing agent) that inhibited 50% of the CMCase activity of endoglucanase without LAS or sodium hypochlorite (50% inhibitory concentration [IC50]) was determined. Resistance to the anionic surfactant or oxidizing agent was represented by the IC50 of endoglucanase.
Cloning and sequence analysis of the stce1 gene.
When S. coccosporum genomic DNA digested with EcoRI was Southern blotted with the egl4 gene from H. grisea as a probe (38), two signals at about 5 and 10 kbp appeared (the hybridized membrane was washed two times with 0.6× SSC buffer at 42°C [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], and the positive bands were detected with an ECL direct DNA/RNA labeling detection system). This result suggested that two genes homologous to egl4 were present. Since the amino acid residues deduced from the partial sequence of the 10-kbp DNA fragment coincided with the internal amino acid sequences of the purified STCE1 from S. coccosporum, the gene homologous to egl4 located in the 10-kbp DNA fragment was designated the stce1 gene.
To clone the stce1 gene, plaque hybridization was performed with the egl4 gene as a probe against the genomic library using the DNA fragments of 8- to 23-kbp DNA fragments produced by EcoRI digestion of the S. coccosporum genomic DNA. Four positive phage clones were obtained from about 50,000 phage plaques. These clones were considered to contain the stce1 gene detected by Southern hybridization. The phage DNA extracted from a positive clone was digested with SalI, and the resultant 4.4- and 2.5-kbp fragments were subcloned into the SalI site of pUC119. Nucleotide sequencing of these two fragments indicated the presence of a single open reading frame coinciding with the N-terminal amino acid sequence of STCE1. However, it was presumed that this sequence contained introns, and therefore, cDNA of the stce1 gene was isolated by reverse transcription-PCR. Oligonucleotide primers for the N and C termini (stce1-CN [5′-GCGGATCCATGCGTTCCTCCCCCGTC-3′] and stce1-CC [5′-GCGGATCCTTAAAGGCACTGCGAGTACC-3′]) were prepared from the information for the genomic stce1 sequence. As a result of reverse transcription-PCR, the amplified fragment appeared at about 0.95 kbp on an agarose gel electrophoresis gel. The fragment was subcloned into the BamHI site of pUC18. Nucleotide sequencing of the 0.95-kbp fragment indicated the presence of a single open reading frame encoding a predicted 316-residue protein and the presence of one intron between positions 333 and 334 for the stce1 gene.
Expression of STCE1 by H. insolens.
The family 45 endoglucanase NCE4 gene from H. insolens strain FERM BP-5799 was ligated between the cellobiohydrolase NCE2 promoter and NCE2 terminator from H. insolens as described previously (18). The resulting plasmid was designated pJND-NCE4. The NCE4 gene was replaced by the open reading frame of the stce1 cDNA without the signal sequence next to the signal sequence of the NCE4 gene of pJND-NCE4 (18). H. insolens strain FERM BP-5799 was transformed with this plasmid as described previously (18). H. insolens transformants were selected on a selective medium containing 1% glucose, 0.4% yeast extract, 0.2% malt extract, 0.02% hygromycin, and 1.0% agar (pH 6.8). Transformants overexpressing a 45- to 49-kDa protein detected by SDS-PAGE were grown at 37°C for 96 h with shaking in a medium containing 5.0% Avicel, 2.0% yeast extract, 0.1% polypeptone, and 0.03% Mg2SO4 (pH 6.8).
Quantitative analysis of recombinant STCE1 in the culture supernatant.
Culture supernatant of H. insolens was diluted 20-fold with water and centrifuged at 27,000 × g for 30 min. The supernatant (0.1 ml) was subjected to HPLC on a TSKgel TMS-250 column (4.6 mm [inside diamter] by 7.5 cm; TOSOH) eluted with a linear 0 to 80% acetonitrile gradient in 0.05% trifluoroacetic acid. The peak of STCE1 concentration was monitored with a UV detector at 280 nm. Protein concentrations were determined by comparison with the peak area for purified STCE1 from S. coccosporum, for which the concentration was previously determined using bovine serum albumin as the standard.
