Abstract
The gene encoding a novel alcohol dehydrogenase (ADH) that belongs to the short-chain dehydrogenase/reductase (SDR) superfamily was identified in the extremely thermophilic, halotolerant gram-negative eubacterium Thermus thermophilus HB27. The T. thermophilus ADH gene (adhTt) was heterologously overexpressed in Escherichia coli, and the protein (ADHTt) was purified to homogeneity and characterized. ADHTt is a tetrameric enzyme consisting of identical 26,961-Da subunits composed of 256 amino acids. The enzyme has remarkable thermophilicity and thermal stability, displaying activity at temperatures up to ∼73°C and a 30-min half-inactivation temperature of ∼90°C, as well as good tolerance to common organic solvents. ADHTt has a strict requirement for NAD(H) as the coenzyme, a preference for reduction of aromatic ketones and α-keto esters, and poor activity on aromatic alcohols and aldehydes. This thermophilic enzyme catalyzes the following reactions with Prelog specificity: the reduction of acetophenone, 2,2,2-trifluoroacetophenone, α-tetralone, and α-methyl and α-ethyl benzoylformates to (S)-(−)-1-phenylethanol (>99% enantiomeric excess [ee]), (R)-α-(trifluoromethyl)benzyl alcohol (93% ee), (S)-α-tetralol (>99% ee), methyl (R)-(−)-mandelate (92% ee), and ethyl (R)-(−)-mandelate (95% ee), respectively, by way of an efficient in situ NADH-recycling system involving 2-propanol and a second thermophilic ADH. This study further supports the critical role of the D37 residue in discriminating NAD(H) from NADP(H) in members of the SDR superfamily.
Alcohol dehydrogenases (ADHs) are of interest for the synthesis of the (S) or (R) enantiomers of alcohols from prochiral ketones (11). Since their early application in asymmetric synthesis using horse liver and yeast ADHs (13) and Thermoanaerobacter brockii ADH (ADHTb) (15), screening efforts have been directed at various species of microorganisms, which has resulted in new ADHs that have distinctive substrate specificity, good efficiency, and high enantioselectivity. Representative examples of enzymes from mesophilic microorganisms are the NADP-dependent (R)-specific ADH from Lactobacillus brevis (RADHLb), which is active on aryl ketones and whose crystal structure has recently been solved (29), the NAD-dependent (S)-specific 1-phenylethanol dehydrogenase from the denitrifying bacterium strain EbN1 (PED), which was characterized and crystallized (10), and the NAD-dependent ADH from Leifsonia sp. strain S749 (ADHLs), which was found to be active on (R)-sec alcohols, aryl ketones, aldehydes, and keto esters and whose gene has recently been cloned for protein expression in Escherichia coli (12). These enzymes are homotetrameric and belong to the short-chain dehydrogenase/reductase (SDR) superfamily (14), which is characterized by ∼250-residue subunits, a Gly motif in the coenzyme-binding regions, and a catalytic triad formed by the highly conserved residues Tyr, Lys, and Ser, to which an Asn residue has been added based on the proposal of Filling et al. (2), which was recently supported by the RADHLb structure (29). Representative examples of ADHs from thermophilic microorganisms are medium-chain enzymes, including ADHTb (18), the ADH from Bacillus stearothermophilus (ADHBs) (1), and two archaeal enzymes, the ADH from Aeropyrum pernix (8) and the well-known ADH from Sulfolobus solfataricus (ADHSs) (27, 6). The latter is a tetrameric, S-specific, NAD-dependent zinc enzyme, which is more active on primary alcohols than on secondary alcohols and is poorly active on arylketones. By contrast, the thermophilic S-specific enzyme ADHTb is NADP dependent and complements horse liver ADH by preferentially accepting acyclic ketones and secondary alcohols rapidly (15).
In order to find a dehydrogenase/reductase that is both functionally stable and NAD dependent, we focused on the genomes of thermophilic microorganisms containing genes encoding putative ADHs belonging to the SDR superfamily. An open reading frame coding for a protein belonging to the SDR superfamily with high sequence identity to an RADHLb open reading frame was found in the genome of the extremely thermophilic, halotolerant gram-negative eubacterium Thermus thermophilus HB27 (9). The amino acid sequence (Fig. 1) revealed the presence of an N-terminal nucleotide-binding fingerprint motif, TGXXXGXG; these residues are spaced like those seen in typical SDRs (14). The protein also possesses an aspartate residue (D37) located 18 amino acids C terminal of the third glycine residue; structural studies of the RADHLb G37D mutant have shown that this residue plays a critical role in determining the preference of SDRs for NAD(H) (29). Further evidence of this role is provided by the ADHLs (12) and PED (10) proteins, which are strictly NAD-dependent SDRs possessing an aspartate residue at position 37. Thus, the alignment shown in Fig. 1 suggests that the putative dehydrogenase/reductase from T. thermophilus (ADHTt) should show a preference for NAD(H) over NADP(H). This preference, which is advantageous because of the lower cost of NADH, and intrinsic protein thermostability are two desirable features of oxidoreductases used in biotechnology applications (11).
FIG. 1.
Multiple-sequence alignment of the T. thermophilus ADH (TtADH) and ADHs belonging to the SDR family, including Leifsonia sp. strain S749 ADH (LSADH) (NCBI accession no. BAD99642), L. brevis ADH (LB-RADH) (PDB code 1ZK4), and (S)-1-phenylethanol dehydrogenase from denitrifying bacterial strain EbN1 (PED) (PDB code 2EWM). The sequences were aligned using the BioEdit program. Gray shading indicates residues highly conserved in the SDR family. The four members of the catalytic tetrad are indicated by a black background. The following positions are indicated by bold type: the glycine-rich consensus sequence and the sequence motif Dhx(cp) that (in all SDRs) have a structural role in coenzyme binding (14). The star indicates the major determinant of the coenzyme specificity. The RADHLb G37D mutant shows preference for NAD+ over NADP+ (29).
This report describes cloning and heterologous expression of the T. thermophilus HB27 adhTt gene, which encodes the SDR ADHTt. The purified enzyme was characterized in terms of substrate specificity, kinetics, and stability. ADHTt was found to be a thermophilic and thermostable dehydrogenase/reductase active on both aromatic alcohols and a discrete spectrum of carbonyl compounds, including substituted benzaldehydes, aromatic ketones, diketones, and α-keto esters. The enantioselectivity exhibited toward prochiral carbonyl compounds led us to define ADHTt as a novel thermophilic SDR that has Prelog specificity (25).
MATERIALS AND METHODS
Chemicals.
NAD(P)+ and NAD(P)H were obtained from AppliChem (Darmstadt, Germany). Alcohols, aldehydes, ketones, and keto esters were obtained from Sigma-Aldrich. MES [2-(N-morpholino)ethanesulfonic acid] was obtained from Sigma Chemical Co. (St. Louis, MO). Other chemicals were grade A substances obtained from AppliChem. 1-Butyl-3-methylimidazolium tetrafluoroborate was a kind gift from S. Cacchi. Recombinant ADHBs was prepared as previously described (3). Solutions of NADH and NAD+ were prepared as previously reported (27). All solutions were made with MilliQ water.
Amplification and cloning of the adhTt gene.
