Abstract
Marine sponges in the genus Ircinia are known to be good sources of secondary metabolites with biological activities. A major obstacle in the development of sponge-derived metabolites is the difficulty in ensuring an economic, sustainable supply of the metabolites. A promising strategy is the ex situ culture of sponges in closed or semiclosed aquaculture systems. In this study, the marine sponge Ircinia strobilina (order Dictyoceratida: family Irciniidae) was collected from the wild and maintained for a year in a recirculating aquaculture system. Microbiological and molecular community analyses were performed on freshly collected sponges and sponges maintained in aquaculture for 3 months and 9 months. Chemical analyses were performed on wild collected sponges and individuals maintained in aquaculture for 3 months and 1 year. Denaturing gradient gel electrophoresis was used to assess the complexity of and to monitor changes in the microbial communities associated with I. strobilina. Culture-based and molecular techniques showed an increase in the Bacteroidetes and Alpha- and Gammaproteobacteria components of the bacterial community in aquaculture. Populations affiliated with Beta- and Deltaproteobacteria, Clostridia, and Planctomycetes emerged in sponges maintained in aquaculture. The diversity of bacterial communities increased upon transfer into aquaculture.
Sponges harbor diverse microorganisms, and various symbiotic relationships between sponges and microorganisms may contribute to the sponge's health and nutrition. Sponge-associated microbes can constitute up to 60% of the sponge biomass (24, 66, 67, 72). Important roles of the symbionts include photosynthetic carbon fixation (73), nitrification (6, 9), nitrogen fixation (59, 74, 75), and anaerobic metabolism (27). Another important role of sponge-associated bacteria is the production of potential secondary metabolites, such as antibiotics, antifungal compounds, and compounds that prevent predation or fouling (45). Sponges are sessile filter feeders with numerous tiny pores on the body surface and channels within the body that allow water to enter and circulate, with microorganisms and organic matter being removed through filtration (35, 68). Sponges contain assemblages of symbiotic bacteria that are distinct from the bacteria that are being filtered out of the surrounding water during the sponges' feeding process (62).
Marine sponges are a rich source of pharmacologically active compounds that can potentially be used as medicines to cure human diseases, and the isolation of bioactive compounds from sponges has been reviewed extensively (17, 25, 30, 35, 49, 55, 58). Bacteria isolated from sponges have also been sources of novel bioactive compounds. In some cases, symbiotic bacteria may be the producers of promising compounds first found in the extracts of sponges. Circumstantial evidence for the microbial origin of a sponge-derived compound may be supplied by the structure of the compound (31). The onnamides and theopederins are polyketides that structurally resemble pederin, the defensive polyketide in Paederus fuscipes beetles. Strong evidence was provided for a bacterial producer of the onnamides and theopederins found in the sponge Theonella swinhoei (49). Genes closely resembling those encoding pederin were found in the complex metagenome of T. swinhoei and had characteristic prokaryotic signatures (49).
Several bioactive compounds from sponges have passed the preclinical stage (44). The low yields of these compounds, known as the supply problem, are a major obstacle limiting successful transition of marine-derived compounds through clinical studies and into commercial production (42, 46). Harvesting sponges from the environment generally will not provide a reliable, large-scale supply and could lead to the extinction of the particular sponge species. Several techniques may produce large quantities of sponge biomass needed for the extraction of bioactive metabolites that are not viable candidates for total synthesis. Such techniques include sponge farming (10, 11) and the culture of primmorphs. Primmorphs are aggregates of sponge cells that still contain bacteria (reviewed by Müller et al. [41]). The NOMATEC (Novel Marine Technologies) project showed that Ircinia variabilis is suitable for mariculture (69). Furthermore, De Rosa et al. (7) reported the development of cell cultures from Ircinia muscarum. There were major differences in the composition of secondary metabolites between the wild sponge and its cell cultures, with a lower concentration of lipids and a loss of sterols and volatile compounds in cell cultures (8). When the bioactive compounds are produced by a sponge-associated bacterium, options include isolation and cultivation of this producer bacterium as well as using molecular approaches, including transfer of symbiont biosynthetic genes into cultivable bacteria (20, 48). Another possibility is to grow the entire sponge and its microbial community in self-contained aquaculture systems for the economic, sustainable supply of important metabolites. The advantage of the latter strategy compared with growth of sponges in the wild or in open-water mariculture is better control of environmental conditions, such as temperature, light, food supply, and possibly precursors of important bioactive metabolites. In addition, aquaculture of sponges provides less perturbation of the bacterium-host association over growth of bacterial “producer” strains in pure culture, which could be very important for maintaining production of compounds of interest.
Aquaculture in tanks might be preferable to open-water mariculture because it is generally more reliable and may also be less expensive, partly because it is possible to shift from seasonal growth to continuous growth during the year (10-12, 46, 60). To examine the potential of ex situ culture of sponges in closed aquaculture systems, it is crucial to determine whether the microbial communities change upon transfer into aquaculture. The aim of our study was to address the following two questions. (i) Does transferring the sponge into aquaculture affect the stability of sponge-microbe associations? (ii) Are changes in the microbial communities correlated with changes in the chemistry of the sponge? Ircinia strobilina (Lamarck, 1816) was chosen for this study as a representative of the genus Ircinia (order Dictyoceratida: family Irciniidae). This genus includes several species that are very rich sources of secondary metabolites with a variety of biological activities and structural classes (18). I. strobilina contains variabilin, a furanosesterterpene that has been identified as a fish feeding deterrent (16). A diverse group of metabolites has been found from other sponges in the genus Ircinia and includes other sesterterpenes (1, 3, 29, 38, 52, 65, 76). The compounds ircinal A and B, precursors of antimalarial manzamine alkaloids, were isolated from the Okinawan marine sponge Ircinia sp. (32). Ircinamine, an alkaloid with moderate activity against the murine leukemia cell line P388, was purified from a marine Ircinia sp. (33). Tedanolide C, a cytotoxic macrolide, was isolated from the Papua New Guinea sponge Ircinia sp. (5). Other compounds of potential biomedical importance from the genus Ircinia include two murine and human cancer cell growth inhibitors, irciniastatin A and B (47), and a ceramide (77).
MATERIALS AND METHODS
Sponge and water sample collection.
The marine sponge I. strobilina was collected by scuba diving at Conch Reef, Key Largo, FL, in June 2004 at a depth of ca. 18 m (latitude, 24°57.11′N; longitude, 80°27.57′W). The water salinity was 36 ppt, and the temperature was 26.7°C. Voucher samples were preserved in 70% ethanol immediately after collection for taxonomic identification. Sponge samples were frozen at −20°C for later molecular and chemical characterization. Three water samples were collected from the vicinity of the sponge in sterile 20-liter containers and filtered through 0.22-μm-pore-size Sterivex filters (Millipore, Billerica, MA). The Sterivex filters were frozen immediately and stored at −20°C for isolation of nucleic acids. Three individuals of I. strobilina were collected for the aquaculture study and kept in containers filled with seawater that was replaced every 2 to 6 h during road transportation from Florida to Baltimore. Histology sections were examined using light microscopy of thin sections for taxonomic identification. In the wild under moderate current conditions, the sponge generally forms a squat rubbery mass with distinctive webs extending between large blunt conules set well apart on the sponge surface. The oscules are typically grouped together on the apex of the sponge. In reef areas with high current activity, sponges are taller and more cylindrical, with oscules raised on an apical ridge. The sponge is dark brownish black under full illumination; shaded portions are cream. The consistency is tough and spongy, and it is very difficult to tear or cut; upon being cut, the sponge emits a fetid odor. The primary fibers are large and form coarse trellises with only rare connecting fibers.