Nucleotide sequence accession number.
The nucleotide sequence of the stce1 gene from S. coccosporum NBRC 31817 has been accessioned in the DDBJ database under number AB248917.
RESULTS AND DISCUSSION
Purification and characterization of endoglucanase STCE1.
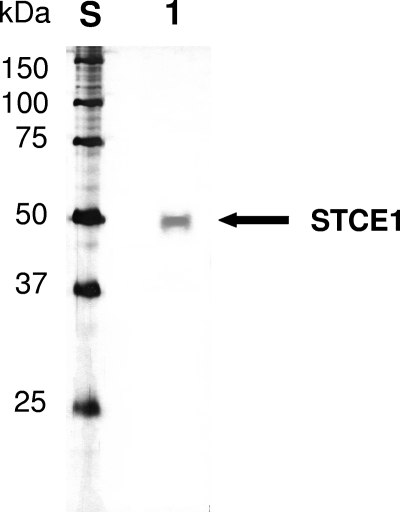
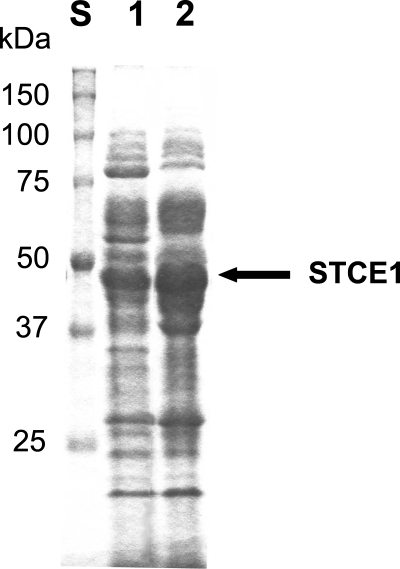
About 1,600 fungal culture supernatants were screened for endoglucanases having a neutral or alkaline optimum pH, high defibrillation activity, and high levels of resistance to anionic surfactants and oxidizing agents. As a result, we found a new endoglucanase, designated STCE1, in the culture supernatant of S. coccosporum. By using two-step chromatographic fractionation, STCE1 was purified to apparent homogeneity by SDS-PAGE as a single band with a mobility corresponding to an apparent molecular mass of 49 kDa (Fig. 1). The N-terminal amino acid sequence of STCE1 (STCE1-N) was determined to be ADGKSTRYWDCCKPSCSWPGKASVN. The consensus amino acid residues of family 45 glycoside hydrolase, (S, T, or A)-T-R-Y-(F, Y, or W)-D-X-X-X-X-X-(C or A) (http://www.cazy.org/fam/GH45.html), were conserved in the N-terminal amino acid sequence of STCE1. Hence, STCE1 was considered to be a family 45 glycoside hydrolase protein.
FIG. 1.
SDS-PAGE of purified STCE1 from S. coccosporum. Lane S, standard proteins; lane 1, purified STCE1 obtained by chromatographic fractionation in two steps.
The purified STCE1 was digested with lysylendopeptidase, and each peptide fragment (STCE1a to STCE1e) obtained was sequenced. The amino acid sequences of these peptide fragments are as follows: STCE1a, ASVNQPVFAC; STCE1b, TMVVQST; STCE1c, PGCYWRF; STCE1d, NADNPTFTFR; and STCE1e, QNDWYSQCL.
Comparison of the defibrillation activities of various endoglucanases.