Chromosomal DNA was extracted by cesium chloride purification as described by Sambrook et al. (28). Ethidium bromide and cesium chloride were removed by repeated extraction with isoamyl alcohol and by extensive dialysis against 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, respectively. The DNA concentration was determined spectrophotometrically at 260 nm, and the molecular weight was determined by electrophoresis on a 0.8% agarose gel in 90 mM Tris-borate (pH 8.0), 20 mM EDTA, using DNA molecular size markers. The adhTt gene was amplified by PCR using oligonucleotide primers based on the adhTt sequence of T. thermophilus HB27 (GenBank accession no. YP_003977). The following oligonucleotides were used: 5′-GGTTGGGGTTCATATGGGCCTTTTCGCTGGCAAAGGGGTGCTG-3′ as the forward primer (the NdeI restriction site is underlined) and 5′-GGTTGGTTGAATTCCTACACCGGCCGCCCCGCCATCATGAAGCT-3′) as the reverse primer. The latter oligonucleotide introduced a translational stop following the last codon of the ADH gene, followed by an EcoRI restriction site (underlined). The PCR product was cloned into the expression vector pET29a (Novagen, Madison, WI) and digested with the appropriate restriction enzymes to create pET29a-ttADH. The insert was sequenced in order to verify that mutations had not been introduced during PCR.
Expression and purification of recombinant ADHTt.
Recombinant protein was expressed in E. coli BL21(DE3) cells (Novagen) transformed with the corresponding expression vector. Cultures were grown at 37°C in 2 liters of LB medium containing 30 μg/ml kanamycin. When the A600 of a culture reached 1.4, protein expression was induced by addition of isopropyl-β-d-1-thiogalactopyranoside (IPTG) to a concentration of 1.0 mM. The bacterial culture was incubated at 37°C for a further 24 h. Cells were harvested by centrifugation, and the pellet was stored at −20°C until use. The cells obtained from 2 liters of culture were suspended in 20 mM Tris-HCl buffer (pH 7.5) containing 0.1 mM phenylmethylsulfonyl fluoride and were lysed using a French pressure cell (Aminco Co., Silver Spring, MD) at 2,000 lb/in2 (13.8 × 103 kPa). The lysate was centrifuged, and the supernatant was incubated in the presence of DNase I (50 μg per ml of solution) and 5 mM MgCl2 for 30 min at 37°C, followed by protamine sulfate (1 mg per ml of solution) at 4°C for 30 min. The nucleic acid fragments were removed by centrifugation, and the supernatant was incubated at 75°C for 15 min. The host protein precipitate was removed by centrifugation. The supernatant was dialyzed overnight at 4°C against 20 mM Tris-HCl (pH 8.4) (buffer A) containing 1 mM phenylmethylsulfonyl fluoride. The dialyzed solution was applied to a DEAE-Sepharose Fast Flow column (1.6 by 12 cm) equilibrated in buffer A. After washing with 1 bed volume of the same buffer, elution was performed with a linear gradient of 0 to 0.06 M NaCl (80 ml of each concentration) in buffer A at a flow rate of 60 ml·h−1. The active pool was dialyzed against buffer A, concentrated fivefold with a 30,000-molecular-weight-cutoff centrifugal filter device (Millipore), and applied to a Sephadex G-75 column equilibrated in buffer A containing 0.15 M NaCl. The active pool was dialyzed against buffer A and concentrated to obtain 2.5 mg protein·ml−1 as described previously. ADHTt was stored at −20°C, and there was no loss of activity following several months of storage. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) and nondenaturing PAGE were carried out by using the Laemmli method (19), with minor modifications (27). The subunit molecular mass was determined by electrospray ionization mass spectrometry with a QSTAR Elite instrument (Applied Biosystems, United States).
Determination of protein concentration.
The protein concentration was determined with a Bio-Rad protein assay kit using bovine serum albumin as a standard (A280 of 1% bovine serum albumin in 50 mM sodium phosphate [pH 7.0]-0.9% NaCl, 6.6).
Enzyme assay.
ADHTt activity was assayed spectrophotometrically at 65°C by measuring the change in absorbance at 340 nm of NADH using a Cary 1E spectrophotometer equipped with a Peltier effect-controlled temperature cuvette holder. The standard assay for the reduction reaction was performed by adding 5 to 25 μg of the enzyme to 1 ml of preheated assay mixture containing 20 mM methyl benzoylformate (MBF) and 0.3 mM NADH in 50 mM potassium phosphate (pH 6.0). The standard assay for the oxidation reaction was performed using a mixture containing 20 mM (S)-(−)-1-phenylethanol and 3 mM NAD+ in 100 mM glycine-NaOH (pH 10.5). Screening of the substrates was performed using 1 ml of assay mixture containing either 20 mM alcohol and 1 mM NAD+ in 100 mM glycine-NaOH (pH 10.5), 0.1 M KCl or 5 mM carbonyl compound and 0.3 mM NADH in 50 mM potassium phosphate (pH 6.0). The activity of ADHBs was determined at 60°C using 1 ml of an assay mixture containing either 8 mM alcohol and 18 mM NAD+ in 200 mM glycine-NaOH (pH 9.0) or 8 mM carbonyl compound and 0.3 mM NADH in 100 mM Tris-HCl (pH 7.7); 0.5- and 1.0-μg aliquots of ADHBs were used for the forward and reverse reactions, respectively.
One unit of ADHTt and 1 U of ADHBs represented 1 μmol of NADH produced or utilized per min at 65 and 60°C, respectively, on the basis of an absorption coefficient at 340 nm for NADH of 6.22 mM−1.
Effect of pH on activity.
The optimum pH values for the reduction and oxidation reactions were determined at 65°C under the conditions used for MBF and (S)-(−)-1-phenylethanol, respectively, except that different buffer systems were used. The pH was controlled in each assay mixture at 65°C.
Kinetics.
The ADHTt kinetic parameters were calculated from measurements determined in duplicate or triplicate and by analyzing the kinetic results using the program GraFit (20). The turnover value (kcat, expressed in s−1) for ADHTt was calculated on the basis of a molecular mass of 27 kDa, assuming that the four subunits are catalytically active.
Thermophilicity and thermal stability.
ADHTt was assayed in the temperature range from 30 to 95°C using standard assay conditions and 25 μg of protein·ml−1 of assay mixture. The stability at various temperatures was studied by incubating 1-mg·ml−1 protein samples in 50 mM potassium phosphate (pH 6.0) at temperatures between 25 and 95°C for 30 min. Each sample was then centrifuged at 5°C, and the residual activity was assayed as described above. Long-term stability was studied by incubating protein samples (0.1 and 1.0 mg·ml−1) in 50 mM MES (pH 6.0), 100 mM KCl at 50, 60, and 70°C, and the residual activity was assayed after 24 h as described above.
Effects of compounds on enzyme activity.
The effects of salts, metal ions, ionic liquids, and chelating agents on ADHTt activity were investigated by assaying the enzyme in the presence of an appropriate amount of each compound in the standard assay mixture.
The effects of organic solvents were investigated by measuring the activities in enzyme samples (0.12 mg·ml−1 in 100 mM MES [pH 6.0], 5 mM 2-mercaptoethanol, and 100 mM KCl) immediately after the addition of organic solvents at different concentrations and after incubation for 5 and 24 h at 50 and 60°C. The percentage of activity for each sample was calculated by comparison with the value measured prior to incubation. The volume of the solution in a tightly capped test tube did not change during incubation.
Effects of chelating agents.
The effects of chelating agents were studied by measuring the activities before and after exhaustive dialysis of the enzyme against buffer A containing 1 mM EDTA and then against buffer A alone. An aliquot of the dialyzed enzyme was then incubated at 70°C in the absence and presence of 1 mM EDTA, and the activity was assayed at different times. Another aliquot was treated with 0.5 M guanidinium HCl at 30°C in the absence and presence of 1 mM EDTA or o-phenanthroline, and the activity was assayed at different times.
Enantioselectivity.