Sponge aquaculture.
In an attempt to maintain healthy sponges in captivity, a recirculating aquaculture system was designed to house the collected I. strobilina sponges as described by Mohamed et al. (39). Sponges were fed the microalga Nanochloropsis sp., with the addition of 40 ml of culture (4 × 106 cells ml−1) every 2 to 3 days. Three I. strobilina sponges were maintained in the aquaculture system. Sponges were inspected visually during this period to monitor their health. Viability assays (2, 40) were used to check that the sponges were alive immediately before sacrificing them for microbiological studies. Manually dispersed sponge tissues reaggregated spontaneously, indicating the viability of sponge cells. Two sponges were sacrificed after 3 months. A third sponge was subsampled for microbiology at 9 months and retained in aquaculture for an additional 3 months prior to sacrifice at 1 year for chemical analysis.
Sponge processing for isolation of culturable bacteria.
Sponge samples were rinsed with sterile artificial seawater immediately after collection from the field and after being harvested from the aquaculture system to remove any transient bacteria. Sponge tissue (1 cm3) was ground in artificial seawater, and 10-fold serial dilutions were plated in triplicate onto Difco marine agar 2216 (BD Biosciences, Franklin Lakes, NJ). Plates were incubated at 30°C for 1 week, at which time the plate counts were determined. To determine the total bacterial counts, a defined volume of the tissue homogenate was fixed with 37% (wt/vol) paraformaldehyde to a final concentration of 2 to 4% (wt/vol) and stored at 4°C until use. DAPI (4′,6′-diamidino-2-phenylindole) was added to the fixed samples to a final concentration of 20 μg ml−1, as described previously (51). Ten milliliters of DAPI-stained homogenates was filtered under slow vacuum onto a 25-mm-diameter, 0.1-μm polycarbonate membrane (GE Osmonics, Minneapolis, MN) that was supported with a 45-μm GF-F-type membrane (Whatman International Ltd., Maidstone, England). The filters were air dried and mounted with immersion oil onto a microscope slide. Bacterial numbers were determined using an epifluorescence microscope (Axioplan microscope; Zeiss, Germany). Three, two, and one independent sample was processed from wild, 3-month, and 9-month sponges, respectively. For each sample, an average bacterial number was determined by counting 10 fields.
Identification of isolates by 16S rRNA gene sequence analysis.
All cultured bacteria from initial isolation plates were subcultured to obtain isolates for each distinctive morphology. A representative of each colony type was selected from each sample for sequencing. Single pure colonies of each isolate were transferred to 20 ml of marine broth and incubated overnight at 30°C in a shaking incubator. DNAs were extracted from isolates by use of an Ultra-Clean microbial kit (MoBio Laboratories, Carlsbad, CA). Isolates were cryopreserved at −80°C in marine broth 2216 supplemented with 30% glycerol for long-term storage. 16S rRNA gene fragments were PCR amplified using universal primers 27F and 1492R (34).
DNA extraction from sponges and water samples.
To extract DNA from sponges, sponge tissue (1 cm3) was lyophilized and ground by use of a sterile mortar and pestle. Total genomic DNA was extracted mechanically using the method described by Pitcher et al. (50), modified for sponge tissues according to the method of Enticknap et al. (15). To recover nucleic acids from seawater and aquarium water samples, the protocol described by Somerville et al. (61) was used. Briefly, bacteria were concentrated by filtration of water through Sterivex filters, stored frozen in SET buffer (20% sucrose, 50 mM EDTA, 50 mM Tris-HCl, pH 7.6), and processed later in the laboratory for DNA extraction within the intact filters (61). The extracted supernatant was precipitated with 2 volumes of 100% ethanol on ice, followed by centrifugation at 10,000 × g for 30 min. The pellets were washed with 70% ethanol, air dried, and suspended in 100 μl of Tris-EDTA buffer.
DGGE.
16S rRNA denaturing gradient gel electrophoresis (DGGE) was used to analyze total bacterial communities present in sponges. A 195-bp region corresponding to positions 341 to 534 in the 16S rRNA gene of Escherichia coli was amplified from the genomic DNAs extracted from sponges and water samples, using the P2 and P3 primers (43). DGGE was performed using a Bio-Rad DCode system (Bio-Rad, Hercules, CA) and a 6% (wt/vol) polyacrylamide gel with a denaturing gradient of 40 to 70% in 1× Tris-acetate-EDTA buffer. Electrophoresis was performed for 17 h at 60 V and 60°C, and gels were stained in a staining bath of SYBR green in 1× Tris-acetate-EDTA. DGGE gels were run twice to confirm the reproducibility of the overall pattern.
PCR amplification of genomic DNA, cloning, and sequencing.
16S rRNA gene fragments from the total genomic DNA were PCR amplified using the same general protocol described for the culturable isolates. PCR was terminated after 15, 20, 25, and 30 cycles, with 30 cycles used for the negative control sample. Amplification products were visualized by agarose gel electrophoresis. Visible bands of approximately 1,500 bp, corresponding in size to the expected 16S rRNA gene products, were excised from the reaction products with the smallest number of cycles that gave visible bands and then gel purified. The corresponding position of the negative control was also excised. PCR products were ligated into PCR-XL-TOPO vector and transformed into OneShot TOP 10 chemically competent E. coli cells, using a TOPO XL PCR cloning kit (Invitrogen Life Technologies, Carlsbad, CA). Plasmid DNA was isolated from individual clones and purified using a SprintPrep 384 HC kit (Agencourt Bioscience, Beverly, MA). Sequencing was done using an ABI Prism 3130xl genetic analyzer (Applied Biosystems, Foster City, CA) and the universal M13 forward primer.
Phylogenetic analysis.
16S rRNA gene sequences derived from isolates and the three clone libraries were edited using PreGap4 and Gap4 in the Staden Package and analyzed initially using the BLASTN tool at the National Center for Biotechnology Information website to aid in the selection of the closest reference sequences. Chimeric sequences were identified using the CHECK_CHIMERA program of the Ribosomal Database Project (37). All of the sequences were imported into the ARB software package (36), which was used to align homologous regions of 16S rRNA gene sequences, using the PT server, with a data set containing the nearest relative matches. This database was supplemented with relevant environmental sequences that were submitted recently to GenBank. Multiple alignments were checked manually and improved by the ARB editor tool. Phylogenetic trees including novel sequences and reference taxa were constructed using the neighbor-joining algorithm (Jukes-Cantor correction) implemented in ARB (53). The robustness of the inferred tree topologies was evaluated after 1,000 bootstrap replicates of the neighbor-joining data. Phylip, version 3.6, was used to generate bootstrap values (19). Short sequences (<500 bp) were analyzed with the BLASTN algorithm for initial identification. The identification of partial sequences was confirmed by adding them to the inferred tree without changing the tree topology by using the ARB parsimony interactive method.
