Abstract
IncU plasmids are a distinctive group of mobile elements with highly conserved backbone functions and variable antibiotic resistance gene cassettes. The IncU archetype is conjugative plasmid RA3, whose sequence (45,909 bp) shows it to be a mosaic, modular replicon with a class I integron different from that of other IncU replicons. Functional analysis demonstrated that RA3 possesses a broad host range and can efficiently self-transfer, replicate, and be maintained stably in alpha-, beta-, and gammaproteobacteria. RA3 contains 50 open reading frames clustered in distinct functional modules. The replication module encompasses the repA and repB genes embedded in long repetitive sequences. RepA, which is homologous to antitoxin proteins from alpha- and gammaproteobacteria, contains a Cro/cI-type DNA-binding domain present in the XRE family of transcriptional regulators. The repA promoter is repressed by RepA and RepB. The minireplicon encompasses repB and the downstream repetitive sequence r1/r2. RepB shows up to 80% similarity to putative replication initiation proteins from environmental plasmids of beta- and gammaproteobacteria, as well as similarity to replication proteins from alphaproteobacteria and Firmicutes. Stable maintenance functions of RA3 are most like those of IncP-1 broad-host-range plasmids and comprise the active partitioning apparatus formed by IncC (ParA) and KorB (ParB), the antirestriction protein KlcA, and accessory stability components KfrA and KfrC. The RA3 origin of transfer was localized experimentally between the maintenance and conjugative-transfer operons. The putative conjugative-transfer module is highly similar in organization and in its products to transfer regions of certain broad-host-range environmental plasmids.
Conjugative plasmids contribute greatly to the global spread of genetic information and gene exchange, as in some cases they can self-transfer even between distant bacterial species. Conjugative R factors assigned to the IncU incompatibility group have been isolated from a number of Aeromonas spp. and Escherichia coli strains from seawater fish hatcheries and diseased fish, as well as from clinical environments (2, 4, 6, 15, 26, 30, 31, 42). Members of the IncU plasmid group are implicated particularly in the dissemination of antibiotic resistance in Aeromonas strains associated with aquatic environments (15).
IncU representatives pAr-32 and pRAS1 contain resistance genes encoded within integrons, and on the basis of restriction enzyme analysis of both plasmid molecules, Sørum et al. (35) postulated a highly conserved backbone structure of IncU group members with variability in the region coding for antibiotic resistance. Similar observations were made with plasmids pASOT and pFBAOT (2, 26, 27). Plasmid pFBAOT6 (84,749 bp) has been sequenced recently and analyzed in silico (27). Plasmid Rms149 of the Pseudomonas IncP-6 group was assigned to the IncU group on the basis of incompatibility tests (12). Apart from homology between the replication genes of pFBAOT6 and Rms149, no conservation of the backbone structure was observed, in contrast to previous suggestions (11, 27).
As the main focus has been on their resistance gene cassettes, little is known about the biology of plasmids from the IncU group, of which RA3 is the archetype (3). This study describes the initial step in a complete functional analysis of plasmid RA3. First, the sequence of RA3 (45,909 bp) had to be established (accession no. DQ401103). It proved to be almost identical to pFBAOT6 in the backbone region. RA3 possesses a 10.5-kb antibiotic resistance region that contains a class I integron carrying sulI, aadA2, and catA2 determinants. This differs from a similar insertion in pFBAOT6, the RA3 sequence possessing duplicated sulI and truncated qac genes in the 3′ conserved segment (3′CS). The insertion site of the integron in RA3 corresponds to the insertion site of the main variable region (54 kb) in pFBAOT6. Plasmid RA3 does not contain an IS630 element, which in pFBAOT6 is inserted in the putative conjugative transfer region. Functional analysis demonstrated that RA3 was capable of high-frequency conjugal transfer to beta- and gammaproteobacteria, with a 1,000-fold lower frequency of transfer to alphaproteobacteria, revealing that the plasmid has a broad host range. The segregational stability of RA3 was confirmed in diverse bacterial host strains, the minimal region required for replication was defined, and the origin of conjugative transfer was localized.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains used were K-12 strains DH5α [F− (Φ80dlacZΔM15) recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U19] (13), CV601 (gfp Rifr Kmr), kindly supplied by K. Smalla (Braunschweig, Germany), and C600K (thr-1 leu-6 thi-1 lacY1 supE44 ton-21 galK) (23). E. coli C strain C2110 (polA1 his rha) (D. R. Helinski, University of California San Diego) was used for minireplicon analysis. Pseudomonas putida KT2442 Rifr was provided by C. M. Thomas (University of Birmingham, Birmingham, United Kingdom). Agrobacterium tumefaciens UBAPF2 (gfp Kmr Rifr Smr) and Ralstonia eutropha 7MP228r (gfp Rifr Kmr Pnr) were kindly supplied by K. Smalla (Braunschweig, Germany).
Bacteria were grown in L broth or L agar (L broth with 1.5% [wt/vol] agar) at 37°C or 28°C (18) supplemented with appropriate antibiotics. For E. coli, we used benzylpenicillin sodium salt at 150 μg ml−1 in liquid medium and 300 μg ml−1 in agar plates for penicillin resistance, kanamycin sulfate (50 μg ml−1) for kanamycin resistance, streptomycin sulfate (20 to 30 μg ml−1) for streptomycin resistance, and chloramphenicol (10 μg ml−1) for chloramphenicol resistance. For R. eutropha, P. putida, and A. tumefaciens, chloramphenicol (10 μg ml−1) was used. L agar used for blue/white screening contained isopropyl-β-d-thiogalactopyranoside (IPTG; 0.1 mM) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg ml−1).
Gene amplification.
Standard PCRs (24) were performed with RA3 DNA as the template and the following pairs of primers: ant1 5′-CGGATCCGCGGGCCTGATCTATTGTTG-3′ (coordinates 45365 to 45384) and ant2 5′-CGGATCCGCATGCTTTCTATGCCGCTAACGGC-3′ (coordinates 45741 to 45722) for the repAp fragment, rep1 5′-CGGATCCCCGGAAACCAACTTGGCGGGC-3′ (coordinates 45750 to 45769) and rep2 5′-CGGATCCGCATGCCGCATAAACTCGGCCTGT-3′ (coordinates 203 to 186) or rep5 5′-CGCATGCCCCCGGAAAACCAACTTGGCG-3′ (coordinates 45750 to 45769) and rep7 5′-CGGATCCGCCGCATAAACTCGGCCTGT-3′ (coordinates 203 to 186) for repB1p/repB2p, rep6 5′-CGCATGCTTTCGGTGAGCTTGGCGGC-3′ (coordinates 45800 to 45818) and rep2 for repB2p, ant3 5′-CGAGCTCCACATCATTCTGATGAGGGC-3′ (coordinates 45414 to 45433) and ant2 for repA, and rep3 5′-CGAGCTCTTGGCAAGTTAGGGGTGCAT-3′ (coordinates 45885 to 45904) and rep4 5′-CCGTCGACGCCATCTAAACGGCTTTAC-3′ (coordinates 1385 to 1365) for repB. The restriction sites introduced for cloning are in italics. PCR steps were as follows: an initial denaturation at 95°C for 5 min and then 30 cycles of denaturation (95°C for 30 s), annealing (52°C for 30 s), and elongation (72°C for 90 s). Reactions ended with a final elongation step (72°C for 7 min). The minireplicon fragments tested were amplified with primers str7 5′-CGCAGATAAGCCAGCGCATC-3′ (coordinates 44251 to 44270) and 6L2 5′-AAGCGGGTAGTGATAGACTG-3′ (coordinates 1497 to 1478), str7 and inp5 5′-AGTGATTCCGTACGGGGCCA-3′ (coordinates 45512 to 45493), str7 and rep2, and miniR1 5′-GCGGATCCGCGTTGCGTAGTCGTCTAG-3′ (coordinates 45693 to 45711) and snab2 5′-CGGCATGCCGATCACGCTCCCAGGTCAA-3′ (coordinates 2347 to 2329). PCRs were performed as described above with an annealing temperature of 54°C and an elongation time of 3 min in each cycle. All PCR-derived clones were analyzed by DNA sequencing to check their fidelity.
Plasmid DNA isolation, analysis, cloning, and manipulation.
Plasmid RA3 DNA was extracted with a midi kit used according to the manufacturer's instructions (Qiagen, West Sussex, United Kingdom). Plasmid DNA was isolated and manipulated by standard procedures (29). For the cloning of PCR fragments, either promoter-probe vector pPTO1 (39) or expression vector pGBT30 (17) with lacIq and tacp was used. The plasmids used and constructed in this study are presented in Table 1.