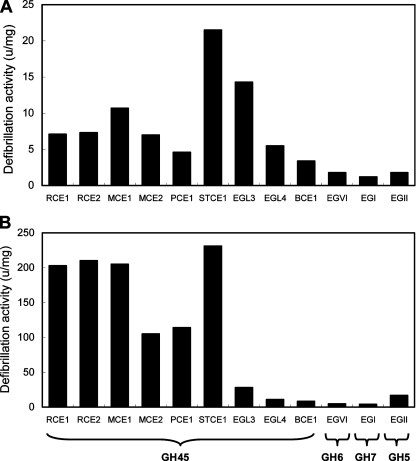
To estimate the properties for laundry use, the defibrillation activities of the purified STCE1 on the dyed cotton and lyocell fabrics were compared with those of other purified representative endoglucanases used with textiles or for laundry. These endoglucanases, including family 45 endoglucanases RCE1 and RCE2 from R. oryzae, MCE1 and MCE2 from M. circinelloides, PCE1 from P. nitens, EGL3 and EGL4 from H. grisea, BCE1 from Beltraniella portoricensis, family 6 endoglucanase EGVI (Cel6B) from H. insolens, family 7 endoglucanase EGI (Cel7B) from Trichoderma viride, and family 5 endoglucanase EGII from Trichoderma reesei, were purified as described previously (2, 3, 10, 24, 25, 27, 32, 34, 37, 38). As shown in Fig. 2A, the defibrillation activity of STCE1 (21.5 U/mg) on the cotton fabric was higher than that of the other endoglucanases (Fig. 2A). The defibrillation activity of STCE1 (231 U/mg) on the lyocell fabric was almost the same as the activities of RCE1, RCE2, and MCE1 and was higher than the activities of MCE2, PCE1, EGL3, EGL4, BCE1, EGVI, EGI, and EGII (Fig. 2B). These results indicate that family 45 endoglucanases had higher defibrillation activity than family 5, 6, and 7 endoglucanases and that, in particular, STCE1 had the highest defibrillation activities on the dyed cotton and lyocell fabrics among the family 45 endoglucanases tested.
FIG. 2.
Comparison of the defibrillation activities of representative endoglucanases on dyed cotton fabric (A) and on dyed lyocell fabric (B). The defibrillation activity was evaluated by determining the amount of microfibrils released by measuring the turbidity of the reaction mixture as OD595 as described previously (12). One unit of activity was defined as the amount of enzyme that released microfibrils that increased the OD595 to 0.3 in 60 min. The defibrillation activity of endoglucanase was expressed as units per milligram of enzyme.
Comparison of the resistance of family 45 endoglucanases to an anionic surfactant and an oxidizing agent.
First, the pH and temperature profiles for the CMCase activity of STCE1 were examined. The optimal pH for activity of STCE1 was 6.0, and the optimum temperature for STCE1 was 60°C. Because a neutral or alkaline optimum pH for enzymes is in demand for laundry use, STCE1 may be appropriate for laundry use.
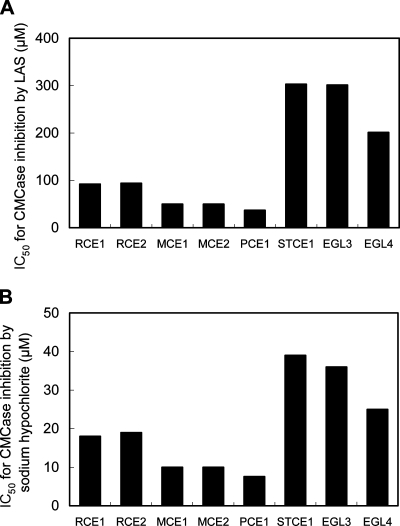
Because BCE1, EGI, EGII, and EGVI had much lower defibrillation activities on the dyed cotton and lyocell fabrics (Fig. 2A and B) and the optimal pH for BCE1, EGI, and EGII was acidic (3, 32, 37), these endoglucanases are not suitable for laundry use. Hence, we concluded that further characterization of BCE1, EGI, EGII, and EGVI for laundry use is not required. Therefore, the resistances to LAS (an anionic surfactant) or sodium hypochlorite (an oxidizing agent), the main components in detergents, were compared using only STCE1, RCE1, RCE2, MCE1, MCE2, PCE1, EGL3, and EGL4 (Fig. 3A and B). The CMCase activity of STCE1 was inhibited by LAS, with an IC50 of 303 μM, which was almost the same as the IC50 of EGL3 and was higher than the IC50s of RCE1, RCE2, MCE1, MCE2, PCE1, and EGL4. The CMCase activity of STCE1 was inhibited by sodium hypochlorite, with an IC50 of 39 μM, which was almost the same as the IC50 of EGL3 and was higher than the IC50s of RCE1, RCE2, MCE1, MCE2, PCE1, and EGL4. These results indicate that STCE1 was the most resistant to the anionic surfactant and oxidizing agent among the family 45 endoglucanases tested. Based on the results described here, STCE1 is considered to be an all-around suitable enzyme that has all the properties required for laundry use.