Estimation of microbial diversity and statistical analysis of clone libraries.
To compare libraries statistically, S-LIBSHUFF was used (57). S-LIBSHUFF compares more than two libraries at once with the same distance matrix to determine whether two libraries were drawn from the same population. DOTUR (distance-based OTU and richness) was used to assign sequences to operational taxonomic units (OTUs) (56). It was also used to calculate collector's curves for observed unique OTUs, Chao1 and abundance-base coverage estimator (ACE) richness estimators, and Shannon's and Simpson's indices. Rarefaction analysis was done to determine the number of observed OTUs as a function of the distance between sequences and the number of sequences sampled.
Profiles of small molecules.
Overall profiles of small molecules, including primary and secondary metabolites extracted from sponge samples, were determined in order to detect any gross shifts in the chemistry of sponges upon transfer into aquaculture. Three I. strobilina individuals were used as control samples, whereas test samples comprised two I. strobilina individuals maintained for 3 months and one individual maintained for 1 year in the aquaculture system. Two grams of frozen sponge tissue was lyophilized and extracted with ethanol. The dried ethanol extract (100 mg) was dissolved in methanol and passed through a C18 column. Liquid chromatography-mass spectrometry (LC-MS) analysis was performed on a Bruker micro-time-of-flight spectrometer with electrospray ionization. Sample solutions were prepared in methanol and subjected to LC-MS analysis using a reverse-phase C18 column (5 μm by 4.6 mm by 150 mm; Phenomenex, Torrance, CA) eluting at 0.4 ml/minute with a 15-min linear gradient from 20% to 100% phase B. Phase A was water and phase B was acetonitrile. Electrospray ionization-MS of the samples (eluates) was carried out in positive mode on a mass spectrometer equipped with an electrospray ion source and a micro-time-of-flight data system.
Nucleotide sequence accession numbers.
16S rRNA gene sequences from isolates were submitted to GenBank under accession numbers EF629549 to EF629580. 16S rRNA gene sequences from clone libraries were submitted to GenBank under accession numbers EF629581 to EF629828.
RESULTS
Sponge aquaculture.
The marine sponge I. strobilina was maintained in the recirculating aquaculture system for 1 year. The growth of the sponges was observed visually in aquaculture. Growth rates of the sponges were not quantified, but significant growth was not visually apparent. The health of sponges was assessed visually and judged to be good because no necrosis was observed and all three Ircinia individuals remained unfouled for the entire course of the study. In addition, when sponges were removed for analysis, the area beneath and immediately adjacent to each sponge was unfouled, whereas the rest of the sediment in tanks was covered by a thin algal film. All sponges in aquaculture also retained the black coloration typical of these sponges in the wild.
Bacterial enumeration.
Total (DAPI-stained) and culturable (plate) bacterial counts were determined for samples from wild I. strobilina sponges and sponges maintained in a closed aquaculture system for 3 months and 9 months. For the three wild sponges, the total count was 2.8 × 109 ± 0.4 × 109 cells ml−1 (mean ± standard error) and the plate count was 1.1 × 106 ± 0.4 × 106 CFU ml−1. In the case of the two 3-month individuals, the total count was 5.8 × 109 ± 0.8 × 109 cells ml−1 and the plate count was 9.7 × 106 ± 3.3 × 106 CFU ml−1. In the case of the 9-month sponge, the total count was 7.3 × 109 cells ml−1 and the plate count was 8.0 × 105 CFU ml−1. Based on these counts, the percentages of culturable bacteria in the sponge samples ranged from 0.01% to 0.17%, indicating the importance of assessing these communities by using molecular techniques.
DGGE.
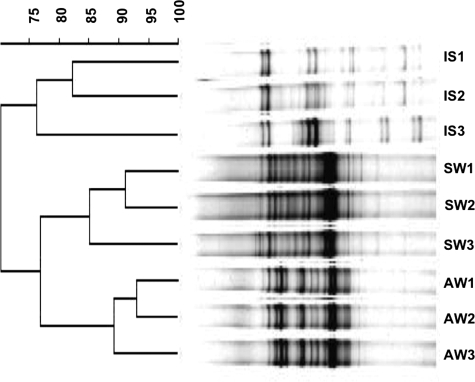
Bacterial communities varied substantially between wild sponges and surrounding seawater and water in the aquaculture system (Fig. 1). This indicates that the sponges harbor different assemblages of bacteria from those found in the surrounding seawater. The diversity of the microbial community, inferred by the complexity of the banding patterns, increased upon maintenance of I. strobilina in aquaculture for 3 months (see Fig. S1 in the supplemental material). A replicate 3-month sample gave a similar banding pattern to that of the 3-month sample shown in Fig. S1, lane 2, in the supplemental material (data not shown). The amount of community diversity then decreased in the sample from the sponge maintained for 9 months in aquaculture, reverting to a similar pattern (judged on the basis of the four dominant bands for the 9-month sample, marked by black arrows in Fig. S1, lane 3, in the supplemental material) to that of the wild sponge, although this should be interpreted with caution because the weak banding pattern of the 9-month sponge sample might be due to poor PCR amplification and only a single sponge was processed after 9 months in aquaculture, so it was not possible to obtain a replicate for this time point. The shifts that occurred in sponge-associated bacterial communities, as indicated by DGGE analysis, are consistent with the findings from statistical analysis of clone library data (below).
FIG. 1.
Dendrogram constructed from DGGE profiling of PCR-amplified 16S rRNA genes of bacterial communities associated with I. strobilina sponges collected from the wild (lanes IS1 to -3), from seawater (lanes SW1 to -3), and from water samples from the aquaculture system (lanes AW1 to -3). Jukes and Cantor's model was used for distance calculation, and the unweighted-pair group method using average linkages was used for dendrogram construction.
Phylogenetic analysis of bacterial isolates.