TABLE 1.
Plasmids used and constructed in this study
| Plasmid | Relevant features | Copy no. | Reference or source |
|---|---|---|---|
| Used | |||
| pAKE600 | oriMB1oriTRK2 PnrsacB | High | 7 |
| pBGS18 | oriMB1 Kmr | High | 36 |
| pET28 | oriMB1 Kmr T7p lacO His6 tag, T7 tag | Medium | Novagen |
| pET28mod | oriMB1 Kmr T7p lacO His6 tag, T7 tag deleted, no BamHI site | Medium | G. Jagura-Burdzy |
| pGBT30 | oriMB1 PnrlacIqtacp expression vector | High | 17 |
| pGEM-T Easy | oriMB1Pnr | High | Promega |
| pPTOI | oriSC101 Kmr promotorless xylE | Medium | 39 |
| pUC18 | oriMB1 Pnr | High | 43 |
| RA3 | IncU Cmr Smr Sur | Low | F. Hayes |
| Constructed | |||
| pAKB1.101 | pGEM-T with PCR product (6L2 and str7)a corresponding to 3,155-bp fragment of RA3 | ||
| pAKB1.102 | pGEM-T with PCR product (inp5 and str7) corresponding to 1,261-bp fragment of RA3 | ||
| pAKB1.103 | pGEM-T with PCR product (rep2 and str7) corresponding to 1,861-bp fragment of RA3 | ||
| pAKB7.5 | pBGS18 with 460-bp HincII-BamHI fragment from RA3 (coordinates 9395-9854) | ||
| pAKB9.5 | pAKE600 with BamHI-PstI fragment from pAKB7.5 | ||
| pMOB1.4 | pBGS18 with 2,563-bp PCR product (miniR1 and snaB2) | ||
| pMOB1.5 | pBGS18 with 327-bp PCR product (ant3 and ant2) | ||
| pMOB1.5.1 | pGBT30 with SacI-SalI fragment of pMOB1.5 to create transcriptional fusion tacp-repA | ||
| pMOB1.6 | pBGS18 with 1,409-bp PCR product (rep3 and rep4) | ||
| pMOB1.6.1 | pGBT30 with SacI-SalI fragment of pMOB1.6 to create transcriptional fusion tacp-repB | ||
| pMOB1.7 | pBGS18 with 362-bp PCR product (rep5 and rep7) | ||
| pMOB1.7.1 | pPTO1 with SphI-BamHI fragment of pMOB1.7 to create transcriptional fusion repBp1/p2-xylE | ||
| pMOB1.8 | pBGS18 with 312-bp PCR product (rep6 and rep7) | ||
| pMOB1.8.1 | pPTO1 with SphI-BamHI fragment of pMOB1.8 to create transcriptional fusion repBp2-xylE | ||
| pMOB1.9 | pBGS18 with 362-bp PCR product (rep1 and rep2) | ||
| pMOB1.9.1 | pPTO1 with SphI-BamHI fragment of pMOB1.9 to create transcriptional fusion repXp-xylE | ||
| pMOB1.10 | pBGS18 with 376-bp PCR product (ant1 and ant2) | ||
| pMOB1.10.1 | pPTO1 with BamHI fragment of pMOB1.10 to create transcriptional fusion repAp-xylE |
Primers used in PCR.
Sequencing and sequence assembly.
Sequencing reactions were performed with custom primers with RA3 as the template. A library of EcoRI fragments of RA3 was created in pUC18, and partially HincII-digested DNA was also inserted into pUC18 to assist the sequencing of problematic regions with the use of universal primers. Sequences were generated with the ABI Big Dye terminator kit. Sequences were assembled with the DNASTAR Lasergene 99 package. Restriction enzyme analysis verified the assembly. Open reading frame (ORF) predictions were made with Glimmer 2.0 according to standard parameters. The function of each ORF was predicted by BLAST searches against the EMBL/SWISSPROT databases, and the function was annotated based on homology. The predicted proteins were characterized by isoelectric point and molecular weight and searched for coiled-coil regions, conserved domains, and transmembrane helices with the use of bioinformatics on-line software on the EXPASY server (pI, MolMass, Coils, InteroScan [EMBL-EBI], and TMHMM v2.0 [CBS], respectively).
Determination of catechol 2,3-oxygenase activity (XylE).
Catechol 2,3-oxygenase activity (the product of xylE) was assayed (44) in exponentially growing bacteria. Plasmid content of all assayed cultures was monitored to ensure that differences in XylE activity were not due to variations in plasmid copy number. One unit of catechol 2,3-oxygenase is defined as the amount needed to convert 1 μmol of catechol in 1 min under standard conditions. Protein concentration was determined by the Bradford method (5).
Transformation and conjugation procedures.
E. coli competent cells were prepared by CaCl2 treatment (29). Samples (0.2 ml) of overnight cultures of the donor and recipient strains grown under selection were pelleted at 5,000 × g for 2 min, and the pellets were resuspended in 100 μl of fresh medium. Recipient and donor cell suspensions were mixed (2:1), spread on L agar, and incubated overnight at 37°C (for E. coli) or 28°C (for P. putida, R. eutropha, and A. tumefaciens). Cells were suspended in 1 ml of L broth. Cell suspensions were serially diluted, plated on L agar with appropriate selection, and allowed to grow for 48 h.
oriT mapping.
A restriction fragment of 458 bp (coordinates 9397 to 9854) predicted to contain oriT was cloned into pAKE600 to create pAKB9.5. E. coli DH5α harboring RA3 was transformed with pAKE600 or pAKB9.5. The double transformants were used as donors in conjugal-transfer mating experiments with E. coli K-12 strain CV601 as the recipient, with selection for either Pnr Knr (pAKE600) or Cmr Knr (RA3) transconjugants. Transconjugants were then tested for the presence of the mobilized or conjugative plasmids (DNA analysis).
Stability assays.
E. coli, A. tumefaciens, R. eutropha, and P. putida harboring RA3 were grown overnight in liquid cultures with selective pressure and analyzed for plasmid content. The cultures were then diluted 105-fold and grown without selection over 20 generations, and dilutions were plated onto L agar plates with and without chloramphenicol to estimate plasmid retention. The procedure was repeated over 60 generations. Plasmid content was also checked at the end of the experiment.
Nucleotide sequence accession number.
The annotated sequence of plasmid RA3 has been deposited in the GenBank database under accession number DQ401103.
RESULTS AND DISCUSSION
RA3 host range.
Conjugative plasmid RA3 was originally isolated from Aeromonas hydrophila and was also stably maintained in E. coli (3). The host range of RA3 was examined by conjugal-transfer mating experiments. RA3 was transferable to a wide spectrum of gram-negative bacteria, among them P. putida (gammaproteobacteria), R. eutropha (betaproteobacteria), and A. tumefaciens (alphaproteobacteria). The frequency of transfer between E. coli and either P. putida or R. eutropha was very high (1 × 10−1 transconjugant per donor). The frequency of transfer between E. coli and A. tumefaciens was much lower (1 × 10−4 transconjugant per donor under the conditions tested). RA3 retention in populations of transconjugants of A. tumefaciens, R. eutropha, and P. putida was evaluated every 20 generations during growth without selection for approximately 60 generations. RA3 replicated and was stably maintained in almost 100% of the cells after 60 generations of nonselective growth in all three subgroups of proteobacteria. The restriction enzyme profiles of plasmid DNA isolated from transconjugants at the end of the stability experiments were similar to the profiles of the original plasmid indicating that these were not chromosomal integrants (data not shown).
General features of RA3.
The 45,909-bp sequence of plasmid RA3 was determined by primer walking; when necessary, subcloned restriction fragments were sequenced with universal primers.
The number of identified coding sequences within the RA3 plasmid is 50, based on a Glimmer 2.0 cutoff of 250 bp, followed by visual inspection. Among the 50 ORFs, only 1 (orf11) has no homologs in public databases and 10 have homologs of unknown function (Table 2; for the full version, see Table S1 in the supplemental material). Forty-four of the ORFs show 100% identity to ORFs found in pFBAOT6 from the IncU incompatibility group (accession no. NC_006143; 27). Hence, the annotation of RA3 arbitrarily follows that for pFBAOT6 in the region from the first base pair to 33 kb, with the exception of orf01, which is designated repB instead of rep. The equivalent of orf50 (coordinates 45441 to 45737) has not been identified in the pFBAOT6 annotation, but there is strong experimental evidence that this locus is a part of the replication cassette (see below) and was thus annotated in RA3 as repA.