FIG. 3.
Comparison of the levels of resistance to LAS (A) or sodium hypochlorite (B) of family 45 endoglucanases. The resistance to the anionic surfactant or oxidizing agent of each endoglucanase is represented by the IC50 for LAS or sodium hypochlorite for CMCase activity, respectively. The IC50 is the concentration of LAS or sodium hypochlorite that inhibited 50% of the CMCase activity of endoglucanase without LAS or sodium hypochlorite.
Although the catalytic domain of RCE1 had high defibrillation activities on dyed cotton and lyocell fabrics, this enzyme had low resistance to anionic surfactants and oxidizing agents (29, 35). By contrast, the catalytic domain of EGL3 was highly resistant to anionic surfactants and oxidizing agents, whereas it had low defibrillation activity on dyed lyocell fabrics (35). In a previous study, to identify the amino acids involved in the various properties needed for laundering, we compared the characteristics of RCE1, EGL3, and three chimeric enzymes (ER1, ER2, and ER3) in which each of the three regions of the catalytic domain of EGL3 was replaced with the corresponding region of the catalytic domain of RCE1 (26). We found that amino acids in the N-terminal region were involved in resistance to anionic surfactants and oxidizing agents and that amino acids in the region adjacent to the N-terminal region were involved in defibrillation of dyed cotton and lyocell fabrics (36). Considering this, STCE1 might be composed of a chimeric structure that has both regions involved in the high level of resistance and high defibrillation activity. However, the properties of STCE1 were superior to those of the chimeric endoglucanase ER2, which had two regions involved in the high level of resistance and high defibrillation activity (i.e., defibrillation activities of 17.1 U/mg on the cotton fabric and 91.0 U/mg on the lyocell fabric; IC50s of 202 μM for LAS and 31 μM for sodium hypochlorite [36]), suggesting that the amino acids of STCE1 are more optimized for laundry use than those of ER2. The identification of such amino acids is one of challenges in our studies.
Structural characterization of the stce1 gene.
The stce1 gene was cloned from the S. coccosporum genomic library using the egl4 gene from H. grisea as a probe (38). The amino acid residues (residues 22 to 52, 123 to 129, 185 to 191, 196 to 205, and 308 to 316) deduced from the stce1 cDNA gene perfectly coincided with the N-terminal and internal amino acid sequences of the purified STCE1 from S. coccosporum (Fig. 4). This coincidence suggests that the stce1 cDNA gene encodes STCE1, and residues 1 to 21 are a signal peptide. The calculated molecular mass of mature STCE1 was 32,645 Da.
FIG. 4.
Nucleotide and deduced amino acid sequences for the stce1 cDNA from S. coccosporum. The arrows indicate the borders of each domain, including the signal peptide, catalytic domain, linker domain, and CBM. The N-terminal and internal amino acid sequences of the purified STCE1 from S. coccosporum are underlined. The consensus amino acid residues of family 45 glycoside hydrolase are double underlined. A potential N-linked glycosylation site (N-X-S/T) is shaded.