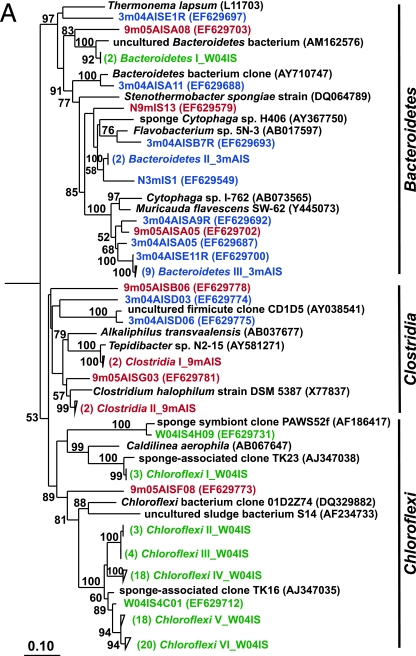
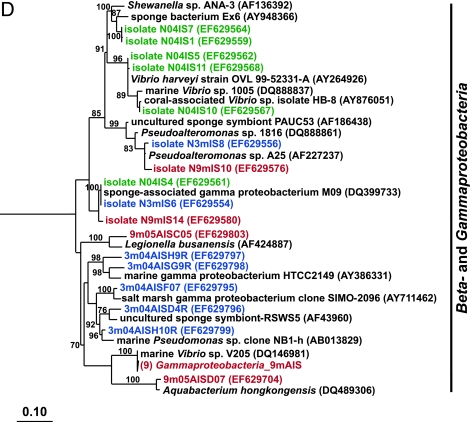
Traditional culturing techniques were used to isolate heterotrophic bacteria from sponges. Ten strains from wild sponges and sponges maintained for 3 months in aquaculture were characterized by 16S rRNA sequence analysis, and 12 strains were characterized from sponges maintained for 9 months. Alpha- and gammaproteobacterial strains dominated the culturable bacterial assemblages of both wild sponges and those maintained in aquaculture (Fig. 2C and D). Bacteria belonging to the Bacteroidetes appeared only in the culturable bacterial communities of sponges maintained in aquaculture (Fig. 2A).
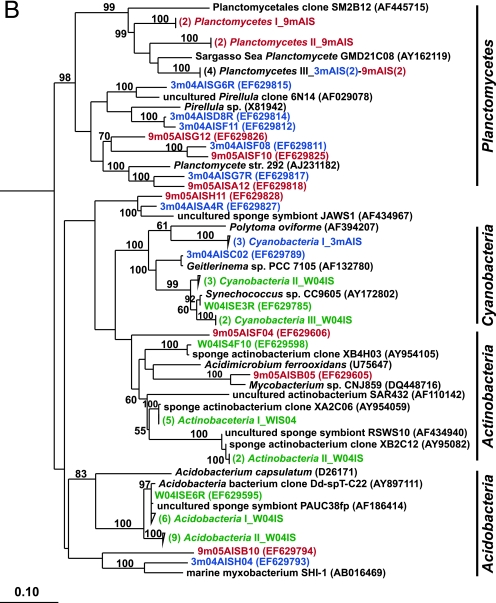
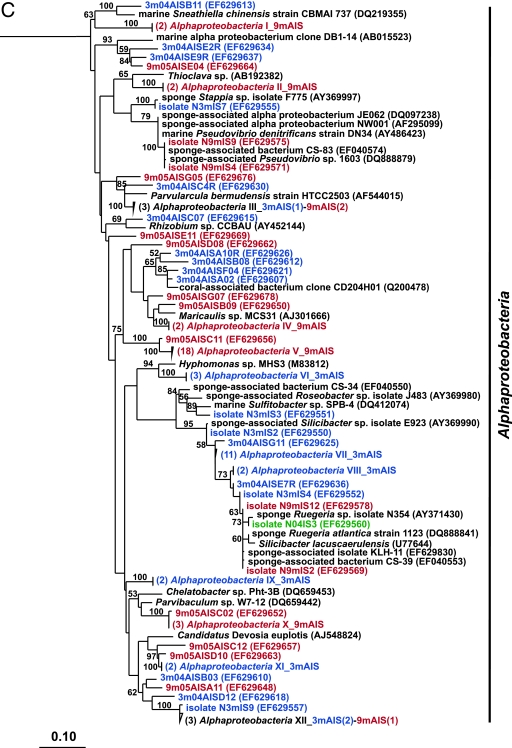
FIG. 2.
Rooted neighbor-joining phylogenetic trees of partial 16S rRNA genes of isolates and clones in the Bacteroidetes, Chloroflexi, and Clostridia (A); Acidobacteria, Actinobacteria, Cyanobacteria, and Planctomycetes (B); Alphaproteobacteria (C); and Beta- and Gammaproteobacteria (D). Isolates and clones were recovered from I. strobilina sponges collected from the wild (prefixed N04IS for isolates and W04IS for clones and shown in green) and maintained for 3 months (prefixed N3mIS for isolates and 3m04AIS for clones and shown in blue) and 9 months (prefixed N9mIS for isolates and 9m05AIS for clones and shown in red) in the aquaculture system. Bootstrap confidence values of >50% are shown at the nodes. The polygons represent clones that are ≥98% similar. The composition of each of these groups is shown in Table S1 in the supplemental material. The numbers listed in bold before the group names indicate the numbers of clones. Thermotoga maritima was used as an outgroup. The scale bar indicates 0.10 substitution per nucleotide position. Reference sequences are shown in bold, with GenBank accession numbers listed after each sequence name.
Phylogenetic analysis of 16S rRNA gene clone libraries.
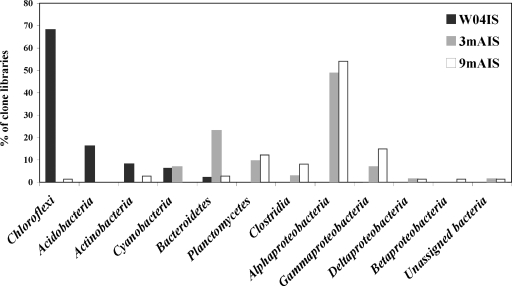
To determine the stability of the microbial community upon transfer of I. strobilina into aquaculture, 16S rRNA gene clone libraries were generated from community DNAs obtained from a representative wild sponge and sponges maintained for 3 months and 9 months in aquaculture. After elimination of a small number of chimeric clones from each library, 100, 74, and 74 clones were analyzed from the wild, 3-month, and 9-month libraries, respectively. The 16S rRNA gene clones from the wild sponge corresponded to 35 unique OTUs, which fell into the following five bacterial lineages: Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, and Cyanobacteria (Fig. 2). The 16S rRNA gene clones from the 3-month sponge corresponded to 48 unique OTUs that encompassed the following eight bacterial lineages: Bacteroidetes, Clostridia, Cyanobacteria, Planctomycetes, Proteobacteria (Alpha-, Gamma-, and Deltaproteobacteria), and unassigned bacteria (Fig. 2). The 16S rRNA gene clones from the 9-month sponge corresponded to 47 unique OTUs, which fell into the following 10 bacterial lineages: Actinobacteria, Bacteroidetes, Chloroflexi, Clostridia, Planctomycetes, Proteobacteria (Alpha-, Beta-, Gamma-, and Deltaproteobacteria), and unassigned bacteria (Fig. 2). The relative distribution of the major phylogenetic groups from each clone library is shown in Fig. 3.
FIG. 3.
Distribution of bacterial 16S rRNA gene clones from I. strobilina sponges collected from the wild (W04IS) and maintained for 3 months (3mAIS) and 9 months (9mAIS) in the aquaculture system within the major phylogenetic groups detected in the three libraries. We determined percentages for each group from sequence data.