The enantioselectivity of ADHTt was determined by examining the reduction of aryl ketones, bicyclic ketones, and α-keto esters using an NADH regeneration system consisting of ADHBs and 2-propanol. The reaction mixture contained 1 mM NAD+, 20 mM carbonyl compound, 11 U of ADHBs, 260 mM (2%) 2-propanol, and 50 μg ADHTt in 1 ml of 100 mM MES (pH 6.0), 5 mM 2-mercaptoethanol, and 100 mM KCl. The reaction mixtures were incubated at 50 and 60°C for different times in a temperature-controlled water bath. Upon termination of the reaction, each reaction mixture was extracted twice with ethyl acetate. The enantiomeric excess (ee) of the product and the level of conversion were determined by gas-liquid chromatography (Agilent 6850) using a dimethylpentyl, β-cyclodextrin MEGA column (25 m; inside diameter, 0.25 mm; Legnano, Italy). The conditions used for 1-phenylethanol and α-(trifluoromethyl)benzyl alcohol were was follows: the oven temperature was increased from 90°C (initial time, 10 min) to 110°C (final time, 5 min) at a rate of 2.5°C/min. The conditions for ethyl and methyl mandelate were as follows: the oven temperature was increased from 100°C (initial time, 5 min) to 130°C (final time, 5 min) at a rate of 2.5°C/min. And the conditions for α-tetralol were as follows: the oven temperature was increased from 100°C (initial time, 5 min) to 130°C (final time, 5 min) at a rate of 2.0°C/min. The conversion yield was determined on the basis of the peak areas of the carbonyl substrate and alcohol products obtained in the same gas chromatography (GC) chromatogram.
Size exclusion chromatography.
Molecular masses were determined by size exclusion chromatography using a Superdex 200 10/300 GL column (Amersham) equilibrated with 50 mM Tris-HCl (pH 8.5), 1 mM 2-mercaptoethanol containing 0.15 M NaCl at a flow rate of 0.5 ml·min−1. The following molecular mass standards were used for calibration: horse myoglobin (17.5 kDa), chicken ovalbumin (44 kDa), beef γ-globin (158 kDa), and tyroglobulin (670 kDa) from Bio-Rad. In order to calculate the distribution coefficient, the void and total volumes of the column were determined with tryptophan and blue dextran.
RESULTS AND DISCUSSION
Expression and protein purification.
Analysis of the T. thermophilus genome (9) for genes encoding short-chain ADHs resulted in identification of a putative oxidoreductase gene. Based both on the presence of an aspartic residue at position 37 (Fig. 1) determined by the multiple-sequence alignment analysis of Kallberg et al. (14) and on kinetic and structural studies of the L. brevis ADH G37D mutant (29), the oxidoreductase protein was expected to be NAD dependent and was designated ADHTt. ADHTt is a 26,961.1-Da protein whose sequence showed the highest levels of identity to three typical SDRs, ADHLs (38% identity), RADHLb (34% identity), and PED (32% identity) (Fig. 1). The adhTt gene was successfully expressed in E. coli cells, which yielded an active enzyme accounting for more than 7.5% of the total protein in the cell extract (Table 1). Host protein precipitation at 75°C was the most effective purification step. The enzyme was enriched 11-fold with a 47% yield and was deemed homogeneous by denaturing and nondenaturing PAGE (data not shown).
TABLE 1.
Purification of recombinant ADHTta
| Step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 714 | 12,940 | 18.1 | 100 | 1 |
| Thermal step | 159 | 12,100 | 76.1 | 93.5 | 4.2 |
| DEAE FF | 63 | 9,040 | 143.5 | 69.8 | 7.9 |
| Gel filtration | 54 | 8,159 | 151.1 | 63.0 | 8.3 |
The data are for a 2-liter culture. The substrate was (S)-(−)1-phenylethanol, and the assay temperature was 65°C.
Protein separation by sodium dodecyl sulfate-PAGE resulted in a single band corresponding to a molecular mass of ∼26 kDa. Gel filtration performed in 50 mM Tris-HCl buffer (pH 8.5), 1 mM 2-mercaptoethanol containing 0.15 M NaCl yielded a profile consisting of a single peak corresponding to a molecular mass of ∼71 kDa. The corresponding value was ∼105 kDa when the same buffer containing 25 mM NaCl was used. These data are consistent with the hypothesis that ADHTt has a tetrameric structure, which adopts a more compact structure in the presence of a relatively high salt concentration, resulting in greater permeation into the packing pores of the gel matrix. The molecular mass of the subunit determined by electrospray ionization mass spectrometry analysis was 26,830.0 Da (average mass), in agreement with the theoretical value of the sequence lacking the N-terminal methionine.
Optimal pH and thermophilicity.
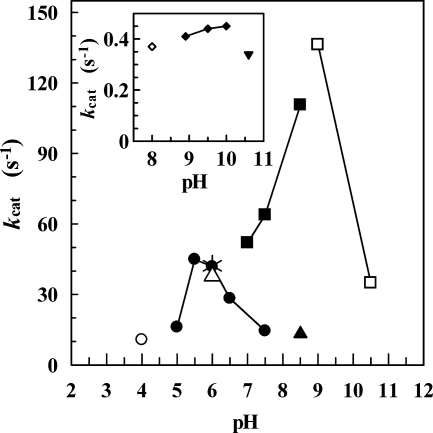
Figure 2 shows the pH dependence of ADHTt in the reduction and oxidation reaction and the pH dependence of ADHBs in the oxidation reaction alone. The ADHTt activity was found to be dependent on the pH during the reduction reaction, and there was a narrow peak of maximum activity at approximately pH 6.0. The oxidation reaction showed a less marked dependence on pH, and there was a broad peak with maximum activity at around pH 10.0. The pH profile of ADHBs (Fig. 2) shows that the activity on 2-propanol at pH 6.0 is about 30% of the optimal activity at pH 9 and is comparable to the activity of ADHTt in the reduction reaction. The data suggest that pH could control both the ADHTt activity for the desired reduction reaction and the ADHBs activity in NADH recycling, depending on the oxidation of 2-propanol.
FIG. 2.
ADHTt activity as a function of pH in the reduction (main plot) and oxidation (inset) reactions and ADHBs activity as a function of pH in the oxidation reaction (main plot). The following 50 mM buffer solutions containing 0.1 M KCl were used: sodium acetate (○), sodium phosphate (•), MES (*), and Tris-HCl (▴) in the reduction reaction and sodium phosphate (⋄), glycine-NaOH (⧫), and disodium phosphate-NaOH (▾) in the oxidation reaction. The mixture for the reduction reaction contained 20 mM methyl benzoylformate and 0.3 mM NADH, and the mixture for the oxidation reaction contained 20 mM (S)-(−)-1-phenylethanol and 3 mM NAD+. The activity of the ADHBs oxidation reaction was measured in the following 50 mM buffer solutions containing 0.1 M KCl: MES (▵), Tris-HCl (▪), and glycine-NaOH (□). The mixture consisted of 260 mM 2-propanol and 18 mM NAD+. The assays were performed under the conditions described in Materials and Methods. The kcat values for ADHBs were calculated on the basis of the monomer molecular mass (32.25 kDa).
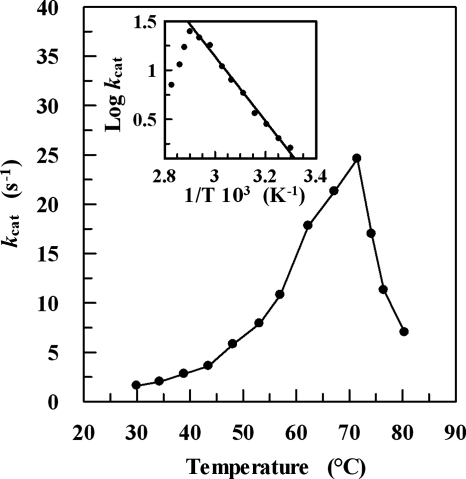
The effect of temperature on ADHTt activity is shown in Fig. 3. The reaction rate increases up to a temperature of about 73°C and then decreases rapidly due to thermal inactivation. This optimal temperature is comparable to those of other ADHs from thermophilic organisms, such as ADHSs (88°C) (27), ADHTb (85°C) (18), and ADHBs (65°C) (7), which are medium-chain, zinc-containing ADHs, and lower than that of the recently described Pyrococcus furiosus NAD(P)H-dependent ADH that belongs to the aldo-keto reductase superfamily (100°C) (22). The activation energy for oxidation is 62.9 ± 2.6 kJ mol−1, which is slightly higher than that determined for ADHBs (52.6 kJ mol−1) (data not shown) or ADHSs (46.7 kJ mol−1) (27) and 1.5-fold lower than that reported for ADHTb (94.1 kJ mol−1) (18).