The consensus amino acid residues of family 45 glycoside hydrolase, (S, T, or A)-T-R-Y-(F, Y, or W)-D-X-X-X-X-X-(C or A) (http://www.cazy.org/fam/GH45.html), were conserved in the amino acid sequences (residues 26 to 37) of STCE1. Therefore, STCE1 was considered to belong to glycoside hydrolase family 45. The consensus amino acid residues of the carbohydrate-binding module (CBM) in family 1 (7) were conserved in the amino acid sequences deduced from the stce1 gene (Fig. 4). STCE1 consisted of three distinct domains: an N-terminal catalytic domain (family 45), a linker domain, and a C-terminal CBM (family 1) (Fig. 4). The amino acid sequence of STCE1 contained one potential N-linked glycosylation site (N-X-S/T) at position 87.
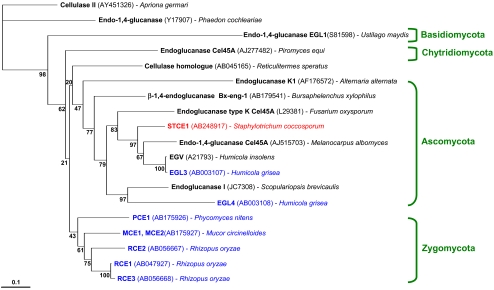
A phylogenetic tree analysis of the amino acid sequences of catalytic domains of family 45 endoglucanases suggested that the catalytic domain of STCE1 is evolutionarily close to the catalytic domains of enzymes from organisms classified as members of phylum Ascomycota, including those of EGL3 and EGL4 from H. grisea, EGV from H. insolens, and Cel45A from Fusarium oxysporum; however, they are evolutionarily far from the catalytic domains of MCE1, MCE2, RCE1, RCE2, RCE3, and PCE1 from members of the phylum Zygomycota (Fig. 5). Of the various family 45 endoglucanases, MCE1 and MCE2 (2), RCE1, RCE2, and RCE3 (23), and PCE1 (34) from members of the phylum Zygomycota had the CBM at the N terminus, whereas STCE1, EGL3 and EGL4 (38), EGV (11), and Cel45A from F. oxysporum (33) from members of the phylum Ascomycota had the CBM at the C terminus. Furthermore, MCE1, MCE2, RCE1, RCE2, RCE3, and PCE1 from members of phylum Zygomycota exhibited low levels of resistance to anionic surfactants and oxidizing agents, whereas STCE1, EGL3, and EGL4 from members of the phylum Ascomycota exhibited high levels of resistance to anionic surfactants and oxidizing agents (Fig. 3). It is very interesting that the taxonomic classification perfectly coincided with the phylogenetic classification, the classification based on the CBM location, and the classification based on the detergent resistance.
FIG. 5.
Phylogenetic tree for the catalytic domains of family 45 endoglucanases. The analysis was performed based on a comparison of amino acid sequence data using the Clustal W ver.1.83 program. Scale bar = 0.1% amino acid substitutions. Bootstrap percentages from 1,000 resamplings are indicated at the internal nodes. Family 45 endoglucanases tested in this study are indicated by blue or red.
Expression of STCE1 by H. insolens.
The amino acid sequences of the catalytic domain of STCE1 were phylogenetically close to those of EGL3 and EGL4 from H. grisea and of EGV from H. insolens, suggesting that the recombinant STCE1 might be overproduced in Humicola species. Therefore, H. insolens strain FERM BP-5799 was transformed with the plasmid containing the stce1 cDNA, and transformants that had a new band at around 49 kDa in SDS-PAGE gels were screened.
As shown in Fig. 6, it is evident that an abundant new protein with a band around 49 kDa was present only in the transformant with the stce1 cDNA. Next, to examine whether the abundant new protein is STCE1, we analyzed the supernatant of the transformant with the stce1 cDNA and the supernatant of the transformant without the stce1 cDNA using HPLC. A peak corresponding to the retention time for the purified STCE1 from S. coccosporum appeared only for the transformant with the stce1 cDNA (data not shown). The N-terminal amino acid sequence (ADGKSTRYWDCCKPSCSWPG) of the protein obtained from the peak coincided with the N-terminal amino acid sequence of STCE1, and one of the internal amino acid sequences was QNDWYSQCL, which is the C-terminal amino acid sequence of STCE1. These results clearly show that the recombinant STCE1 was secreted in a mature form. Furthermore, extremely high levels (0.90 mg protein per ml of culture supernatant, 27% of the total proteins) of the recombinant STCE1 were secreted in the culture supernatant. This level of production was about 100-fold higher than that in the culture supernatant of S. coccosporum (0.0085 mg STCE1 protein per ml of culture supernatant).