TABLE 2.
Predicted coding sequences in RA3
| ORF | Gene | Coordinatesa | Size in aa (mass in kDa) | % Similarity, homolog (protein ID) | pFBAOT6 (protein ID) |
|---|---|---|---|---|---|
| 01 | repB | 1-1380 | 459 (52.27) | 82, RepARms149 (CAI46990) | CAG15048 |
| 02 | RA3.02 | 2248-2526 | 92 (11.41) | CAG15049 | |
| 03 | klcA | 2637-3062 | 141 (16.35) | 55, KlcAR751 (NP044227) | CAG15050 |
| 04 | RA3.04 | 3190-3345 | 51 (5.71) | CAG15051 | |
| 05 | korC | 3391-3687 | 98 (10.84) | 55, KorCRK2 (C35387) | CAG15052 |
| 06 | kfrC | 3692-4759 | 355 (38.57) | 69, KfrCR751 (Upf54.4) (AAC64416) | CAG15053 |
| 07 | kfrA | 4889-5956 | 355 (38.68) | 30, KfrAR751 (AAC64418) | CAG15054 |
| 08 | korA | 6037-6348 | 104 (11.3) | 53, KorARK2 (P03052) | CAG15055 |
| 09 | incC | 6353-7105 | 250 (27.11) | 63, IncCRK2 (P07673) | CAG15056 |
| 10 | korB | 7102-8460 | 452 (47.81) | 53, KorBRK2 (P07674) | CAG15057 |
| 11 | RA3.11 | 8463-8690 | 75 (7.93) | Not annotated | |
| 12 | mpR | 8816-9547c | 243 (27.3) | 61, MpRR46 (AAD17385) | CAG15059 |
| 13 | mobC | 9837-10367 | 176 (19.97) | 57, MobApXF51 (AAF85616) | CAG15060 |
| 14 | nic | 10360-11355 | 331 (36.9) | 63, MobBpXF51 (AAF85615) | CAG15061 |
| 15 | traC3 | 11394-13088c | 564 (62.19) | 59, TraC2R751 (AAC64468) | CAG15062 |
| 16 | traD | 13091-13237c | 48 (5.3) | 60, TraDR751 (AAC64471) | CAG15063 |
| 17 | traC4 | 13227-16292c | 1021 (111.9) | 51, TraC4R751 (AAC64469) | CAG15064 |
| 18 | virD4 | 16305-18230c | 641 (72) | 55, TraNpIPO2 (CAC82760) | CAG15065 |
| 19 | virB11 | 18196-19188c | 330 (37.3) | 66, VirB11pAT (AAL45862) | CAG15066 |
| 20 | virB10 | 19202-20416c | 404 (42.3) | 55, TraLpIPO2 (CAC82758) | CAG15067 |
| 21 | virB9 | 20416-21249c | 277 (30.54) | 63, VirB9pXF51 (AAF85582) | CAG15068 |
| 22 | virB8 | 21252-22001c | 249 (28.1) | 61, VirB8pXF51 (AAF85581) | CAG15069 |
| 23 | RA3.23 | 22001-22207c | 68 (7.4) | 51, OPRF from Brucella melitensis 16M (AAL53277) | CAG15070 |
| 24 | virB6 | 22322-23332c | 336 (35.4) | 54, VirB6pXF51 (AAF85580) | CAG15071 |
| 25 | RA3.25 | 23344-23613c | 89 (9.73) | 65, TraGpIPO2 (CAC82753) | CAG15072 |
| 26 | virB5 | 23648-24292c | 214 (23.44) | 53, TraFpIPO2 (CAC82752) | From 24286 identical to CAG15073 |
| 27 | virB4 | 24289-26736c | 815 (91.70) | 68, VirB4pXF51 (AAF85576) | CAG15074 |
| 28 | virB3 | 26733-27053c | 106 (11.90) | 61, VirB3pXF51 (AAF85575) | CAG15075 |
| 29 | virB2 | 27071-27394c | 107 (11.21) | 54, VirB2pXF51 (AAF85574) | CAG15076 |
| 30 | RA3.30 | 27372-27728c | 118 (13.6) | 67, XF_a0004pXF51 (AAF85573) from Xylella fastidiosa 9a5c | CAG15077 |
| 31 | top | 27793-29769c | 658 (72.29) | 73, TopApXF51 (AAF85572) | CAG15078 |
| 32 | RA3.32 | 29766-30503c | 245 (26.23) | 45, LysM from Desulfotomaculum reducens MI-1 (EAR43569) | CAG15079 |
| 33 | RA3.33 | 30514-31008c | 164 (17.97) | 56, TraL from Dichelobacter nodosus (AAW31839) | CAG15081 |
| 34 | RA3.34 | 31194-31583 | 129 (14.06) | CAG15082 | |
| 35 | RA3.35 | 31603-31863 | 86 (9.52) | 71, Ypesb_01003249 from Yersinia pestis (ZP_01175336) | CAG15083 |
| 36 | RA3.36 | 31856-32251 | 131 (14.89) | 44, Nham_0207 from Nitrobacter hamburgensis X14 (ABE61107) | CAG15084 |
| 37 | rec | 32316-32885c | 189 (20.89) | 79, PinRpMAQU01 from Marinobacter aquaeolei VT8 (ABM21007) | CAG15085 |
| 38 | tnp | 33177-33971c | 264 (30.71) | 100, TnppRAS1 (CAD57187) | CAG15086 |
| 39 | orf6 | 34138-34425c | 95 (9.77) | 100, Orf6pRAS1 (CAD57186) | CAG15087 |
| 40 | orf5 | 34449-34949c | 166 (18.33) | 99, Orf5pRAS1 (CAD57185) | CAG15088 |
| 41 | sulI | 35076-35915c | 279 (30.12) | 100, SulIpAr-32 (CAD57201) | CAG15089 |
| 42 | qacEdelta1 | 35909-36115c | 68 (7.42) | 97, truncated QacEdelta1pAr-32 (CAD57200) | |
| 43 | catA2 | 36861-37502c | 213 (24.77) | 99, CatA2pAr-32 (CAD57199) | |
| 44 | tnp513 | 38253-39794c | 513 (58.49) | 100, TnppAr-32 (CAD57198) | |
| 45 | sulI | 40199-41038c | 279 (30.12) | 100, SulIpAr-32 (CAD57201) | |
| 46 | qacEdelta1 | 41032-41379c | 115 (12.33) | 100, QacEdelta1pAr-32 (CAD57196) | CAG15090 |
| 47 | aadA2 | 41543-42334c | 263 (29.54) | 100, AadA2pY2 from Klebsiella pneumoniae (AAF87686) | CAG15091 |
| 48 | int1 | 42480-43493 | 337 (38.38) | 100, IntIpRAS1 (CAD57181) | CAG15092 |
| 49 | res | 43836-44411c | 191 (20.28) | 100 (in 174 aa), PinpRSB101 (CAG27808) | CAG15093b |
| 50 | repA | 45441-45737 | 98 (10.91) | 58, VapI from Rhodopseudomonas palustris BisB18 (YP_531137); 54, HigARts1 (AAC43983) | Not annotated |
The letter c follows the coordinates of ORFs on the complementary strand. The hypothetical presence of transmembrane helices within the predicted proteins was analyzed with TMHMM v 2.0 software.
93% homology.
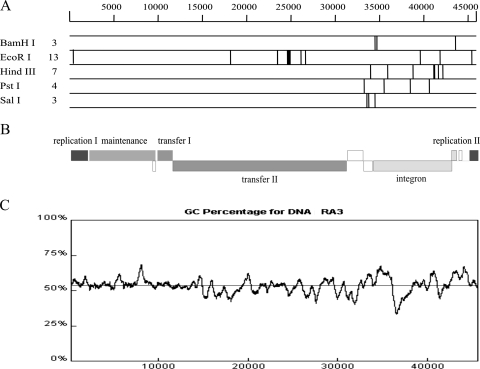
Plasmid RA3 has an uneven distribution of restriction endonuclease recognition sites (Fig. 1A and B). The backbone region seems to be inaccessible to most of the common restriction endonucleases, with the exception of EcoRI; 11 GAATTC palindromic sequences are found in the region coding for replication and conjugative-transfer functions. This observation was made previously with IncU plasmid pRAS1 (35) and may indicate that E. coli is a relatively new host for RA3. Aside from EcoRI, the majority of sites sensitive to digestion by class II restriction enzymes localize to the integron (coordinates 34138 to 43493), suggesting recent acquisition of this DNA.