Rarefaction analysis.
Rarefaction analysis was performed to determine whether the total diversity in the bacterial communities was well represented by the number of clones sequenced in each library. The rarefaction curves were obtained with DOTUR, using 10,000 random iterations. For clones from wild I. strobilina, the rarefaction curves at the phylum (distance = 0.20) and species (distance = 0.03) levels reached saturation, indicating sufficient sampling of this clone library (see Fig. S2 in the supplemental material). The rarefaction curves for the 3-month and 9-month sponges reached saturation at the phylum level, where richness reached an asymptotic maximum, but not at the species level, indicating that further sampling of the clone library would have revealed additional diversity. The wild sponge had less bacterial richness than sponges maintained in aquaculture did, especially at the species level. Maintaining I. strobilina in aquaculture clearly increased the bacterial richness, as demonstrated by steeper rarefaction curves than those for wild sponges.
Statistical analysis of bacterial diversity.
Additional measures of diversity and richness were obtained with the statistical richness estimators and diversity indices shown in Table 1. These indices were calculated using DOTUR. The input files were in the form of distance matrices generated by ARB. The total number of OTUs in a bacterial population was calculated using nonparametric estimators. Chao1 richness estimates were based on singletons and doubletons, as described by Chao (4), while ACE was based on the distribution of abundant (>10) and rare (≤10) species. Shannon's index and the reciprocal of Simpson's index were used as diversity indices, where higher numbers indicate greater diversity. Consistent with the rarefaction curves, both statistical indices suggested that the community diversity in sponges maintained in aquaculture was greater than that in the wild sponge. LIBSHUFF was used to quantitatively compare the three libraries. Evolutionary distances were calculated using the neighbor-joining algorithm in ARB, and the three libraries were significantly different (P < 0.0001/0.0100).
TABLE 1.
Richness and diversity estimates for bacterial 16S rRNA gene clone libraries from I. strobilina samples collected from the wild and maintained in aquaculture
| Sponge (I. strobilina) source (n)a | Distanceb | Richnessc | ACEd | Chao1 estimatore | Shannon indexf | 1/Simpson indexf |
|---|---|---|---|---|---|---|
| Wild (100) | 0.2 | 7 | 8 | 7 | 1.2 | 2.3 |
| 0.03 | 14 | 15 | 14 | 2.2 | 8.3 | |
| 3 Mo of aquaculture (74) | 0.2 | 12 | 17 | 17 | 1.8 | 4 |
| 0.03 | 41 | 200 | 107 | 3.3 | 17.8 | |
| 9 Mo of aquaculture (74) | 0.2 | 14 | 21 | 19 | 1.8 | 3.8 |
| 0.03 | 37 | 130 | 67 | 3.1 | 12.8 |
n, number of 16S rRNA gene sequences analyzed.
Eighty percent identity was estimated as the phylum-level distance (D = 0.20), and 97% identity was estimated as the species-level distance (D = 0.03).
Richness is based on observed unique OTUs.
Nonparametric statistical predictions of the total richness of different OTUs were based on the distribution of abundant (>10) and rare (≤10) OTUs.
Nonparametric statistical predictions of the total richness of OTUs were based on the distribution of singletons and doubletons.
A higher number represents more diversity.
LC-MS profiles of small molecules.
LC-MS analysis was performed to determine whether gross changes in overall metabolic profiles of I. strobilina occurred upon transfer into aquaculture. Minor changes in profiles of small molecules were observed in the sponges maintained in aquaculture compared to control wild samples, but overall patterns remained consistent, indicating no major shift in the profiles of secondary metabolites (see Fig. S3 in the supplemental material).
DISCUSSION
The aquaculture of sponges in closed or semiclosed systems is a promising strategy to overcome the supply problem for sponge-derived natural products. Although it might be possible in some cases to make bioactive metabolites produced by sponges by chemical synthesis and transgenic techniques, direct extraction from the sponge biomass may be the best option in other cases (43). A recirculating aquaculture system was used to examine the possibility of maintaining I. strobilina in ex situ closed systems under controlled environmental conditions with ecological parameters similar to those in the sponge's natural habitat. Using different biotechnological methods to produce sponge biomass may affect the microbial assemblages of the sponge host. In cases where microbes are the producers of compounds of interest, changes in the microbial communities may affect the production of natural products. The bacteria associated with sponges are also likely to be important for the health of the sponges (26). For these reasons, it is important to study the effects of these methods on the microbial communities associated with sponges. The present study is one of few studies that have looked at the changes in microbial communities associated with sponges following aquaculture or sponge transplantation. In this long-term study of the bacterial communities associated with I. strobilina in a self-contained, recirculating aquaculture system, an increase in diversity of the bacterial communities after 3 months and a return to an intermediate level of diversity after 9 months was found, suggestive of acclimation to aquaculture conditions.
We previously showed that the bacterial community of the marine sponge Mycale laxissima changed substantially upon transfer into flowthrough and recirculating aquaculture systems (39). Hoffmann and coworkers (28) used fluorescence in situ hybridization to study the stability and specificity of microbes associated with the cold-water marine sponge Geodia barretti during cultivation for 8 months in an open recirculation system where members of the Alpha- and Gammaproteobacteria were maintained during the period of cultivation. Friedrich and coworkers (21) found that a large fraction of the microbial community of the Mediterranean sponge Aplysina aerophoba remained stable upon starvation of sponges or after antibiotic exposure for 11 days in recirculating seawater aquaria.
Phylogenetic and statistical analyses of small-subunit-rRNA gene libraries were used to monitor structural shifts in the bacterial communities associated with I. strobilina following cultivation in aquaculture. Bacterial communities associated with I. strobilina were different from bacterioplankton communities found in the surrounding seawater. DGGE analysis of sponges maintained in aquaculture and the surrounding water indicated substantial differences in these bacterial communities. This was confirmed by community analysis of these samples by 16S rRNA gene sequencing studies. The data for bacterial community analysis of the I. strobilina samples are presented here, and those for bacterial community analysis of the surrounding water were reported by Mohamed et al. (39). This suggests that the bacterial community associated with wild I. strobilina is sponge specific rather than simply comprising transient bacteria from the water column. The fact that marine sponges harbor different bacteria from those in the water column has been shown in previous reports (23, 39, 54, 62-64, 71).
The culturable bacteria isolated from wild sponges included isolates found only in marine sponges, which indicates that they might be sponge specific (14, 23, 70). A comparison was made between a subset of bacteria isolated from I. strobilina and the surrounding seawater. Sixty percent of the top BLAST hits of isolates from I. strobilina were for bacteria found only in sponges. Bacteria isolated from the seawater had only 16% of top BLAST hits matching sponge bacteria (data not shown).