FIG. 6.
SDS-PAGE of STCE1 expressed by H. insolens. Lane S, standard proteins; lane 1, a culture supernatant of H. insolens transformant with vector plasmid alone; lane 2, a culture supernatant of H. insolens transformant with the stce1 cDNA.
We deduced that such a high level of production of STCE is due to the following two reasons. First, the codon usage of genes derived from Staphylotrichum was very similar to that of genes derived from Humicola, and the stce1 cDNA gene was not spliced unexpectedly. Second, the linker region between the CBM and catalytic domain of STCE1 could not be cleaved easily by proteases from H. insolens. In fact, it has been reported that the linker region of recombinant RCE1 endoglucanase or chimeric RCE1 and EGL3 endoglucanases was cleaved by proteases produced from the host fungus H. insolens, and subsequently only the catalytic domain was secreted in a truncated form (26, 28). Although even the catalytic domain alone had defibrillation activity, the CBM at the C terminus enhanced the defibrillation activity of the catalytic domain (1, 36). From this point of view, the high levels of production of recombinant STCE1 in a mature form are of great importance in terms of reducing the cost of enzymatic treatment for the detergent industry.
Endoglucanases for laundry use constitute a large world market (about 100 million dollars), and EGV from H. insolens (Carezyme; Novozymes) has dominated this market for several years (13, 17, 20). Because the level of similarity in the amino acid sequences of EGV from H. insolens (11) and EGL3 from H. grisea (38) is 99.0%, it has been deduced that these two endoglucanases have the same properties. Hence, we are convinced that the properties of STCE1 for laundry use should be superior to those of EGV.
Footnotes
Published ahead of print on 11 April 2008.
REFERENCES
- 1.Azevedo, H., D. Bishop, and A. Cavaco-Paulo. 2000. Effects of agitation level on the adsorption, desorption, and activities on cotton fabrics of full-length and core domains of EGV (Humicola insolens) and CenA (Cellulomonas fimi). Enzyme Microb. Technol. 27:325-329. [DOI] [PubMed] [Google Scholar]
- 2.Baba, Y., A. Shimonaka, J. Koga, H. Kubota, and T. Kono. 2005. Alternative splicing produces two endoglucanases with one or two carbohydrate-binding modules in Mucor circinelloides. J. Bacteriol. 187:3045-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, Y., A. Shimonaka, J. Koga, K. Murashima, H. Kubota, and T. Kono. 2005. Purification and characterization of a new endo-1,4-β-d-glucanase from Beltraniella portoricensis. Biosci. Biotechnol. Biochem. 69:1198-1201. [DOI] [PubMed] [Google Scholar]
- 4.Beguin, P., J. Millet, S. Chauvaux, S. Salamitou, K. Tokatlidis, J. Navas, T. Fujino, M. Lemaire, O. Raynaud, M. K. Daniel, and J. P. Aubert. 1992. Bacterial cellulases. Biochem. Soc. Trans. 20:42-46. [DOI] [PubMed] [Google Scholar]
- 5.Bhat, M. K. 2000. Cellulases and related enzymes in biotechnology. Biotechnol. Adv. 18:355-383. [DOI] [PubMed] [Google Scholar]
- 6.Bhat, M. K., and S. Bhat. 1997. Cellulose degrading enzymes and their potential industrial applications. Biotechnol. Adv. 15:583-620. [DOI] [PubMed] [Google Scholar]
- 7.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Cavaco-Paulo, A. 1998. Mechanism of cellulase action in textile processes. Carbohydr. Polym. 37:273-277. [Google Scholar]