FIG. 1.
RA3 sequence analysis. (A) Restriction map of the RA3 sequence generated in silico. The map shown (Lasergene package) represents RA3 restriction patterns for BamHI, EcoRI, HindIII, PstI, and SalI. The number of recognition sequences is indicated at the left. (B) Modular organization of RA3 with plus- and minus-strand genes. (C) GC content of the RA3 DNA sequence. The x axis indicates the plasmid sequence coordinates, while the y axis shows the percentage of GC counted for a 500-bp window.
The average GC content of the RA3 sequence is 53.6% (10). For comparison, two recently sequenced chromosomes of A. salmonicida subsp. salmonicida (NC_009348) and A. hydrophila subsp. hydrophila (NC_008570) show higher GC contents of 58% and 61%, respectively. The GC content of RA3 varies along the plasmid molecule (Fig. 1C), and the fluctuations coincide with the boundaries of functional modules, with a minimum value of 33% around position 37,000 bp (integron) and the maximum reaching 68% at position 8,500 bp in the stability region.
RA3 contains long repetitive sequences that are also evident in some broad-host-range environmental plasmids, e.g., pIPO2 (37) and pSB102 (34). The repetitive sequences in RA3 fall into two categories: intragenic repetitions translated into protein domains (class I) and intergenic long repetitive sequences (class II). Both repetitions show considerable sequence conservation that ranges from 85 to 100% for a particular unit. Moreover, a characteristic feature of these sequences is their compact structure; the units are organized head to tail, forming a repeat module.
The class I repetitions were found in three loci (Fig. 2), orf07, orf10, and orf17, which code for proteins KfrA, KorB, and TraC4, respectively. The lengths of the repeated units vary from 13 amino acids in TraC4 to 19 amino acids in KorB and 35 amino acids in KfrA. The class II repetitions of 345 and 537 bp surround the replication module and are described in detail below.
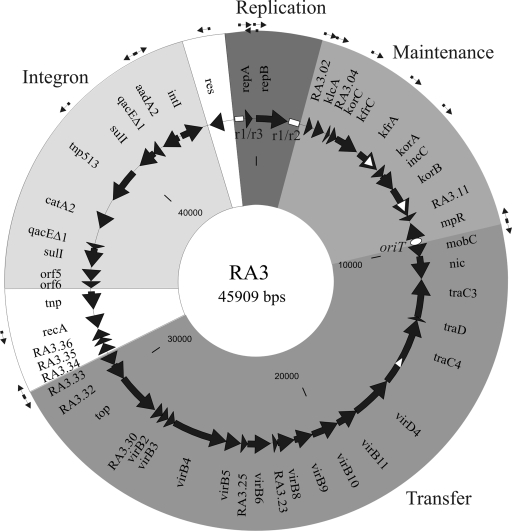
FIG. 2.
Circular representation of plasmid RA3. All identified ORFs are represented by solid arrows indicating the direction of transcription. Functional gene clusters are highlighted with different shades of grey. The genes which are not obvious parts of the modules are left white. The predicted promoters are marked with thin black arrows. The class I intragenic repetitions in kfrA, korB, and traC and the class II repetitive regions r1/r3 and r1/r2 are represented by white triangles and boxes, respectively. The white oval marks the position of oriT.
Modular structure and transcriptional organization.
Genes in the plasmid backbone (coordinates 45019 to 31194) are organized in blocks associated with particular functions such as replication, stable maintenance, and conjugative transfer (Fig. 2). These three functional modules seem to be of distinct origins showing homology to unrelated groups of plasmids. The genetic organization of the RA3 backbone reveals a compact structure (Fig. 2). The coding sequences cover 91% of the RA3 backbone and 96% of the plasmid when the two long repetitive regions surrounding the rep genes and the intergenic region between maintenance and transfer modules encoding oriT (see below) (1,400 bp) are subtracted. The region of RA3 encompassing the integron (coordinates 31194 to 45019) shows only 72% coding sequences.
There are 17 predicted promoters in the RA3 sequence. Twelve of the promoters potentially drive the expression of the majority of backbone functions, with nine oriented clockwise and three counterclockwise (Table 3 and Fig. 2). There are three putative divergent promoter regions: repBp/repXp within the replication module, mpRp/mobCp between the maintenance and conjugative transfer-associated genes, and RA3.33p/RA3.34p in the conjugative transfer region.
TABLE 3.
Predicted promoters in the RA3 sequencea
| Locus | −35 motif (coordinates) | Spacer size (bp) | −10 motif (coordinates) |
|---|---|---|---|
| repB | TTGGCG (45762-45767) | 17 | TAAATT (45785-45790) |
| RA3.02 | TTGCCA (2168-2173) | 18 | TATAAT (2192-2197) |
| klcA | TTGACA (2558-2563) | 18 | TATTAT (2582-2587) |
| korC | TTGAGG (3175-3180) | 20 | TATTGT (3201-3206) |
| kfrA | TTGTAT (4829-4834) | 18 | TAAAAT (4853-4858) |
| korA | TTGACG (5967-5972) | 16 | TATCTT (5989-5994) |
| mpR | TGGCTC (9641-9646c) | 12 | TAACTT (9623-9628c) |
| mobC | TTGCTA (9761-9766) | 16 | TAAAAT (9787-9792) |
| RA3.33 | TGTCCA (31079-31084c) | 17 | TATATT (31056-31061c) |
| RA3.34 | TGGCCT (31136-31141) | 18 | TACCAT (31160-31165) |
| rec | TCCCCG (32958-32964c) | 18 | TAAAAT (32935-32940c) |
| tnp513 | CTGATA (39994-39999c) | 16 | TATTGT (39972-39977c) |
| aadA2 | TGGACA (42590-42595c) | 17 | TAAACT (42567-42572c) |
| int1 | TTGCTG (42425-42430) | 17 | TAGACT (42448-42453) |
| res | TTGCGC (44457-44472c) | 17 | TAAGAT (44444-44449c) |
| repA | TTGTTG (45379-45384) | 17 | TACACT (45402-45407) |
| repX | TTGCCA (45886-45891c) | 16 | TATAAT (45864-45869c) |
The putative −35 and −10 motifs are accompanied by the sequence coordinates in plasmid RA3 (DQ401103). The letter c follows the coordinates of promoters on the complementary strand.
Replication functions.
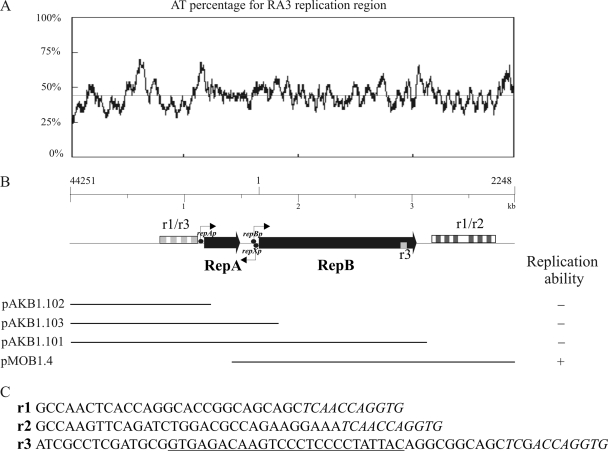
The predicted replication region of RA3 spans >3 kb (coordinates 45019 to 2069) and encompasses two ORFs, designated repA and repB (Fig. 3B). As repetitive sequences upstream and downstream of the repAB genes are homologous, the arbitrarily identified separate units building both regions were designated r1 (38 bp), r2 (42 bp), and r3 (60 bp) (Fig. 3C) to differentiate between sequences that are common or distinct in these two regions. The final 11 nucleotides (nt) are conserved in all three units (in italics in Fig. 3C). The repetitive sequences (346 bp) upstream of repA (designated r1/r3) are organized as (r3r1)3r3. The repetitions (557 bp) located downstream of repB (designated r1/r2) are organized as (r1r2)3r1(r1r2)3r1. Interestingly, 25 bp of the r3 unit is present within the 3′ terminus of the repB coding sequence (Fig. 3B; underlined in panel C).
FIG. 3.