Total communities included representatives primarily clustered within the Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, and Cyanobacteria groups. Interestingly, a large number of the clones were affiliated with uncultured Chloroflexi (59% of the total wild sponge library). Phylotypes related to Actinobacteria were no longer detectable in sponges maintained for 3 months in aquaculture and then were detected again in the 9-month sponge. Since Actinobacteria organisms associated with marine sponges may be sources of bioactive compounds, the resumption of actinobacterial diversity may play a role in the production of specific bioactive compounds that are derived from this group.
Both culture-based and molecular techniques showed an increase in the Bacteroidetes community in aquaculture, with the highest representation in the 3-month sponge. Populations affiliated with Clostridia, Planctomycetes, and Proteobacteria (Alpha-, Gamma-, and Deltaproteobacteria) emerged in aquaculture. This indicates that adaptation to aquaculture conditions favored the abundance of these populations. These increasingly large populations may have originally existed in lower abundances in the wild sponge and/or have been acquired from the surrounding water in the aquaculture system. The diversity of the bacterial community associated with I. strobilina increased in aquaculture. This trend of increasing complexity of the bacterial community upon transfer to aquaculture tanks was observed with the marine sponge Mycale laxissima in our previous study (36). Statistical analyses revealed a significant shift in the bacterial communities in sponges maintained for both 3 and 9 months in aquaculture compared to the community in wild sponges. Based on the observed diversity indices, more bacterial diversity was present in the 3-month sponge samples than in wild sponges, possibly due to the stress of the sponge when it was first transferred from its natural habitat into aquaculture. It is not clear whether this is a result of stress, the presence of different bacteria in the surrounding water, the light regimen, or some other parameter that differs in the wild and in aquaculture. On the basis of all five statistical tests used to compare clone libraries at the species level, the level of diversity of the bacterial community associated with the sponge maintained for 9 months in aquaculture was intermediate between those for wild and three-month sponges. This may indicate an acclimation in this bacterial community after the 9-month period in aquaculture.
Metabolomics, defined as the study of the nonproteinaceous, endogenously synthesized small molecules present in an organism, is an emerging strategy in drug discovery and development (13, 22). The combination of chromatography and mass spectrometry allows the separation of individual metabolites and their identification based on mass. The change in the metabolome of an organism can be used to understand what has changed in the system. Applying this tool to our study showed that the environmental stress following the transfer of I. strobilina into aquaculture produced no detectable effect on the overall profile of small molecules associated with the sponges. LC-MS chemical fingerprinting revealed no major changes in the natural product profiles of I. strobilina, although the composition of the bacterial community changed substantially following transfer into aquaculture. This suggests that bacterial symbionts associated with I. strobilina may not be involved in the production of the major metabolites or that these metabolites are produced by a stable bacterial fraction that was maintained in aquaculture. Candidates are members of the Actinobacteria, Bacteroidetes, and Chloroflexi. In this case, the stability of the metabolites in aquaculture may imply that these symbionts constitutively produce essential metabolites. Another possible explanation for the stability of metabolites in aquaculture is that they could have been synthesized by the sponge or its associated microbes while the sponge was in the wild, stored, and remained undegraded when the sponge was transferred into aquaculture.
In this study, the possibility of maintaining the marine sponge I. strobilina alive in a recirculating aquaculture system is demonstrated. Further optimization of the aquaculture system is required for it to be useful in terms of production of sponge biomass for harvesting natural products. Our key finding is that bacterial communities changed upon transfer of the sponge to aquaculture but showed signs of returning to the community present in wild sponges after 9 months of maintenance in aquaculture. This highlights the importance of monitoring the bacterial communities associated with marine sponges when maintaining sponges in aquaculture systems by showing that profound changes may occur in these bacterial communities. Concomitant changes in the overall chemical profile of the sponge were not detected. Additional detailed studies of this type are needed to determine on a case-by-case basis whether changes in sponge-associated microbial communities are linked with changes in overall chemical profiles or specific compounds of interest.
Supplementary Material
Acknowledgments
We give special thanks to Julie Enticknap for her invaluable advice and assistance and to Matthew Anderson for his help in maintaining the sponges in aquaculture and assistance in the statistical analyses. We thank Scott McIntosh for designing and implementing the recirculating aquaculture system. We thank Marcelino Suzuki and Naomi Montalvo for help using ARB. We acknowledge the National Undersea Research Center (NURC), University of North Carolina at Wilmington, for providing sampling opportunities in Key Largo, FL, and the Aquaculture Research Center (ARC) at the Center of Marine Biotechnology for assistance in maintaining sponges in aquaculture.
Funding for this research was provided by the Microbial Observatories Program, National Science Foundation (MCB-0238515), to R.T.H. and by NIH grant RO1 AI 036596 to M.T.H. and R.T.H.
Footnotes
Published ahead of print on 9 May 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
Contribution no. 06-143 from the Center of Marine Biotechnology.
REFERENCES
- 1.Alfano, G., G. Cimino, and S. De Stefano. 1979. Palinurin, a new linear sesterterpene from a marine sponge. Cell. Mol. Life Sci. 35:1136-1137. [Google Scholar]
- 2.Blumbach, B., Z. Pancer, B. Diehl-Seifert, R. Steffen, J. Munkner, I. Muller, and W. E. Muller. 1998. The putative sponge aggregation receptor. Isolation and characterization of a molecule composed of scavenger receptor cysteine-rich domains and short consensus repeats. J. Cell Sci. 111:2635-2644. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan, M. S., A. Edser, G. King, J. Whitmore, and R. J. Quinn. 2001. Cheilanthane sesterterpenes, protein kinase inhibitors, from a marine sponge of the genus Ircinia. J. Nat. Prod. 64:300-303. [DOI] [PubMed] [Google Scholar]
- 4.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:783-791. [Google Scholar]
- 5.Chevallier, C., T. S. Bugni, X. Feng, M. K. Harper, A. M. Orendt, and C. M. Ireland. 2006. Tedanolide C: a potent new 18-membered-ring cytotoxic macrolide isolated from the Papua New Guinea marine sponge Ircinia sp. J. Org. Chem. 71:2510-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corredor, J. E., C. R. Wilkinson, V. P. Vicente, J. M. Morell, and E. Otero. 1988. Nitrate release by Caribbean reef sponges. Limnol. Oceanogr. 33:114-120. [Google Scholar]
- 7.De Rosa, S., S. De Caro, G. Tommonaro, K. Slantchev, K. Stefanov, and S. Popov. 2001. Development in a primary cell culture of the marine sponge Ircinia muscarum and analysis of the polar compounds. Mar. Biotechnol. 3:281-286. [DOI] [PubMed] [Google Scholar]
- 8.De Rosa, S., G. Tommonaro, K. Slantchev, K. Stefanov, and S. Popov. 2002. Lipophylic metabolites from the marine sponge Ircinia muscarum and its cell culture. Mar. Biol. 140:465-470. [DOI] [PubMed] [Google Scholar]
- 9.Diaz, M. C., and B. B. Ward. 1997. Sponge-mediated nitrification in tropical benthic communities. Mar. Ecol. Prog. Ser. 156:97-109. [Google Scholar]
- 10.Duckworth, A. R., and C. N. Battershill. 2003. Developing farming structures for production of biologically active sponge metabolites. Aquaculture 217:139-156. [Google Scholar]
- 11.Duckworth, A. R., and C. N. Battershill. 2003. Sponge aquaculture for the production of biologically active metabolites: the influence of farming protocols and environment. Aquaculture 221:311-329. [Google Scholar]
- 12.Duckworth, A. R., G. A. Samples, A. E. Wright, and S. A. Pomponi. 2003. In vitro culture of the tropical sponge Axinella corrugata (Demospongia): effect of food cell concentration on growth, clearance rate and biosynthesis of stevensine. Mar. Biotechnol. 5:519-527. [DOI] [PubMed] [Google Scholar]
- 13.Dunn, W. B., and D. I. Ellis. 2005. Metabolomics: current analytical platform and methodologies. Trends Anal. Chem. 24:285-294. [Google Scholar]
- 14.Enticknap, J. J., M. K. Shanks, O. Peraud, and R. T. Hill. 2006. Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl. Environ. Microbiol. 72:4105-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enticknap, J. J., R. Thompson, O. Peraud, J. E. Lohr, M. T. Hamann, and R. T. Hill. 2004. Molecular analysis of a Florida Keys sponge: implications for natural products discovery. Mar. Biotechnol. 6:S288-S293. [Google Scholar]
- 16.Epifanio, R. D. A., D. L. Martins, and G. Muricy. 1999. The sesterterpene variabilin as a fish-predation deterrent in the western Atlantic sponge Ircinia strobilina. J. Chem. Ecol. 25:2247-2254. [Google Scholar]
- 17.Faulkner, D. J. 2002. Marine natural products. Nat. Prod. Rep. 19:1-48. [DOI] [PubMed] [Google Scholar]
- 18.Faulkner, D. J., M. K. Harper, M. G. Haygood, C. E. Salomon, and E. W. Schmidt. 2000. Symbiotic bacteria in sponges: sources of bioactive substances, p. 107-119. In N. Fusetani (ed.), Drugs from the sea. Karger, Basel, Switzerland.