FIG. 3.
Temperature dependence of the ADHTt activity. The assay was carried out as described in Materials and Methods, using 2,2,2-trifluoroacetophenone as the substrate. The inset shows the Arrhenius plot for the same data. The activation energy was 62.9 ± 2.6 kJ mol−1. The kcat data for temperatures higher than 65°C were corrected for the change in optical density at 340 nm due to NADH heat lability.
Coenzyme and substrate specificity.
The enzyme showed no activity with NADP(H) and full activity with NAD(H). This preference further supports the evidence that an aspartate residue at position 37 of an SDR enzyme plays a critical role in discriminating NAD(H) from NADP(H) (2, 14). The specificity of ADHTt for various alcohols, aldehydes, and ketones was examined (Tables 2 and 3). The enzyme showed no activity on aliphatic linear, branched, and cyclic alcohols except for a discrete activity on cyclohexanol (Table 2). Benzyl alcohol and 4-methyl- and 4-bromo-benzyl alcohols were found not to be substrates. In contrast, in view of the greater influence that the strong electron-donating methoxy group exerts in the para and ortho positions of the benzene ring compared with the meta position, the three methoxy-substituted benzyl alcohols were found to be highly active substrates. (S)-(−)-1-Phenylethanol was also found to be an active substrate, whereas the S and R enantiomers of α-(trifluoromethyl)benzyl alcohol and methyl and ethyl mandelates showed no apparent activity with ADHTt. Moreover, the activity on (±)-1-phenyl-1-propanol was similar to that observed for racemic 1-phenylethanol, whereas the activities on 1-(4′-fluorophenyl)ethanol, 1-(4′-chlorophenyl)ethanol, and trans-cinnamyl alcohol were 45, 26, and 25%, respectively, of the activity observed for (S)-(−)-1-phenylethanol (Table 2).
TABLE 2.
Substrate specificity of ADHTt in the oxidation reactiona
| Substrate | Relative activity (%)c |
|---|---|
| Ethanol | 0 |
| 1-Propanol | 0 |
| 2-Propyn-1-ol | 0 |
| 1-Butanol | 0 |
| 3-Methyl-1-butanol | 0 |
| 1-Pentanol | 0 |
| 1-Hexanol | 0 |
| 1-Heptanol | 0 |
| 1-Octanolb | 0 |
| 2-Propanol | 0 |
| (S)-2-Butanol | 0 |
| (R)-2-Butanol | 0 |
| 2,3-Butanediol | 0 |
| (S)-2-Pentanol | 0 |
| (R)-2-Pentanol | 0 |
| 3-Pentanol | 0 |
| 2-Hexanolb | 0 |
| 2-Heptanolb | 0 |
| 6-Methyl-5-hepten-2-olb | 0 |
| Cyclopentanol | 0 |
| Cyclohexanol | 13 |
| Cycloheptanol | 0 |
| Cyclohexylmethanolb | 0 |
| 2-Cyclohexylethanol | 0 |
| 3-Methylcyclohexanolb | 0 |
| 3-Cyclohexyl-1-propanolb | 0 |
| Geraniol | 0 |
| Crysanthemyl alcohol | 0 |
| Benzyl alcohol | 0 |
| 2-Methoxybenzyl alcohol | 25 |
| 3-Methoxybenzyl alcohol | 13 |
| 4-Methoxybenzyl alcohol | 99 |
| 4-Methylbenzyl alcohol | 0 |
| 3-Bromobenzyl alcoholb | 0 |
| 4-Bromobenzyl alcoholb | 0 |
| (R,S)-1-Phenylethanolb | 62 |
| (S)-(−)-1-Phenylethanol | 100 |
| (R)-(+)-1-Phenylethanol | 0 |
| 1-(4′-Fluorophenyl)ethanolb | 45 |
| 1-(4′-Chlorophenyl)ethanolb | 26 |
| 1-Phenyl-2-propanol | 16 |
| (±)-1-Phenyl-1-propanolb | 59 |
| (±)-2-Phenyl-1-propanol | 0 |
| (R)-1-Phenyl-2-propen-1-ol | 14 |
| 1-(2-Chlorophenyl)-1-propanolb | 0 |
| (R)-(−)-2-Chloro-1-phenylethanol | 0 |
| (R,S)-α-(Trifluoromethyl)benzyl alcohol | 0 |
| (R)-α-(Trifluoromethyl)benzyl alcohol | 0 |
| (S)-α-(Trifluoromethyl)benzyl alcohol | 0 |
| trans-Cinnamyl alcoholb | 25 |
| Methyl (R)-(−)-mandelate | 0 |
| Methyl (S)-(+)-mandelate | 0 |
| (R)-(−)-Mandelic acid | 0 |
| (S)-(+)-Mandelic acid | 0 |
| Ethyl (R)-(−)-mandelate | 0 |
| Ethyl (S)-(+)-mandelate | 0 |
| (±)-1-Indanold | 3,900 |
| (±)-α-Tetralolb | 2,700 |
The activity was measured at 65°C as described in Materials and Methods. The concentration of each substrate was 20 mM.
The substrate was dissolved in 50% 2-propanol (the only exception was α-tetralol, which was dissolved in 100% 2-propanol).
Relative rates were calculated by defining the activity for (S)-(−)-1-phenylethanol as 100%.
The substrate was dissolved in 100% acetonitrile.
TABLE 3.
Substrate specificity of ADHTt in the reduction reaction
| Substratea | Relative activity (%)b |
|---|---|
| Ketones | |
| 3,3-Dimethyl-2-butanone | 0 |
| 2-Pentanone | 0 |
| 3-Methyl-2-pentanone | 0 |
| 4-Methyl-2-pentanone | 0 |
| 2-Methyl-3-pentanone | 0 |
| 5-Chloro-2-pentanone | 0 |
| 2,5-Hexanedione | 0 |
| 6-Methyl-5-hepten-2-one | 0 |
| Cyclopentanone | 0 |
| 2-Methylcyclopentanone | 0 |
| Cyclohexanone | 0 |
| 2-Methylcyclohexanone | 0 |
| 3-Methylcyclohexanone | 0 |
| 4-Methylcyclohexanone | 0 |
| 2,6-Dimethylcyclohexanone | 0 |
| Cycloheptanone | 0 |
| (±)-Camphor | 0 |
| 4-Methoxyphenylacetone | 0 |
| 2-Acetylfuran | 0 |
| Phenylacetone | 0 |
| Acetophenone | 0 |
| 2-Hydroxyacetophenone | 0 |
| 3′-Methoxyacetophenone | 0 |
| 4′-Methoxyacetophenone | 0 |
| 2,2,2-Trifluoroacetophenone | 100 |
| 2,2-Dichloroacetophenone | 32 |
| 3-Chloropropiophenone | 0 |
| 4-Chlorobutyrophenone | 10 |
| 2′,3′,4′,5′,6′-Pentafluoroacetophenone | 0 |
| Chalcone | 0 |
| Benzil | 0 |
| 1-Phenyl-1,2-propanedione | 146 |
| 1-Indanone | 3 |
| α-Tetralone | 4 |
| Aldehydes | |
| Propionaldehyde | 0 |
| Butyraldehyde | 0 |
| iso-Butyraldehyde | 0 |
| 3-Methyl-butyraldehyde | 0 |
| Valeraldehyde | 0 |
| Hexanal | 0 |
| Benzaldehyde | 14 |
| Salicylaldehyde | 0 |
| 3-Hydroxybenzaldehyde | 0 |
| 4-Hydroxybenzaldehyde | 0 |
| 2-Methoxybenzaldehyde | 13 |
| 3-Methoxybenzaldehyde | 14 |
| 4-Methoxybenzaldehyde | 13 |
| trans-Cinnamaldehyde | 0 |
| Keto esters | |
| Ethyl pyruvate | 0 |
| Ethyl 3-oxohexanoate | 0 |
| MBF | 57 |
| Ethyl benzoylformate | 100 |
| Ethyl benzoylacetate | 0 |
| Keto acids | |
| Phenylglyoxylic acid | 0 |
| Sodium phenylpyruvate | 0 |
The activity was measured at 65°C as described in Materials and Methods. Substrates were dissolved in 2-propanol and added to the reaction mixture (5 mM in 2% [vol/vol] 2-propanol).