Replication region of RA3. (A) AT percentage plot for the region from 44251 to 2248 encompassing the replication module. The x axis indicates the position in the RA3 sequence. (B) Functional mapping of the replication region. The organization of the replication module is depicted with thin black arrows marking the putative transcription start points, black circles marking putative promoters, and combinations of the repetitive sequences indicated by boxes as follows: r1, white; r2, dark grey; r3, light grey. The bottom part of the panel illustrates the DNA fragments cloned into a high-copy-number vector (pMB1 origin) and tested for the ability to replicate in E. coli C2110 (polA). The results of the tests are summarized on the right. (C) Repetition units r1, r2, and r3. The sequence that is the same in all three units is in italics; the fragment of repetition unit r3 found at the 3′ end of repBRA3 is underlined.
RA3 replication is independent of polymerase I, as shown by propagation in the polA mutant E. coli C2110. To examine the minimal requirements for replication activity, PCR fragments corresponding to different parts of the replication region (Fig. 3B) were cloned into the pGEM-T Easy vector and tested for the ability to replicate in the C2110 strain, which is not transformable by pGEM-T Easy alone. A fragment encompassing repB with the r1/r2 repetitive region (coordinates 45693 to 2248) was sufficient for replication (pMOB1.4; Fig. 3B). Neither repA nor the upstream repetitive region r1/r3 was required for minireplicon functionality. A fragment containing repB without the r1/r2 repetitive region was incapable of transforming the C2110 strain (pAKB1.101), suggesting that the isolated r3 unit in the 3′ end of repB is insufficient for oriV activity. The AT percentage plot for the RA3 replication region (coordinates 44251 to 2248) revealed the existence of several AT-rich stretches, with two (>60% AT) within the minimal replicon (Fig. 3A and B). The first of these regions is located between the end of repB and the downstream r1/r2, and the second AT-rich stretch is immediately after the r1/r2 boxes. The RA3 sequence was also inspected for the presence of the highly conserved DnaA box sequence (5′TTA/TTNCACA) (32), but no motifs were identified. This may suggest that the DnaA initiator protein is not necessary for replication initiation in RA3.
Plasmid-encoded toxins-antitoxins eliminate plasmid-free cells that emerge due to segregation or replication defects. Chromosomal homologs of toxin-antitoxin genes are widely distributed in bacteria and induce programmed cell death or reversible cell cycle arrest in response to starvation or other adverse conditions (14). The predicted RepA protein is a polypeptide of 98 amino acids (10.91 kDa) that shows 34% identity to the HigA antitoxin from plasmid Rts1 of Proteus vulgaris (40) (Fig. 4). Similarly, RepA is homologous to a number of chromosomally encoded antidote proteins from alphaproteobacteria that are characterized by a VapI-like domain: the protein shares 41 to 44% identity with putative antitoxins from Mesorhizobium loti, Sinorhizobium meliloti, and Rhodospirillum rubrum. No obvious candidate for a toxin gene is evident in RA3. Protein structure predictions for RepA revealed the existence of a Cro/cI-type DNA-binding domain (residues 15 to 68) present in the XRE family of transcriptional regulators with an α-helix-turn-α-helix (HTH) motif spanning residues 25 to 42 (Fig. 4), and regulatory studies (see below) confirmed its role in the repression of repAp.
FIG. 4.
Amino acid sequence conservation of RepARA3 (ABD64877) and homologous antidote proteins in M. loti MAFF303099 (BAB48919), S. meliloti strain 1021 (CAC45622), R. rubrum (YP_426153), and plasmid Rts1 of P. vulgaris HigA (AAC43983) (the GenBank/EMBL database protein accession numbers are given in parentheses). Identical or functionally similar residues in the five sequences are shown with a black background, and those that are conserved in three or four representatives are shown with two shades of grey background. The HTH motif identified in RepARA3 (residues 25 to 42) is indicated.
The putative RA3 replication initiation protein RepB (459 amino acids, 52.27 kDa) is encoded downstream of repA. RepB shows >99% identity with the putative replication protein of IncU plasmid pFBAOT6: the proline at position 450 of RepB is changed to alanine in the pFBAOT6 homolog. RepB is 70% identical and 82% similar to RepA from Rms149. RepB of RA3 also shows high levels of similarity (69 to 81%) to putative replication initiation proteins of plasmids isolated from species belonging to alpha-, beta-, and gammaproteobacteria (Fig. 5). Intriguingly, RepB also has lower, but significant, similarity to putative replication proteins from plasmids of gram-positive bacteria, i.e., 47% and 50% similarity to Rep43 of Bacillus sp. strain KSM-KP43 plasmid (28) and to Rep of pIP404 from Clostridium perfringens (8), respectively. The conservation in the RepB homologs is sustained over the N-terminal 380 amino acids, whereas the C termini of the polypeptides are variable (Fig. 5). The predicted HTH motif in RepB is positioned in the C-terminal domain between residues 406 and 427, so the recognition specificity for the replication origins apparently is determined by the most variable protein segment.
FIG. 5.
Sequence alignment of the RepBRA3 and homologous replication initiation proteins. Conserved residues in replication initiation proteins from RA3 (ABD64829), Rms149 (YP_245484), pRSB105 (ABI20460), pAb5S9 (YP_001220601), pMATVIM-7 (YP_001427363), pKBB4037 (CAI47016), and Plasmid2 (YP_743798) (GenBank/EMBL database protein accession numbers are given in parentheses) are shown by a black background. Residues identical or functionally similar in five polypeptides are marked with a dark grey background, and residues conserved in four representatives are shown with a light grey background. The predicted HTH motif in the RepB polypeptide of RA3 is indicated.
As described above, the 3′ end of repB of RA3 includes a partial r3 repetition, a variant of which is present in two copies in the putative replication gene of Rms149, where it was proposed to act as an origin of replication (12). Interestingly, Rms149 does not possess the extended r1/r2 and r1/r3 repetitive sequences found in RA3. A homologous part of the r3 motif also is present in two copies in pKBB4037, one in a position at the 3′ end of rep, similar to that in RA3 and Rms149, and a second located intergenically downstream of rep. Plasmid2 from Nitrosomonas eutropha C91 also possesses a partial r3 repeat located downstream of its putative replication gene. As the r3 motif seems to have different roles in Rms149 and RA3, it would be interesting to ascertain the incompatibility types of these sequenced plasmids, along with their host ranges, building on the initial functional minireplicon analysis performed with RA3 (see above), Rms149 (12), and pRSB105 (33). Rms149 has been classified into the IncP-6 group of Pseudomonas plasmids and IncU (IncG) groups of E. coli plasmids (12); however, the host range of Rms149 has not been fully examined. The pRSB105 plasmid encodes two functional replication systems, one with a narrow host range which is active in several gammaproteobacteria and a second homologous to Rms149 which is also active in gammaproteobacteria and which extends the plasmid's host range to betaproteobacteria but not to alphaproteobacteria. The host range of RA3 seems to be the broadest among the three plasmids because, as noted above, it can replicate and be stably maintained in alpha-, beta-, and gammaproteobacteria.
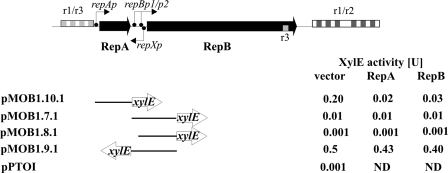
To further understand the function of the rep locus in RA3, a transcriptional analysis of this region was initiated. Putative promoters were found in silico in the replication module preceding the repA and repB coding sequences (Table 3), and an additional promoter, repXp, is evident on the complementary strand in the repA-repB intergenic region (see Fig. S1 in the supplemental material). The region between the end of the upstream repetitive module and the repA start covers 77 bp (coordinates 45364 to 45441) and includes repAp and two imperfect inverted repeats (IRs) with 12- and 7-bp arms that may have a potential regulatory role. The region immediately upstream of repB includes two putative promoter sequences designated repBp1 and repBp2 and divergently oriented repXp, which overlaps repBp2. Fragments bearing these putative promoters were PCR amplified and cloned into promoter-probe vector pPT01 upstream of a promoterless xylE cassette (39). The relative transcriptional activities of the analyzed regions were assessed by measuring levels of catechol oxygenase XylE expressed from the transcriptional fusions (Fig. 6). The repAp promoter upstream of repA showed moderate transcriptional activity (pMOB1.10.1; 0.2 U of XylE). No promoter activity was detected for repBp2 (pMOB1.8.1), whereas a fragment containing both repBp1 and repBp2 showed low transcriptional activity (pMOB1.7.1; 0.01 U of XylE). This activity seems to be sufficient for the expression of repB in the minireplicon tested in the polA mutant (pMOB1.4), although the possibility cannot be excluded that readthrough transcription from a vector promoter might fortuitously drive repB. Transcription from repBp1 is expected to produce a 113-nt 5′ untranslated region that may be highly structured since it includes a long imperfect IR spanning 46 bp (see Fig. S1 in the supplemental material).