- 19.Felsenstein, J. 2004. PHYLIP (phylogenetic inference package), version 3.6. Department of Genetics, University of Washington, Seattle, WA.
- 20.Fortman, J. L., and D. H. Sherman. 2005. Utilizing the power of microbial genetics to bridge the gap between the promise and the application of marine natural products. Chem. Biochem. 6:960-978. [DOI] [PubMed] [Google Scholar]
- 21.Friedrich, A. B., I. Fischer, P. Proksch, J. Hacker, and U. Hentschel. 2001. Temporal variation of the microbial community associated with the Mediterranean sponge Aplysina aerophoba. FEMS Microbiol. Ecol. 38:105-113. [Google Scholar]
- 22.Harrigan, G. G., and L. A. Yates. 2006. High-throughput screening, metabolomics and drug discovery. IDrugs 9:188-192. [PubMed] [Google Scholar]
- 23.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hentschel, U., K. M. Usher, and M. W. Taylor. 2006. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55:167-177. [DOI] [PubMed] [Google Scholar]
- 25.Hildebrand, M., L. E. Waggoner, G. E. Lim, K. H. Sharp, C. P. Ridley, and M. G. Haygood. 2004. Approaches to identify, clone, and express symbiont bioactive metabolite genes. Nat. Prod. Rep. 21:122-142. [DOI] [PubMed] [Google Scholar]
- 26.Hill, R. T. 2004. Microbes from marine sponges: a treasure trove of biodiversity for natural products discovery, p. 177-190. In A. T. Bull (ed.), Microbial diversity and bioprospecting. ASM Press, Washington, DC.
- 27.Hoffmann, F., O. Larsen, V. Thiel, H. T. Rapp, and J. Reitner. 2005. An anaerobic world in sponges. Geomicrobiol. J. 22:1-10. [Google Scholar]
- 28.Hoffmann, F., H. Rapp, and J. Reitner. 2006. Monitoring microbial community composition by fluorescence in situ hybridization during cultivation of the marine cold-water sponge Geodia barretti. Mar. Biotechnol. 8:373-379. [DOI] [PubMed] [Google Scholar]
- 29.Issa, H. H., J. Tanaka, and T. Higa. 2003. New cytotoxic furanosesterterpenes from an Okinawan marine sponge, Ircinia sp. J. Nat. Prod. 66:251-254. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi, J., and M. Ishibashi. 1993. Bioactive metabolites of symbiotic marine microorganisms. Chem. Rev. 93:8305-8308. [Google Scholar]
- 31.Kobayashi, M. 2000. Search for biologically active substances from marine sponges, p. 46-58. In N. Fusetani (ed.), Drugs from the sea. Karger, Basel, Switzerland.
- 32.Kondo, K., H. Shigemori, Y. Kikuchi, M. Ishibashi, T. Sasaki, and J. Kobayashi. 1992. Ircinals A and B from the Okinawan marine sponge Ircinia sp.: plausible biogenetic precursors of manzamine alkaloids. J. Org. Chem. 57:2480-2483. [Google Scholar]
- 33.Kuramoto, M., H. Arimoto, and D. Uemura. 2004. Bioactive alkaloids from the sea: a review. Mar. Drugs 1:39-54. [Google Scholar]
- 34.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 35.Lee, Y. K., J.-H. Lee, and H. K. Lee. 2001. Microbial symbiosis in marine sponges. J. Microbiol. 39:254-264. [Google Scholar]
- 36.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martínez, A., S. Robledo, D. L. Muñoz, S. Blair, E. Higuita, E. Echeverri, J. Bedoya, S. Zea, and I. Vitae. 2001. Antiparasitic activity of methanol extracts and isolated fractions from Caribbean sponges. Rev. Fac. Quím. Farm. 8:71-81. [Google Scholar]
- 39.Mohamed, N. M., J. J. Enticknap, J. E. Lohr, S. M. McIntosh, and R. T. Hill. 2008. Changes in bacterial communities of the marine sponge Mycale laxissima on transfer into aquaculture. Appl. Environ. Microbiol. 74:1209-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller, W. E., I. Muller, and R. K. Zahn. 1974. Two different aggregation principles in reaggregation process of dissociated sponge cells (Geodia cydonium). Experientia 30:899-902. [DOI] [PubMed] [Google Scholar]
- 41.Müller, W. E. G., V. A. Grebenjuk, G. Le Pennec, H. C. Schroder, F. Brummer, U. Hentschel, I. M. Müller, and H. J. Breter. 2004. Sustainable production of bioactive compounds by sponges—cell culture and gene cluster approach: a review. Mar. Biotechnol. 6:105-117. [DOI] [PubMed] [Google Scholar]
- 42.Munro, M. H., J. W. Blunt, E. J. Dumdei, S. J. Hickford, R. E. Lill, S. Li, C. N. Battershill, and A. R. Duckworth. 1999. The discovery and development of marine compounds with pharmaceutical potential. J. Biotechnol. 70:15-25. [DOI] [PubMed] [Google Scholar]
- 43.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newman, D. J., and G. M. Cragg. 2004. Marine natural products and related compounds in clinical and advanced preclinical trials. J. Nat. Prod. 67:1216-1238. [DOI] [PubMed] [Google Scholar]
- 45.Osinga, R., E. Armstrong, J. G. Burgess, F. Hofmann, J. Reitner, and G. Schumann-Kindel. 2001. Sponge-microbe associations and their importance for sponge bioprocess engineering. Hydrobiologia 461:55-62. [Google Scholar]
- 46.Osinga, R., J. Tramper, and R. H. Wijffels. 1999. Cultivation of marine sponges. Mar. Biotechnol. 1:509-532. [DOI] [PubMed] [Google Scholar]
- 47.Pettit, G. R., H. Hoffmann, D. L. Herald, J. McNulty, A. Murphy, K. C. Higgs, E. Hamel, N. E. Lewin, L. V. Pearce, P. M. Blumberg, R. K. Pettit, and J. C. Knight. 2004. Antineoplastic agents 491. Synthetic conversion of aaptamine to isoaaptamine, 9-demethylaaptamine, and 4-methylaaptamine. J. Org. Chem. 69:2251-2256. [DOI] [PubMed] [Google Scholar]