The percentages were determined by comparison to 2,2,2-trifluoroacetophenone for ketones and aldehydes and to ethyl benzoylformate for keto esters and keto acids.
The enzyme was not active on aliphatic aldehydes and showed discrete activity with aryl aldehydes, such as benzaldehyde and 2-methoxy-, 3-methoxy-, and 4-methoxybenzaldehydes (Table 3). No activity was observed on aliphatic linear, branched, and cyclic ketones. However, the enzyme showed a high reduction rate with halogenated aryl ketones, such as 2,2,2-trifluoroacetophenone, 2-chloroacetophenone, and 4-chlorobutyrophenone, and with aryl diketones, such as 1-phenyl-1,2-propanedione, although it was not active on benzil (diphenylethanedione) (Table 3). Although the CF3 group is sterically more demanding than the CH3 group (van der Waals volumes, 42.7 and 24.5 Å3, respectively [30]), the electron-withdrawing character of fluorine favors hydride transfer, inductively decreasing the electron density at the acceptor carbon, C-1. This electronic factor accounts for the apparent catalytic inactivity observed with the corresponding (S)-α-(trifluoromethyl)benzyl alcohol compared to the high activity observed with the nonhalogenated alcohol (S)-(−)-1-phenylethanol (Table 2).
Interestingly, ADHTt proved to be very effective in reducing aryl α-keto esters, although it was not active on aliphatic α-keto esters and aryl β-keto esters. The evidence presented here shows that ADHTt is a strictly NAD(H)-dependent oxidoreductase that has discrete substrate specificity. Thus, ADHTt is distinct from the three SDR superfamily mesophilic, tetrameric ADHs to which it shows high sequence similarity in specific regions throughout the primary structure (i.e., ADHLs, RADHLb, and PED) and which are active on a variety of aliphatic as well as aromatic alcohols ketones, diketones, and keto esters (Fig. 1).
Kinetic studies.
The kinetic parameters of ADHTt determined for the most active substrates are shown in Table 4. Based on the specificity constant (kcat/Km), this enzyme shows the greatest preference for ethyl benzoylformate compared with MBF, 3-methoxybenzaldehyde, 2,2,2-trifluoroacetophenone, and 1-phenyl-1,2-propanedione in the reduction reaction and relatively lower preference for (S)-(+)-1-indanol and (S)-(+)-α-tetralol in the oxidation reaction. Moreover, the kcat and kcat/Km values are considerably higher for NADH than for NAD+. These results suggest that the enzyme is S stereospecific and that the physiological direction of the catalytic reaction is reduction rather than oxidation. However, the natural substrate and function of ADHTt are unknown. Quintela and coworkers (26) highlighted the presence of muropeptides with phenylacetic acid residues as a distinctive feature of T. thermophilus peptidoglycan. It is tempting to speculate that ADHTt may play a role in a hypothetical phenylacetate synthesis via benzoylformate to mandelate.
TABLE 4.
Steady-state kinetic constants of ADHTta
| Substrate | kcat (s−1) | Km (mM) | kcat/Km (s−1 mM−1) |
|---|---|---|---|
| 4-Methoxybenzyl alcohol | 1.6 ± 0.1 | 61.0 ± 7.1 | 0.026 |
| (S)-(−)-1-Phenylethanol | 1.1 ± 0.1 | 18.1 ± 3.3 | 0.06 |
| 3-Methoxybenzaldehyde | 3.1 ± 0.2 | 4.40 ± 0.25 | 0.70 |
| Ethyl benzoylformate | 50.1 ± 3.3 | 1.0 ± 0.1 | 50.1 |
| MBF | 38.1 ± 3.7 | 2.7 ± 0.6 | 14.1 |
| 2,2,2- Trifluoroacetophenone | 25.5 ± 1.8 | 11.2 ± 2.1 | 2.3 |
| 1-Phenyl-1,2-propanedione | 17.1 ± 3.2 | 5.90 ± 1.1 | 2.9 |
| 1-Indanone | 8.30 ± 0.5 | 27.6 ± 4.2 | 0.30 |
| (±)-1-Indanol | 45.7 ± 2.3 | 5.1 ± 0.4 | 8.9 |
| (S)-(+)-1-Indanol | 61.4 ± 4.3 | 4.2 ± 0.5 | 14.6 |
| α-Tetralone | 7.70 ± 0.6 | 5.8 ± 0.6 | 1.3 |
| (±)-α-Tetralol | 48.1 ± 4.9 | 5.3 ± 0.6 | 9.1 |
| (S)-(+)-α-Tetralol | 57.0 ± 4.5 | 4.2 ± 0.5 | 13.6 |
| NAD+ | 0.84 ± 0.06 | 0.24 ± 0.04 | 3.50 |
| NADH | 52.4 ± 4.3 | 0.035 ± 0.003 | 1,490 |
The activity was measured at 65°C as described in Materials and Methods. Kinetic constants for NAD+ and NADH were determined with 35 mM (S)-(−)-1-phenylethanol and 7 mM ethyl benzoylformate, respectively. The kcat and Km data are the means ± standard deviations.
It is noteworthy that the specific activity of ADHTt (60 and 210 U/mg with 2,2,2-trifluoroacetophenone and MBF, respectively) is comparable to the specific activities of ADHLs (77 U/mg with 2,2,2-trifluoroacetophenone) (12), ADHTb (30 to 90 U/mg with the best substrates) (11), and RADHLb (450 U/mg with acetophenone) (11) and higher than the specific activity of ADHSs (5.5 U/mg with benzyl alcohol) (27).
Thermal stability.
The thermal stability of ADHTt was determined by measuring the residual enzymatic activity after 30 min of incubation over a temperature range from 25 to 95°C. ADHTt was shown to be quite stable up to a temperature of 80°C, above which its activity decreased abruptly, and the temperature at which there was 50% inactivation after 30 min of incubation was ∼90°C (data not shown). In 0.1-mg ml−1 protein samples, the residual activities measured after 24 h of incubation at 50, 60, and 70°C were 142, 134, and 107%, respectively; the residual activities in 1.0-mg ml−1 samples were 97, 105, and 94% for the same temperatures.
Effects of various compounds.
The effects of salts, ions, and reagents on ADHTt activity were studied by adding each compound to the standard assay mixture containing 50 mM Tris-HCl buffer (pH 7.0) (Table 5). The chlorides of Li+, Na+, and K+ activated the enzyme, whereas those of Ca2+ and Mg2+ caused partial inactivation. Iodoacetate did not affect the activity, indicating that the only Cys residue in the monomer, C36, which is adjacent to the functionally important residue D37 (Fig. 1), may not have an essential role, although it is susceptible to reactions with heavy metal ions, such as Cu2+ and Hg2+ (Table 5).