FIG. 6.
Regulation of gene expression in the RA3 replication region. DNA fragments cloned into promoter probe vector pPTO1 and tested for promoter activity are shown. Units of XylE transcriptional activity were measured in extracts from IPTG-induced logarithmic-phase cultures of double transformants of C600K with pPTOI or its derivatives and expression vector pGBT30 and its derivatives pMOB1.5.1 (tacp-repA) and pMOB1.6.1 (tacp-repB). The results shown are averages from at least five experiments. ND, not determined.
The strongest promoter in the rep region is repXp, which generates 0.5 U of XylE activity in a transcriptional fusion with the promoterless xylE cassette (pMOB1.9.1; Fig. 6). The repXp promoter either may be responsible for the synthesis of an antisense regulatory RNA that could interact with repB mRNA over a 71-nt stretch or may direct the expression of the ORF encoding a putative RepX polypeptide of 38 amino acids. The start codon of this ORF overlaps the stop codon for repA (see Fig. S1 in the supplemental material). Thus, RepX may be a part of a regulatory circuit that controls RA3 replication. Additionally, the repXp promoter may also influence the activity of the overlapping repBp promoter by competing for binding of RNA polymerase in this region. Further studies are currently under way to distinguish between these possibilities.
The repA and repB genes were PCR amplified and cloned under control of the inducible tacp promoter in expression vector pGBT30 (17) to generate pMOB1.5.1 and pMOB1.6.1, respectively. The expression plasmids were introduced into C600K cells carrying the repAp-xylE (pMOB1.10.1), repBp-xylE (pMOB1.7.1), or repXp-xylE (pMOB1.9.1) transcriptional fusion. Double transformants were grown in the presence of IPTG to induce the synthesis of RepA or RepB to allow an assessment of whether either protein regulates expression from any of the three promoters. Both RepA and RepB downregulated the expression of repAp 5- to 10-fold (Fig. 6). Further studies are required to assign the operator sites for the two regulators, although it is interesting that there are two IR motifs in the repAp region that are candidate sites for RepA and/or RepB interaction. By contrast, neither repBp nor repXp was sensitive to the presence of excess RepA or RepB (Fig. 6). Nevertheless, the divergent promoter region that includes the repBp2 and repXp promoters contains a 46-bp imperfect IR that overlaps both promoters (see Fig. S1 in the supplemental material).
Maintenance functions.
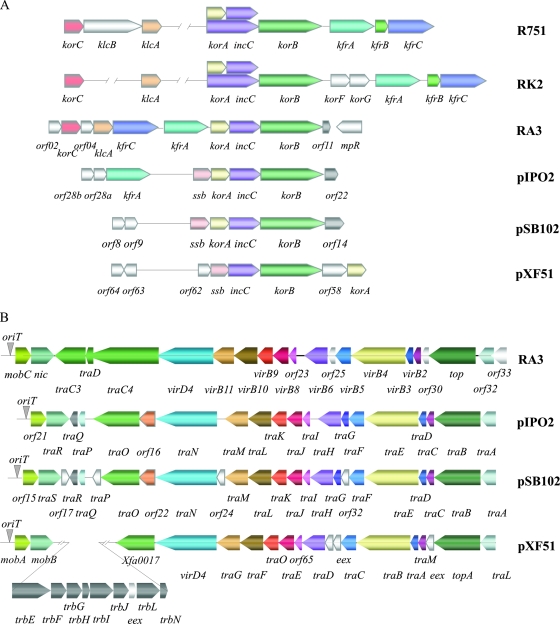
The ORFs related to maintenance functions in RA3 are clustered in a region of 7.7 kb (coordinates 2069 to 9836) located downstream of repB. The region contains 10 ORFs transcribed/translated in the clockwise direction and one oriented in the counterclockwise direction (Fig. 7A). The seven ORFs that are most closely related to IncP-1 counterparts (25, 38) were designated klcA, korC, kfrC, kfrA, korA, incC, and korB. Their products are predicted (on the basis of homology) to form a partitioning apparatus, provide regulatory circuits, and act as accessory stabilization factors. The transcriptional organization of this region suggests that there are promoter regions upstream of five of the genes, orf02, klcA, korC, kfrA, and korA. The putative partitioning operon of RA3 consists of four ORFs, korA, incC, korB, and orf11, with IncC (63% identical to IncC2RK2) belonging to the ParA family of Walker-type ATPases (19) and KorB (53% identical to KorBRK2) acting as the ParB DNA-binding homolog. Unusually among KorB/ParB homologs, KorB of RA3 possesses a class I repetitive sequence in the linker region between the N and C domains. ORF11 has been annotated in the pFBAOT6 sequence, but in a different reading frame. The RA3 partition operon is autoregulated by KorA (A. Kulinska and G. Jagura-Burdzy, unpublished data). Interestingly, the putative partitioning operons of the pIPO2 and pSB102 plasmids from bacteria of the wheat and alfalfa rhizospheres (34, 37) consist of korA-incC-korB preceded by ssb, encoding homologs of single-stranded binding protein, and the downstream genes orf22IPO2 and orf14SB102, which are twice the length of orf11 in RA3 (Fig. 7A). ORF22 and ORF14 seem to be conserved in amino acid sequence, whereas ORF11 of RA3 appears more distant evolutionarily.
FIG. 7.
Organization of maintenance and transfer modules in RA3 and its closest homologs. Comparison of the maintenance (A) and conjugative-transfer (B) regions of IncP-1 plasmids RK2 and R751, IncU plasmid RA3, and environmental broad-host-range plasmids pIPO2, pSB102, and pXF51. Arrows symbolize genes and directions of transcription. The same colors indicate homologous loci; ORFs which do not have a homolog in at least one plasmid are shown in white. The position of oriT is indicated by a grey inverted triangle.
The kfr locus of RA3 encodes two α-helical proteins, KfrA and KfrC, with putative auxiliary function in the segregation process, as shown for the IncP-1 equivalents (1, 16). As noted above, KfrA of RA3 includes a repetitive module of class I (residues 181 to 337) containing four repetitions of 36 amino acids with a fifth, incomplete, copy of 17 amino acids. The coiled-coil structure prediction for KfrARA3 performed with COILS (21) indicated that the repetitive part of KfrA is likely to form an α-helical structure (data not shown). KfrC, another α-helical protein that is 69% identical to the N-terminal part of KfrC from RK2, is encoded in the operon with korC, whereas the three Kfr proteins are encoded in one operon in IncP-1 plasmids.
KorC, KorA, and KorB (53 to 55% similar to the equivalent RK2 proteins) are predicted DNA-binding proteins, their homologs in IncP-1 plasmids acting as global regulators of gene expression in these plasmids (25, 38). The DNA-binding properties of the KorA and KorB proteins of RA3 have been confirmed (Kulinska and Jagura-Burdzy, unpublished).
KlcA, with 54% identity to the equivalent protein of RK2, might provide antirestriction protection, as has been suggested for KlcA of IncP-1α plasmids (20). The products of orf02 and orf04 of RA3 have no known homologs besides counterparts in pFBAOT6. However, BLAST searches revealed 45% identity over a stretch of 60 amino acids between the orf02 product and the putative DnaA protein of A. hydrophila.
Conjugative transfer.
Conjugative transfer functions in RA3 are encoded by a cluster of 21 genes organized in two operons, most of which have a high degree of sequence similarity (Table 2; see Table S1 in the supplemental material) to proposed conjugation gene products of environmental plasmids pXF51 (22), pIPO2 (37), and pSB102 (34) (Fig. 7B). The divergently transcribed tricistronic operon orf34-orf36 encodes a putative integral membrane protein, a putative transcriptional regulator, and a protein with no homologs in the databases. The possibility that they are also involved in conjugative transfer cannot be excluded. By contrast with the IncP-1 tra1 (tra) and tra2 (trb) regions, the RA3 genes for the DNA-processing- and mating pair formation-associated components are not organized in separate blocks. Instead, the organization of the RA3 loci parallels that of pSB102; the relaxase gene (nic) is oriented divergently from the other transfer-associated genes, and a topoisomerase-encoding gene (top) is placed between mating pair formation genes coding for a putative cell wall hydrolase (orf32) and a putative major pilus subunit (virB2) (Fig. 7B). This arrangement also contrasts with the position of the topoisomerase gene traE in IncP-1α plasmid RP4, where traE is encoded between traF and traD in the DNA-processing gene cluster. Thus, RA3, pFBAOT6, pSB102, pXF51, and pIPO2 constitute a new organizational branch of conjugative transfer functions in environmental broad-host-range plasmids.