- 48.Piel, J. 2006. Bacterial symbionts: prospects for the sustainable production of invertebrate-derived pharmaceuticals. Curr. Med. Chem. 13:39-50. [PubMed] [Google Scholar]
- 49.Piel, J., D. Hui, G. Wen, D. Butzke, M. Platzer, N. Fusetani, and S. Matsunaga. 2004. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 101:16222-16227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 51.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 52.Rifai, S., A. Fassouane, P. M. Pinho, A. Kijjoa, N. Nazareth, M. São, J. Nascimento, and W. Herz. 2005. Cytotoxicity and inhibition of lymphocyte proliferation of fasciculatin, a linear furanosesterterpene isolated from Ircinia variabilis collected from the Atlantic Coast of Morocco. Mar. Drugs 3:15-21. [Google Scholar]
- 53.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 54.Santavy, D. L., P. Willenz, and R. R. Colwell. 1990. Phenotypic study of bacteria associated with the Caribbean sclerosponge Ceratoporella nicholsoni. Appl. Environ. Microbiol. 56:1750-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schirmer, A., R. Gadkari, C. D. Reeves, F. Ibrahim, E. F. DeLong, and C. R. Hutchinson. 2005. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl. Environ. Microbiol. 71:4840-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt, E. W., J. T. Nelson, D. A. Rasko, S. Sudek, J. A. Eisen, M. G. Haygood, and J. Ravel. 2005. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 102:7315-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shieh, W. Y., and Y. M. Lin. 1994. Association of heterotrophic nitrogen-fixing bacteria with a marine sponge of Halichondria sp. Bull. Mar. Sci. 54:557-564. [Google Scholar]
- 60.Sipkema, D., R. Osinga, W. Schatton, D. Mendola, J. Tramper, and R. H. Wijffels. 2005. Large-scale production of pharmaceuticals by marine sponges: sea, cell, or synthesis? Biotechnol. Bioeng. 90:201-222. [DOI] [PubMed] [Google Scholar]
- 61.Somerville, C. C., I. T. Knight, W. L. Straube, and R. R. Colwell. 1989. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl. Environ. Microbiol. 55:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor, M. W., R. Radax, D. Steger, and M. Wagner. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor, M. W., P. J. Schupp, I. Dahllof, S. Kjelleberg, and P. D. Steinberg. 2004. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6:121-130. [DOI] [PubMed] [Google Scholar]
- 64.Taylor, M. W., P. J. Schupp, R. de Nys, S. Kjelleberg, and P. D. Steinberg. 2005. Biogeography of bacteria associated with the marine sponge Cymbastela concentrica. Environ. Microbiol. 7:419-433. [DOI] [PubMed] [Google Scholar]
- 65.Tziveleka, L. A., A. P. Kourounakisb, P. N. Kourounakisb, V. Roussisa, and C. Vagias. 2002. Antioxidant potential of natural and synthesised polyprenylated hydroquinones. Bioorg. Med. Chem. 10:935-939. [DOI] [PubMed] [Google Scholar]
- 66.Vacelet, J. 1975. Etude en microscopie electronique de l'association entre bacteries et spongiaires du genre Verongia (Dictyoceratida). J. Microsc. Biol. Cell 23:271-288. [Google Scholar]
- 67.Vacelet, J., and C. Donadey. 1977. Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30:301-314. [Google Scholar]
- 68.Van Soest, R. 1996. Porifera, Schwämme, p. 98-119. In W. Westheide and R. Rieger (ed.), Spezielle Zoologie. Teil 1. Einzeller and wirbellose Tiere. Gustav Fischer Verlag, Stuttgart, Germany.
- 69.van Treeck, P., M. Eisinger, J. Muller, M. Paster, and H. Schuhmacher. 2003. Mariculture trials with Mediterranean sponge species: the exploitation of an old natural resource with sustainable and novel methods. Aquaculture 218:439-455. [Google Scholar]
- 70.Webster, N. S., and R. T. Hill. 2001. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an α-proteobacterium. Mar. Biol. 138:843-851. [Google Scholar]
- 71.Wilkinson, C. R. 1978. Microbial associations in sponges. I. Ecology, physiology and microbial populations of coral reef sponges. Mar. Biol. 49:161-167. [Google Scholar]
- 72.Wilkinson, C. R. 1978. Microbial associations in sponges. II. Numerical analysis of sponge and water bacterial populations. Mar. Biol. 49:169-176. [Google Scholar]
- 73.Wilkinson, C. R. 1983. Net primary productivity in coral reef sponges. Science 219:410-412. [DOI] [PubMed] [Google Scholar]
- 74.Wilkinson, C. R., and P. Fay. 1979. Nitrogen fixation in coral reef sponges with symbiotic cyanobacteria. Nature 279:527-529. [Google Scholar]
- 75.Wilkinson, C. R., R. Summons, and E. Evans. 1999. Nitrogen fixation in symbiotic marine sponges: ecological significance and difficulties in detection. Mem. Queensl. Mus. 44:667-673. [Google Scholar]
- 76.Yang, S. W., T. M. Chan, S. A. Pomponi, W. Gonsiorek, G. Chen, A. E. Wright, W. Hipkin, M. Patel, V. Gullo, B. Pramanik, P. Zavodny, and M. Chu. 2003. A new sesterterpene, Sch 599473, from a marine sponge, Ircinia sp. J. Antibiot. 56:783-786. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, G.-W., X.-Q. Ma, C.-X. Zhang, J.-Y. Su, W.-C. Ye, X.-Q. Zhang, X.-S. Yao, and L.-M. Zeng. 2005. Two new ceramides from the marine sponge Ircinia fasciculata. Helv. Chim. Acta 88:885-890. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.