- 10.Davies, G. J., A. M. Brzozowski, M. Dauter, A. Varrot, and M. Schulein. 2000. Structure and function of Humicola insolens family 6 cellulases: structure of the endoglucanase, Cel6B, at 1.6 A resolution. Biochem. J. 348:201-207. [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, G. J., G. G. Dodson, R. E. Hubbard, S. P. Tolley, Z. Dauter, K. S. Wilson, C. Hjort, J. M. Mikkelsen, G. Rasmussen, and M. Schulein. 1993. Structure and function of endoglucanase V. Nature 365:362-364. [DOI] [PubMed] [Google Scholar]
- 12.Din, N., N. R. Gilkes, B. Tekant, R. C. J. Miller, R. A. J. Warren, and D. G. Kilburn. 1991. Non-hydrolytic disruption of cellulose fibres by the binding domain of a bacterial cellulase. Bio/Technology 9:1096-1099. [Google Scholar]
- 13.Glaser, V. 2000. Steady growth for industrial enzyme market. Genet. Eng. News 20:8-36. [Google Scholar]
- 14.Henrissat, B., M. Claeyssens, P. Tomme, L. Lemesle, and J. P. Mornon. 1989. Cellulase families revealed by hydrophobic cluster analysis. Gene 81:83-95. [DOI] [PubMed] [Google Scholar]
- 15.Henrissat, B., H. Driguez, C. Viet, and M. Schülein. 1985. Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Bio/Technology 3:722-726. [Google Scholar]
- 16.Inoue, T., K. Murashima, J. Azuma, A. Sugimoto, and M. Slaytor. 1997. Cellulose and xylan utilization in the lower termite Reticulitermes speratus. J. Insect Physiol. 43:235-242. [DOI] [PubMed] [Google Scholar]
- 17.Jakobsen, T. S., P. Lindegaard, and M. Chan. 1998. Color-care cellulases: fabric color shield. Inform 9:788-792. [Google Scholar]
- 18.Koga, J., Y. Baba, A. Nakane, S. Hamura, T. Nishimua, S. Gomi, H. Kubota, and T. Kono. September 2006. Endoglucanase STCE and cellulase preparation containing the same. European patent 1700917.
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Miettinen-Oinonen, A. 2007. Cellulases in the textile industry, p. 51-63. In J. Polainoa and A. MacCabe, Industrial enzymes: structure, function and applications. Springer, Dordrecht, The Netherlands.
- 21.Miettinen-Oinonen, A., and P. Suominen. 2002. Enhanced production of Trichoderma reesei endoglucanases and use of the new cellulase preparations in producing the stonewashed effect on denim fabric. Appl. Environ. Microbiol. 68:3956-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, G. L. 1959. Use of dinitrosalicyclic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 23.Moriya, T., K. Murashima, A. Nakane, K. Yanai, N. Sumida, J. Koga, T. Murakami, and T. Kono. 2003. Molecular cloning of endo-β-d-1,4-glucanase genes, rce1, rce2, and rce3, from Rhizopus oryzae. J. Bacteriol. 185:1749-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murashima, K., T. Moriya, T. Hamaya, J. Koga, N. Sumida, K. Aoyagi, T. Murakami, and T. Kono. December 2000. Enzyme endoglucanase and cellulase preparations containing the same. U.S. patent 6159720.
- 25.Murashima, K., T. Nishimura, Y. Nakamura, J. Koga, T. Moriya, N. Sumida, T. Yaguchi, and T. Kono. 2002. Purification and characterization of new endo-1,4-β-d-glucanases from Rhizopus oryzae. Enzyme Microb. Technol. 30:319-326. [Google Scholar]
- 26.Murashima, K., A. Shimonaka, T. Nishimura, Y. Baba, J. Koga, H. Kubota, and T. Kono. 2006. Exploring amino acids responsible for the temperature profile of glycoside hydrolase family 45 endoglucanase EGL3 from Humicola grisea. Biosci. Biotechnol. Biochem. 70:2205-2212. [DOI] [PubMed] [Google Scholar]
- 27.Murashima, K., N. Sumida, A. Nakane, K. Yanai, T. Nishimura, J. Koga, T. Murakami, and T. Kono. November 2006. Endoglucanase enzyme NCE5 and cellulase preparations containing the same. U.S. patent 7138263.