The location of the origin of transfer (oriT) of RA3 was first examined in silico on the basis of sequence homology between IncU representatives RA3 and pFBAOT6 and the oriT sites of pIPO2 (37), pSB102 (34), and pXF51 (22). This analysis suggested that oriT was situated in the long intergenic region upstream of mobC of RA3 (Fig. 2 and 7B). To examine this experimentally, a 458-bp restriction fragment encompassing the putative oriT (coordinates 9397 to 9854) was cloned into pMB1-based replicon pAKE600 (pAKB9.5) and introduced into E. coli DH5α (RA3). Conjugal-transfer mating experiments demonstrated the mobilization of pAKB9.5 into the CV601 recipient at a frequency of 2 × 10−2 transconjugant per donor (4 × 106 transconjugants), whereas mobilization of the pAKE600 vector was not detected under the same conditions. The mobilization frequency of pAKB9.5 was fivefold lower than the conjugation frequency of RA3. Plasmid DNA analysis of transconjugants confirmed the presence of two independent replicons.
Phenotypic traits.
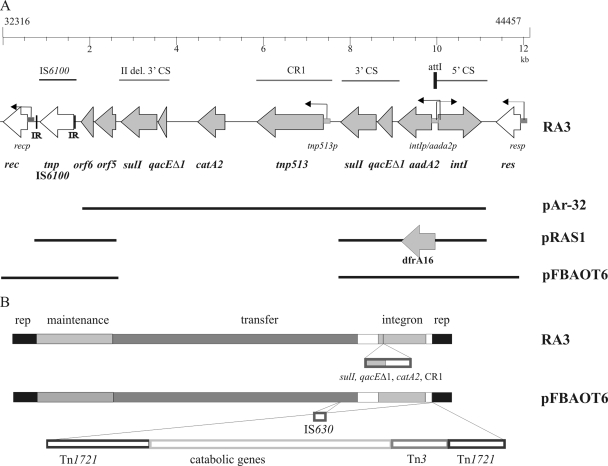
One-quarter of the RA3 plasmid (coordinates 32316 to 44457) constitutes the integration cassette. The module is highly similar to integration cassettes derived from two other IncU representatives, pAr-32 and pRAS1 (35) (Fig. 8A). The backbones (∼35 kb) of IncU plasmids RA3 and pFBAOT6 vary by only seven nucleotide substitutions, one nucleotide deletion, and one nucleotide insertion but differ more dramatically in the amount and properties of more recently acquired genes (Fig. 8B). Thus, a different variable region was inserted into the same genetic context in RA3 and pFBAOT6, between the replication and conjugative-transfer genes. Plasmid RA3 possesses a region of 10.5 kb that contains the class I complex integron with a CR1 element (tnp513 associated with catA2) (41) located between partly duplicated 3′CSs (Fig. 8A). The IS6100 insertion element has been transposed downstream of the 3′CS into a similar position as in pRAS1 (35) (Fig. 8A). In comparison, the variable region of pFBAOT6 encompasses ∼54 kb and comprises IS6100, a class I integron (of a different length than in RA3 as it lacks one copy of the 3′CS and the CR1 element with catA2), and transposable elements, namely, a Tn1721 composite transposon with Tn3 (Fig. 8B). Analysis of sequences of both plasmids revealed that the extra sequence in pFBAOT6 is inserted following position 44968 of RA3, with duplication of the flanking nucleotides (TGTTGG). The transposon insertion in pFBAOT6 occurred in the region between res and about 50 bp before the repetitive region r1/r3, which, as described above, is potentially part of the plasmid replication module. The res genes of RA3 and pFBAOT6 show only 91% identity, which is much lower than for other parts of the plasmid sequences. Moreover, unlike pFBAOT6, RA3 does not include an IS630 element inserted between orf31 and orf33 (note that the equivalent position in RA3 occurs between orf32 and orf33, as traD was named orf16 in RA3 instead of orf15A as in pFBAOT6).
FIG. 8.
Overview of the variable regions of IncU group representatives. (A) Schematic view of class I integrons based on the available sequence data of plasmids RA3 (DQ401103), pAr-32 (AJ517791), pRAS1 (AJ517790), and pFBAOT6 (NC_006143). Grey arrows mark the ORFs contained in the integron; white arrows indicate additional genes. The homologous regions present in different plasmids of the IncU group are marked. (B) Comparative genomic organization of IncU plasmids RA3 and pFBAOT6. Homologous blocks of genes are indicated by the same color; open rectangles represent nonhomologous components.
The comparison of RA3 and pFBAOT6 is a notable example of how a plasmid backbone evolves by the acquisition of resistance and metabolic gene cassettes that enable the plasmid to succeed under variable conditions and under selective pressure after the commencement of broad antibiotic use. Since RA3 is only half of the size of pFBAOT6 with a significantly smaller variable region, it can be anticipated to constitute the incompatibility group U archetype, as previously suggested (6, 35).
Concluding remarks.
Conjugative plasmid RA3 of the IncU incompatibility group is typical of the broad-host-range group of plasmids; it can self-transfer, replicate, and be stably maintained in alpha-, beta-, and gammaproteobacteria. Plasmid RA3 is characterized by a modular and mosaic structure with genes with defined properties clustered in functional blocks that have their closest homologs among plasmids from different incompatibility groups. For example, the maintenance module shows the highest level of relatedness to the maintenance region of IncP-1 plasmids, with 7 of 11 ORFs present in both plasmids, although the two regions differ markedly in gene organization. The partitioning operon of RA3 consists of korA, a short version of incC known as incC2, korB, and orf11 of defined function. Three Kfr proteins specified by IncP-1 are limited to two in RA3, i.e., KfrA and KfrC, which apparently are encoded in separate transcription units. Similarly, KlcA, a putative antirestriction protein, is encoded separately from KorC in RA3, whereas the corresponding genes constitute a single operon in IncP-1 plasmids. Loci for klcA and korC are often found in plasmid replicons of diverse origins, e.g., in extended β-lactamase plasmid pCTXM-3 (9).
It was postulated that IncU plasmids constitute an evolutionarily narrow group in which plasmids possess a conserved backbone structure with a variable region confined to resistance-determining genes (2, 26, 35). The sequence conservation of the backbone functions between RA3 and pFBAOT6 is consistent with those suggestions, as only several nucleotide differences were found in the two plasmid backbones. However, classification of Rms149 into the IncU group (12) based on incompatibility testing and rep gene homology highlights a need to reexamine the notion of such strict conservation in the backbone functions of IncU group plasmids. The identity between RA3 and Rms149 is limited to the rep locus and kfrA (11). Moreover, the different requirements for the minimal replicons of the two plasmids, e.g., independence from DNA polymerase I for RA3 replication and different structures of putative replication origins in the two elements, indicate distinct evolutionary pathways. A wide diversity of beta- and gammaproteobacteria encode RepB homologs with 69 to 81% similarity (Fig. 5). Functional analysis of pRSB105, a dual replicon isolated from activated sludge from a wastewater treatment plant (33), indicated that its rep1 locus provides the ability to replicate in gammaproteobacteria, whereas the rep2 region that is homologous to the Rms149 replicon extends plasmid replication to betaproteobacteria. No ability to replicate in alphaproteobacteria was observed. As incompatibility studies have been pursued only with Rms149, pFBAOT6, and RA3, it is difficult to define the level of replication protein homology sufficient to exert incompatibility among IncU plasmids. However, it is likely that some of these newly sequenced replicons will be classified in the IncU group. Interestingly, all IncU replication proteins have variable C termini in which an HTH motif has been identified for both pFBAOT6 and RA3 (Fig. 5). A role for the intragenic r3 repeat was postulated in the initiation of Rms149 replication (12). However, this repeat is not sufficient for origin activity for RA3 (Fig. 3B). As the host range of RA3 seems to be the broadest among the related replicons tested, it may suggest an important role for the extended repetitive regions surrounding the repA and repB genes that are characteristic only of RA3. In conclusion, a main branch of IncU plasmids possesses a highly conserved core backbone with variable antibiotic resistance cassettes inserted at a common location. The widespread presence of the repB-based replication systems in mosaic plasmids of diverse origins suggests that this replication module is more common than previously realized.
Supplementary Material
Acknowledgments
This project has been partially funded by grant PBZ-MNiSW-04/I/2007 (G.J.-B.). Work in the laboratory of F.H. is supported by grants from the Medical Research Council and The Wellcome Trust.