TABLE 5.
Effects of various compounds on ADHTta
| Compound | Concn (mM) | Relative activity (%) |
|---|---|---|
| None | 100 | |
| LiCl | 1 | 127 |
| 100 | 178 | |
| NaCl | 1 | 102 |
| 100 | 179 | |
| KCl | 1 | 167 |
| 100 | 193 | |
| CaCl2 | 1 | 90 |
| 100 | 70 | |
| MgCl2 | 1 | 96 |
| 100 | 67 | |
| ZnSO4 | 1 | 114 |
| CuCl2 | 1 | 76 |
| HgCl2 | 1 | 33 |
| 2-Mercaptoethanol | 1 | 100 |
| 5 | 100 | |
| Iodoacetate | 1 | 95 |
| o-Phenanthroline | 0.1 | 109 |
| 1 | 108 | |
| EDTA | 1 | 127 |
| 10 | 165 | |
| BMIMBF4b | 2 | 35 |
The activity was measured at 65°C as described in Materials and Methods.
BMIMBF4, 1-butyl-3-methylimidazolium tetrafluoroborate.
The presence of metal-chelating agents did not affect the enzyme activity, suggesting that either the protein does not require metals for activity or the chelating molecule was not able to remove the metals under the assay conditions. Furthermore, the enzyme showed no loss of activity following exhaustive dialysis against EDTA. The EDTA-dialyzed enzyme turned out to be quite stable at 70°C for 4 h, both in the absence and in the presence of EDTA or o-phenanthroline. Moreover, incubation of the dialyzed enzyme at 30°C for 6 h in the presence of a low concentration of denaturant (0.5 M guanidinium HCl) resulted in 20% inactivation in the absence and presence of EDTA or o-phenanthroline added at millimolar concentrations (data not shown). These results indicate that ADHTt does not require metals for activity or for structural stabilization; although this is usual for the typically nonmetal SDR enzymes, the more classical RADHLb-type ADHs show strong Mg2+ dependence (23). The 65% inactivation observed with the ionic liquid was presumably due to competition of the BF4− ion with the coenzyme phosphate moiety for the anion binding site of the enzyme.
Stability in organic solvents.
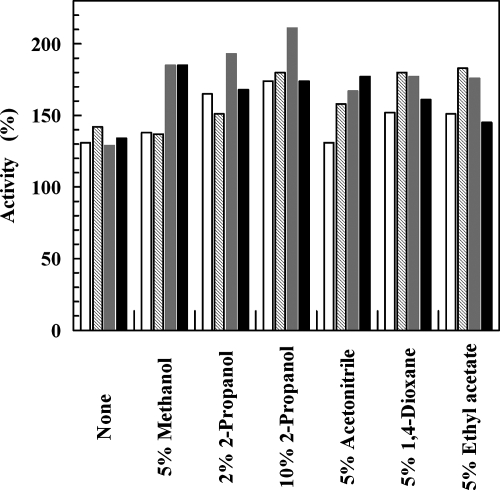
The effects of common organic solvents, such as methanol, 2-propanol, acetonitrile, dioxane, and ethyl acetate, on ADHTt were investigated at two different times and at two different temperatures (Fig. 4). Significant increases in enzyme activity occurred after incubation in aqueous buffer (the values were 130 to 140% of the values prior to incubation) and after incubation in the presence of all the solvents tested. The enzyme activities ranged from 130% of the initial value when 5% acetonitrile was included at 50°C to over 200% of the initial value with 10% 2-propanol at 60°C.
FIG. 4.
Effects of organic solvents on ADHTt. Samples of ADHTt (0.12 mg ml−1) were incubated at 50°C (open and cross-hatched bars) and 60°C (gray and black bars) in the absence and presence of the organic solvents at the indicated concentrations, and assays were performed after 5 h (open and gray bars) and 24 h (cross-hatched and black bars). The activity assays were performed as described in Materials and Methods using ethyl benzoylformate as the substrate. The data obtained in the absence and presence of organic solvents are expressed as percentages of activity relative to the value determined prior to incubation.
The presence of 10% methanol, 2-propanol, acetonitrile, dioxane, ethyl acetate, 1-propanol, and n-hexane in samples of ADHTt incubated for 65 h at 25°C resulted in activities that were 172, 110, 167, 160, 115, 173, and 182%, respectively, of the initial activity measured prior to incubation, whereas the activity of the control remained unchanged. Furthermore, standard assays performed in the presence of 0.01 to 0.5% 2-propanol resulted in no change in activity, suggesting that the enhancement of activity was not due to an immediate effect of solvent on the protein structure. Previous papers have described the activation of thermophilic enzymes by loosening of their rigid structure in the presence of protein perturbants (4, 21). To account for the observed enhancement of ADHTt activity, it is proposed that the organic solvent induces a conformational change in the protein molecule to a more relaxed and flexible conformation that is optimal for activity. Analogously, the activating effect following heating of ADHTt in aqueous buffer at 50 and 60°C for a long time (Fig. 4) may be due to partial loosening of the overall structure, inducing increased flexibility at the active site and consequently increased turnover.
Enantioselectivity.
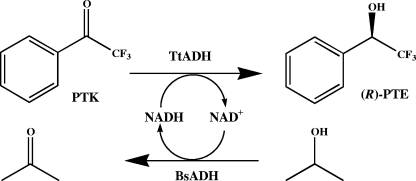
The enantioselectivity of ADHTt was tested using acetophenone, 2,2,2-trifluoroacetophenone, α-methyl and α-ethyl benzoylformates, α-tetralone, and 1-indanone as substrates and an efficient NADH regeneration system (Fig. 5) consisting of Zn-containing, homotetrameric ADH obtained from the moderately thermophilic bacterium Bacillus stearothermophilus strain LLD-R (ADHBs) (7). This ADH has been successfully used in hydrogen tunneling effect studies for a sizeable temperature range (17), as well as in structural and molecular dynamic studies (1, 31); it is NAD(H) dependent and is active mainly on aliphatic and aromatic primary and secondary alcohols and aldehydes (7) but not on aliphatic and aromatic ketones or on the carbonyl substrates of ADHTt and corresponding alcohols (data not shown). Since 2-propanol is not a substrate of ADHTt, it may be a suitable substrate for ADHBs in NADH recycling, as well as for use as a cosolvent. The experimental conditions, including buffer, pH, temperature, and reaction time, were chosen to optimize productivity. Figure 2 shows that MES buffer (pH 6.0) and 60°C were the optimal pH and temperature conditions for catalysis by the two enzymes.
FIG. 5.
Coenzyme recycling in the production of chiral aryl alcohol with ADHTt utilizing B. stearothermophilus ADH (BsADH) and 2-propanol. PTK, 2,2,2-trifluoroacetophenone; (R)-PTE, (R)-(−)-α-(trifluoromethyl)benzyl alcohol; TtADH, T. thermophilus ADH.