- 28.Nakamura, Y., T. Moriya, Y. Baba, K. Yanai, N. Sumida, T. Nishimura, K. Murashima, A. Nakane, T. Yaguchi, J. Koga, T. Murakami, and T. Kono. July 2005. Endoglucanases and cellulase preparations containing the same. U.S. patent 6921655.
- 29.Nakane, A., J. Koga, H. Kubota, and T. Kono. 2005. Specific characteristics of an endoglucanase RCE1 from Rhizopus oryzae in the treatment of the dyed cotton fabrics. Sen'i Gakkaishi 61:229-233. [Google Scholar]
- 30.Ohmiya, Y., T. Takeda, S. Nakamura, F. Sakai, and T. Hayashi. 1995. Purification and properties of wall-bound endo-1,4-β-glucanase from suspension-cultured poplar cells. Plant Cell Physiol. 36:607-614. [PubMed] [Google Scholar]
- 31.Otzen, D. E., L. Christiansen, and M. Schulein. 1999. A comparative study of the unfolding of the endoglucanase Cel45 from Humicola insolens in denaturant and surfactant. Protein Sci. 8:1878-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saloheimo, M., P. Lehtovaara, M. Penttila, T. T. Teeri, J. Stahlberg, G. Johansson, G. Pettersson, M. Claeyssens, P. Tomme, and J. K. Knowles. 1988. EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene 63:11-22. [DOI] [PubMed] [Google Scholar]
- 33.Sheppard, P. O., F. J. Grant, P. J. Oort, C. A. Sprecher, D. C. Foster, F. S. Hagen, A. Upshall, G. L. McKnight, and P. J. O'Hara. 1994. The use of conserved cellulase family-specific sequences to clone cellulase homologue cDNAs from Fusarium oxysporum. Gene 150:163-167. [DOI] [PubMed] [Google Scholar]
- 34.Shimonaka, A., Y. Baba, J. Koga, A. Nakane, H. Kubota, and T. Kono. 2004. Molecular cloning of a gene encoding endo-β-d-1,4-glucanase PCE1 from Phycomyces nitens. Biosci. Biotechnol. Biochem. 68:2299-2305. [DOI] [PubMed] [Google Scholar]
- 35.Shimonaka, A., J. Koga, Y. Baba, T. Nishimura, K. Murashima, H. Kubota, and T. Kono. 2006. Specific characteristics of family 45 endoglucanases from Mucorales in the use of textiles and laundry. Biosci. Biotechnol. Biochem. 70:1013-1016. [DOI] [PubMed] [Google Scholar]
- 36.Shimonaka, A., K. Murashima, J. Koga, Y. Baba, T. Nishimura, H. Kubota, and T. Kono. 2006. Amino acid regions of family 45 endoglucanases involved in cotton defibrillation and in resistance to anionic surfactants and oxidizing agents. Biosci. Biotechnol. Biochem. 70:2460-2466. [DOI] [PubMed] [Google Scholar]
- 37.Shoemaker, S. P., and R. D. Brown, Jr. 1978. Characterization of endo-1,4-β-d-glucanases purified from Trichoderma viride. Biochim. Biophys. Acta 523:147-161. [DOI] [PubMed] [Google Scholar]
- 38.Takashima, S., H. Iikura, A. Nakamura, M. Hidaka, H. Masaki, and T. Uozumi. 1999. Comparison of gene structures and enzymatic properties between two endoglucanases from Humicola grisea. J. Biotechnol. 67: 85-97. [DOI] [PubMed] [Google Scholar]
- 39.Wood, T. M. 1992. Fungal cellulases. Biochem. Soc. Trans. 20:46-53. [DOI] [PubMed] [Google Scholar]