Footnotes
Published ahead of print on 23 May 2008.
REFERENCES
- 1.Adamczyk, M., P. Dołowy, M. Jonczyk, C. M. Thomas, and G. Jagura-Burdzy. 2006. The kfrA gene is the first in a tricistronic operon required for survival of IncP-1 plasmid R751. Microbiology 152:1621-1637. [DOI] [PubMed] [Google Scholar]
- 2.Adams, C. A., B. Austin, P. G. Meaden, and D. McIntosh. 1998. Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida. Appl. Environ. Microbiol. 64:4194-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki, T., S. Egusa, T. Kimura, and T. Watanabe. 1971. Detection of R factors in naturally occurring Aeromonas salmonicida strains. Appl. Microbiol. 22:716-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki, T., T. Kitao, N. Iemura, Y. Mitoma, and T. Nomura. 1983. The susceptibility of Aeromonas salmonicida strains isolated in cultured and wild salmonids to various chemotherapeutants. Bull. Jpn. Soc. Sci. Fish. 49:17-22. [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, D. E., T. Aoki, T. Kitao, T. Arai, and H. Tschape. 1982. Specification of characteristics for the classification of plasmids in incompatibility group U. Plasmid 8:89-93. [DOI] [PubMed] [Google Scholar]
- 7.El-Sayed, A. K., J. Hothersall, and C. M. Thomas. 2001. Quorum-sensing-dependent regulation of biosynthesis of the polyketide antibiotic mupirocin in Pseudomonas fluorescens NCIMB 10586. Microbiology 147:2127-2139. [DOI] [PubMed] [Google Scholar]
- 8.Garnier, T., and S. T. Cole. 1988. Complete nucleotide sequence and genetic organization of the bacteriocinogenic plasmid, pIP404, from Clostridium perfringens. Plasmid 19:134-150. [DOI] [PubMed] [Google Scholar]
- 9.Gołebiewski, M., I. Kern-Zdanowicz, M. Zienkiewicz, M. Adamczyk, J. Zylinska, A. Baraniak, M. Gniadkowski, J. Bardowski, and P. Cegłowski. 2007. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum β-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51:3789-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoriev, A. 1998. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26:2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haines, A. S., K. Jones, M. Cheung, and C. M. Thomas. 2005. The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J. Bacteriol. 187:4728-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haines, A. S., M. Cheung, and C. M. Thomas. 2006. Evidence that IncG (IncP-6) and IncU plasmids form a single incompatibility group. Plasmid 55:210-215. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 14.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 15.Hedges, R. W., and N. Datta. 1971. fi− R factors giving chloramphenicol resistance. Nature 234:220-221. [Google Scholar]
- 16.Jagura-Burdzy, G., and C. M. Thomas. 1992. kfrA gene of broad host range plasmid RK2 encodes a novel DNA-binding protein. J. Mol. Biol. 225:651-660. [DOI] [PubMed] [Google Scholar]
- 17.Jagura-Burdzy, G., J. P. Ibbotson, and C. M. Thomas. 1991. The korF region of broad-host-range plasmid RK2 encodes two polypeptides with transcriptional repressor activity. J. Bacteriol. 173:826-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahn, M. R., R. Kolter, C. M. Thomas, D. Figurski, R. Meyer, E. Remault, and D. R. Helinski. 1979. Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol. 68:268-280. [DOI] [PubMed] [Google Scholar]
- 19.Koonin, E. V. 1993. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 229:1165-1174. [DOI] [PubMed] [Google Scholar]
- 20.Larsen, M. H., and D. H. Figurski. 1994. Structure, expression, and regulation of the kilC operon of promiscuous IncPα plasmids. J. Bacteriol. 176:5022-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 22.Marques, M. V., A. M. da Silva, and S. L. Gomes. 2001. Genetic organization of plasmid pXF51 from the plant pathogen Xylella fastidiosa. Plasmid 45:184-199. [DOI] [PubMed] [Google Scholar]
- 23.McKenney, K., H. Shimatake, D. Court, U. Schmeissner, C. Brady, and M. Rosenberg. 1981. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif. Anal. 2:383-415. [PubMed] [Google Scholar]
- 24.Mullis, K., F. Faloona, S. Scharf, R. Saiki, G. Horn, and H. Erlich. 1986. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp. Quant. Biol. 51:263-273. [DOI] [PubMed] [Google Scholar]
- 25.Pansegrau, W., E. Lanka, P. Barth, D. H. Figurski, D. G. Guiney, D. Haas, R. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncPα plasmids: compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 26.Rhodes, G., G. Huys, J. Swings, P. McGann, M. Hiney, P. Smith, and R. W. Pickup. 2000. Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant TetA. Appl. Environ. Microbiol. 66:3883-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhodes, G., J. Parkhill, C. Bird, K. Ambrose, M. C. Jones, G. Huys, J. Swings, and R. W. Pickup. 2004. Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl. Environ. Microbiol. 70:7497-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saeki, K., J. Hitomi, M. Okuda, Y. Hatada, Y. Kageyama, M. Takaiwa, H. Kubota, H. Hagihara, T. Kobayashi, S. Kawai, and S. Ito. 2002. A novel species of alkaliphilic Bacillus that produces an oxidatively stable alkaline serine protease. Extremophiles 6:65-72. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, NY.
- 30.Sandaa, R. A., and Ø. Enger. 1994. Transfer in marine sediments of the naturally occurring plasmid pRAS1 encoding multiple antibiotic resistance. Appl. Environ. Microbiol. 60:4234-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandaa, R. A., and Ø. Enger. 1996. High frequency transfer of a broad host range plasmid present in an atypical strain of the fish pathogen Aeromonas salmonicida. Dis. Aquat. Org. 24:71-75. [Google Scholar]
- 32.Schaefer, C., and W. Messer. 1991. DnaA protein/DNA interaction. Modulation of the recognition sequence. Mol. Gen. Genet. 226:34-40. [DOI] [PubMed] [Google Scholar]
- 33.Schlüter, A., R. Szczepanowski, N. Kurz, S. Schneiker, I. Krahn, and A. Pühler. 2007. Erythromycin resistance-conferring plasmid pRSB105, isolated from a sewage treatment plant, harbors a new macrolide resistance determinant, an integron-containing Tn402-like element, and a large region of unknown function. Appl. Environ. Microbiol. 73:1952-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneiker, S., M. Keller, M. Droge, E. Lanka, A. Puhler, and W. Selbitschka. 2001. The genetic organization and evolution of the broad host range mercury resistance plasmid pSB102 isolated from a microbial population residing in the rhizosphere of alfalfa. Nucleic Acids Res. 29:5169-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sørum, H., T. M. L'Abee-Lund, A. Solberg, and A. Wold. 2003. Integron containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 47:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spratt, B. G., P. J. Hedge, S. de Heesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, EMBL8 and EMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]
- 37.Tauch, A., S. Schneiker, W. Selbitschka, A. Puhler, L. S. van Overbeek, K. Smalla, C. M. Thomas, M. J. Bailey, L. J. Forney, A. Weightman, P. Ceglowski, T. Pembroke, E. Tietze, G. Schroder, E. Lanka, and J. D. van Elsas. 2002. The complete nucleotide sequence and environmental distribution of cryptic, conjugative, broad-host-range plasmid pIPO2 isolated from bacteria of wheat rhizosphere. Microbiology 148:1637-1653. [DOI] [PubMed] [Google Scholar]
- 38.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 39.Thorsted, P. B., D. S. Shah, D. Macartney, K. Kostelidou, and C. M. Thomas. 1996. Conservation of the genetic switch between replication and transfer genes of IncP plasmids but divergence of the replication functions which are major host-range determinants. Plasmid 36:95-111. [DOI] [PubMed] [Google Scholar]
- 40.Tian, Q. B., M. Ohnishi, A. Tabuchi, and Y. Terawaki. 1996. A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 220:280-284. [DOI] [PubMed] [Google Scholar]
- 41.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tschäpe, H., E. Tietze, and C. Koch. 1981. Characterization of conjugative R plasmids belonging to the new incompatibility group IncU. J. Gen. Microbiol. 127:155-160. [DOI] [PubMed] [Google Scholar]
- 43.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 44.Zukowski, M. M., D. F. Gaffney, D. Speck, M. Kauffmann, A. Findeli, A. Wisecup, and J. P. Lecocq. 1983. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc. Natl. Acad. Sci. USA 80:1101-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.