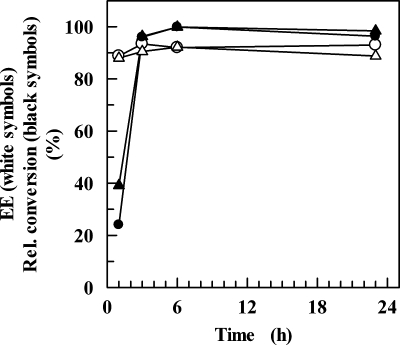
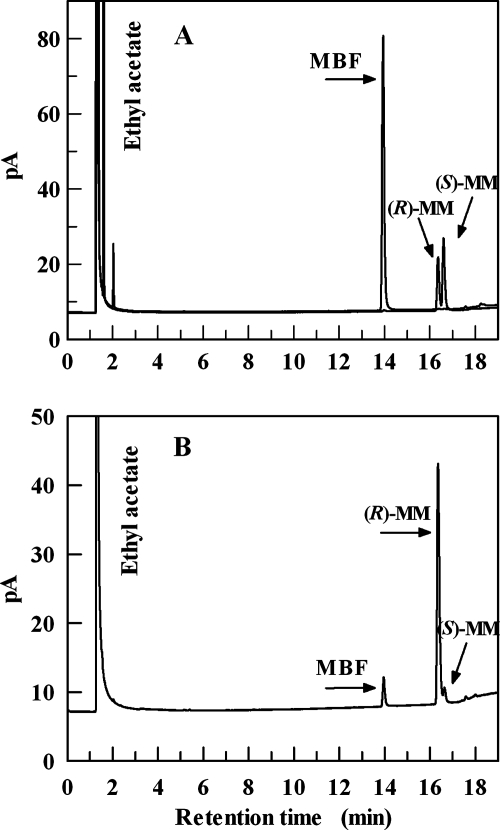
Conversion experiments were carried out at 50 and 60°C, and the reactions were allowed to proceed for 1, 3, 6, and 24 h. Figure 6 shows that the levels of conversion of 2,2,2-trifluoroacetophenone were 20 and 40% in 1 h at 50 and 60°C, respectively, approximating 100% in 6 h at both temperatures. ADHTt preferably reduced this arylketone to (R)-α-(trifluoromethyl)benzyl alcohol with ee of 90 and 93% at 60 and 50°C, respectively, after 3 h of incubation. However, reaction times as long as 24 h did not improve the yield or ee of the biotransformation (Fig. 6). MBF was reduced by ADHTt to methyl (R)-(−)-mandelate with 18, 91, and 99% conversion at 50°C and with 38, 83, and 98% conversion at 60°C after 1, 6, and 24 h, respectively. The highest ee (92%) was observed after 6 h of incubation at 50°C, whereas the ee were 91% at 60°C after 6 h and 89 and 91% at 50 and 60°C, respectively, after 24 h of incubation. Figure 7 shows the results of a GC analysis of the reduction of MBF following 6 h of incubation at 50°C. Similarly, the reaction with ethyl benzoylformate performed for 6 h at 50°C yielded ethyl (R)-(−)-mandelate with a level of conversion of 90% and an ee of 95%. Acetophenone was reduced to (S)-1-phenylethanol following a 6-h reaction at 50°C with a level of conversion of 70% and an ee of 99%, despite the apparent inactivity under different assay conditions (Table 3). This result is remarkable since it shows that the devised NADH regeneration system was able to drive a thermodynamically unfavorable transformation. Analogously, 1-indanone and α-tetralone (data not shown) were reduced to the corresponding (S) alcohols following a 6-h reaction at 50°C, with levels of conversion of 40 and 18%, respectively, and an ee of almost 99%. The results are summarized in Table 6. On the whole, the enantioselectivity data indicate that the hydride ion of NADH is transferred to the re face of the carbonyl of acetophenone, 2,2,2-trifluoroacetophenone, and bicyclic ketones, as well as that of α-methyl and α-ethyl benzoylformates, suggesting that ADHTt exhibits Prelog specificity (25). The ee and yield values obtained for the conversion of the halogenated and nonhalogenated acetophenones are comparable to the values for the SDRs ADHLs (12) and RADHLb (29), both of which possess anti-Prelog selectivity. The activation of ADHTt by 10% (vol/vol) n-hexane proves the sturdiness of the enzyme and suggests promising applications for conversion of poorly water-soluble prochiral substrates at higher concentrations in a biphasic reaction medium. Investigations with water-immiscible organic solvents are in progress to assess the applicability of the reaction system for preparative asymmetric synthesis.
FIG. 6.
Conversion and ee of 2,2,2-trifluoroacetophenone for ADHTt at different reaction times. Biotransformations were carried out at 50°C (circles) and 60°C (triangles) as described in Materials and Methods. The reactions were stopped by addition of ethyl acetate at the times indicated. The dried extracts were analyzed by chiral GC, and the relative conversion was calculated by dividing the area of alcohol products by the total area.
FIG. 7.
Product analysis by GC of the enantioselective reduction of methyl benzoylformate. (A) MBF and racemic methyl mandelate standards. (B) Reaction products. The retention times for MBF, (R)-methyl mandelate [(R)-MM], and (S)-methyl mandelate [(S)-MM] were 13.938, 16.361, and 16.584 min, respectively, for panel A, and 13.958, 16.351, and 16.634 min, respectively, for panel B.
TABLE 6.
Asymmetric reduction of carbonyl compounds by ADHTta
| Substrate | Product | Conversion (%)b | ee (%)b | Configuration of productc |
|---|---|---|---|---|
| MBF | Methyl mandelate | 99 | 91 | R |
| Ethyl benzoylformate | Ethyl mandelate | 90 | 95 | R |
| Acetophenone | 1-Phenylethanol | 70 | >99 | S |
| 2,2,2-Trifluoroacetophenone | α-(Trifluoromethyl)benzyl alcohol | 100 | 93 | R |
| 1-Indanone | 1-Indanol | 40 | >99 | S |
| α-Tetralone | α-Tetralol | 18 | >99 | S |
The reactions were performed at 50°C for 6 h, except for the MBF reaction (24 h), as described in Materials and Methods.
Conversion yields and ee were determined by chiral GC analysis.
The configuration of the product alcohol was determined by comparing the retention time with that of standard samples as described in Materials and Methods.
It is noteworthy that the optically active alcohols produced are used as chiral building blocks in organic synthesis. O-protected methyl (R)-(−)-mandelate is used as an intermediate for the synthesis of pharmaceuticals (16, 24), and optically pure trifluoromethyl-substituted benzyl alcohols are important precursors for specialty chemicals used in electrooptical devices, such as liquid crystal displays (5).
Closing remarks.
The gene encoding a novel ADH from T. thermophilus was successfully expressed in E. coli, and the purified enzyme was shown to possess remarkable thermophilicity, thermal resistance, and tolerance to common organic solvents. ADHTt exhibits Prelog specificity and high enantioselectivity with a discrete spectrum of aromatic carbonyl substrates of interest. Moreover, ADHTt has many advantages with regard to its preparative application, including ease of purification, long-term stability, and absolute preference for NAD(H), which can be efficiently regenerated via a thermophilic bacillus ADH (ADHBs) in a bioconversion process based on coenzyme recycling. ADHTt and ADHBs are particularly amenable to coupling in an efficient synthetic reaction as they possess good functional stability in the presence of 2-propanol at relatively elevated temperatures and only the coenzyme is a cosubstrate. Moreover, the importance of the critical role of the D37 residue mentioned above in SDRs in discriminating NAD(H) from NADP(H) is further supported by the results of this study.
Acknowledgments
This work was funded by FIRB (Fondo per gli Investimenti della Ricerca di Base) grant RBNE034XSW and by the ASI project MoMa n. 1/014/06/0.
Footnotes
Published ahead of print on 2 May 2008.
REFERENCES
- 1.Ceccarelli, C., Z. X. Liang, M. Strickler, G. Prehna, B. M. Goldstein, J. P. Klinman, and B. J. Bahnson. 2004. Crystal structure and amide H/D exchange of binary complexes of alcohol dehydrogenase from Bacillus stearothermophilus: insight into thermostability and cofactor binding. Biochemistry 43:5266-5277. [DOI] [PubMed] [Google Scholar]
- 2.Filling, C., K. D. Berndt, J. Benach, S. Knapp, T. Prozorovski, E. Nordling, R. Ladenstein, H. Jörnvall, and U. Oppermann. 2002. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277:25677-25684. [DOI] [PubMed] [Google Scholar]
- 3.Fiorentino, G., R. Cannio, M. Rossi, and S. Bartolucci. 1998. Decreasing the stability and changing the substrate specificity of the Bacillus stearothermophilus alcohol dehydrogenase by single amino acid replacements. Protein Eng. 11:925-930. [DOI] [PubMed] [Google Scholar]
- 4.Fontana, A., V. De Filippis, P. Polverino de Laureto, E. Scaramella, and M. Zambonin. 1998. Rigidity of thermophilic enzymes, p. 277-294. In A. Ballestreros, F. J. Plou, J. L. Iborra, and P. J. Halling (ed.), Stability and stabilization in biocatalysis, vol. 15. Elsevier Sciences, Amsterdam, The Netherlands. [Google Scholar]
- 5.Fujisawa T., K. Ichikawa, and M. Shimizu. 1993. Stereocontrolled synthesis of p-substituted trifluoromethylbenzylic alcohol derivatives of high optical purity by the baker's yeast reduction. Tetrahedron Asymmetry 4:1237-1240. [Google Scholar]
- 6.Giordano, A., F. Febbraio, C. Russo, M. Rossi, and C. A. Raia. 2005. Evidence for co-operativity in coenzyme binding to tetrameric Sulfolobus solfataricus alcohol dehydrogenase and its structural basis: fluorescence, kinetic and structural studies of the wild-type enzyme and non-co-operative N249Y mutant. Biochem. J. 388:657-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guagliardi, A., M. Martino, I. Iaccarino, M. De Rosa, M. Rossi, and S. Bartolucci. 1996. Purification and characterization of the alcohol dehydrogenase from a novel strain of Bacillus stearothermophilus growing at 70°C. Int. J. Biochem. Cell Biol. 28:239-246. [DOI] [PubMed] [Google Scholar]
- 8.Guy, J. E., M. N. Isupov, and J. A. Littlechild. 2003. The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix. J. Mol. Biol. 331:1041-1051. [DOI] [PubMed] [Google Scholar]
- 9.Henne, A., H. Bruggemann, C. Raasch, A. Wiezer, T. Hartsch, H. Liesegang, A. Johann, T. Lienard, O. Gohl, R. Martinez-Arias, C. Jacobi, V. Starkuviene, S. Schlenczeck, S. Dencker, R. Huber, H. P. Klenk, W. Kramer, R. Merkl, G. Gottschalk, and H. J. Fritz. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22:547-553. [DOI] [PubMed] [Google Scholar]
- 10.Höffken, H. W., M. Duong, T. Friedrich, M. Breuer, B. Hauer, R. Reinhardt, R. Rabus, and J. Heider. 2006. Crystal structure and enzyme kinetics of the (S)-specific 1-phenylethanol dehydrogenase of the denitrifying bacterium strain EbN1. Biochemistry 45:82-93. [DOI] [PubMed] [Google Scholar]
- 11.Hummel, W. 1999. Large-scale applications of NAD(P)-dependent oxidoreductases: recent developments. Trends Biotechnol. 17:487-492. [DOI] [PubMed] [Google Scholar]
- 12.Inoue, K., Y. Makino, T. Dairi, and N. Itoh. 2006. Gene cloning and expression of Leifsonia alcohol dehydrogenase (LSADH) involved in asymmetric hydrogen-transfer bioreduction to produce (R)-form chiral alcohols. Biosci. Biotechnol. Biochem. 70:418-426. [DOI] [PubMed] [Google Scholar]
- 13.Jones, J. B., and J. F. Beck. 1976. Applications of biochemical systems in organic chemistry, p. 248-401. In J. B. Jones, C. J. Sih, and D. Perlman (ed.), Techniques of chemistry series, part I, vol. 10. John Wiley & Sons, New York, NY. [Google Scholar]
- 14.Kallberg, Y., U. Oppermann, H. Jörnvall, and B. Persson. 2002. Short-chain dehydrogenases/reductases (SDRs). Eur. J. Biochem. 269:4409-4417. [DOI] [PubMed] [Google Scholar]
- 15.Keinan, E., E. K. Hafely, K. K. Seth, and R. Lamed. 1986. Thermostable enzymes in organic synthesis. Asymmetric reduction of ketones with alcohol dehydrogenase from Thermoanaerobium brockii. J. Am. Chem. Soc. 108:162-169. [Google Scholar]
- 16.Kobayashi, Y., Y. Takemoto, Y. Ito, and S. Terashima. 1990. A novel synthesis of the (2R,3S)- and (2S,3R)-3-amino-2-hydroxycarboxylic acid derivatives, the key components of a renin inhibitor and bestatin, from methyl (R)- and (S)-mandelate. Tetrahedron Lett. 31:3031-3034. [Google Scholar]
- 17.Kohen, A., R. Cannio, S. Bartolucci, and J. P. Klinman. 1999. Enzyme dynamics and hydrogen tunnelling in a thermophilic alcohol dehydrogenase. Nature 399:496-499. [DOI] [PubMed] [Google Scholar]
- 18.Korkhin, Y., A. J. Kalb(Gilboa), M. Peretz, O. Bogin, Y. Burstein, and F. Frolow. 1998. NADP-dependent bacterial alcohol dehydrogenases: crystal structure, cofactor-binding and cofactor specificity of the ADHs of Clostridium beijerinckii and Thermoanaerobacter brockii. J. Mol. Biol. 278:967-981. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Leatherbarrow, R. J. 1992. GraFit, version 5.0.11. Erithacus Software Ltd., Horley, United Kingdom.
- 21.Liang, Z. X., T. Lee, K. A. Resing, N. G. Ahn, and J. P. Klinman. 2004. Thermal-activated protein mobility and its correlation with catalysis in thermophilic alcohol dehydrogenase. Proc. Natl. Acad. Sci. USA 101:9556-9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machielsen, R., A. R. Uria, S. W. Kengen, and J. van der Oost. 2006. Production and characterization of a thermostable alcohol dehydrogenase that belongs to the aldo-keto reductase superfamily. Appl. Environ. Microbiol. 72:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niefind, K., J. Müller, B. Riebel, W. Hummel, and D. Schomburg. 2003. The crystal structure of R-specific alcohol dehydrogenase from Lactobacillus brevis suggests the structural basis of its metal dependency. J. Mol. Biol. 327:317-328. [DOI] [PubMed] [Google Scholar]
- 24.Pousset, C., M. Haddad, and M. Larchevêque. 2001. Diastereocontrolled synthesis of unit A of cryptophycin. Tetrahedron 57:7163-7167. [Google Scholar]
- 25.Prelog, V. 1964. Specification of the stereospecificity of some oxido-reductase by diamond lattice sections. Pure Appl. Chem. 9:119-130. [Google Scholar]
- 26.Quintela, J. C., E. Pittenauer, G. Allmaier, V. Arán, and M. A. de Pedro. 1995. Structure of peptidoglycan from Thermus thermophilus HB8. J. Bacteriol. 177:4947-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raia C. A., A. Giordano, and M. Rossi. 2001. Alcohol dehydrogenase from Sulfolobus solfataricus. Methods Enzymol. 331:176-195. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Schlieben, N. H., K. Niefind, J. Muller, B. Riebel, W. Hummel, and D. Schomburg. 2005. Atomic resolution structures of R-specific alcohol dehydrogenase from Lactobacillus brevis provide the structural bases of its substrate and cosubstrate specificity. J. Mol. Biol. 349:801-813. [DOI] [PubMed] [Google Scholar]
- 30.Zhao, Y. H., M. H. Abraham, and A. M. Zissimos. 2003. Fast calculation of van der Waals volume as a sum of atomic and bond contributions and its application to drug compounds. J. Org. Chem. 68:7368-7373. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, X., and T. C. Bruice. 2007. Temperature-dependent structure of the E·S complex of Bacillus stearothermophilus alcohol dehydrogenase. Biochemistry 46:837-843. [DOI] [PubMed] [Google Scholar]