Abstract
The hitherto unstudied microbial degradation of the organic disulfide 3,3′-dithiodipropionic acid (DTDP) was investigated with the recently described bacterium Tetrathiobacter mimigardefordensis strain DPN7T (DSM 17166T; LMG 22922T), which is able to use DTDP as the sole carbon source for growth. 3-Mercaptopropionic acid (3MP) and 3-sulfinopropionic acid (3SP) were detected in the growth medium and occurred as intermediates during DTDP degradation. To identify genes coding for enzymes of DTDP catabolism, Tn5::mob-induced mutants of T. mimigardefordensis were generated. Screening of transposon mutant libraries yielded many mutants fully or partially impaired in utilizing DTDP as a carbon source. Mapping of the insertion loci in some mutants identified four disrupted open reading frames (ORFs) with putative metabolic functions. The ORFs were assigned function on the basis of homologies with lpdA (EC 1.8.1.4), cdo (EC 1.13.11.20), sucCD (EC 6.2.1.5), and acnB (EC 4.2.1.3). Tn5::mob insertions occurred additionally in the vicinity of heat shock protein-encoding genes. The predicted function of the LpdA homologue in T. mimigardefordensis is cleavage of the disulfide bond of DTDP to form two molecules of 3MP. Cdo catalyzes the conversion of the sulfhydryl group of 3MP, yielding the corresponding sulfinic acid, 3SP. SucCD exhibits thiokinase activity, ligating coenzyme A (CoA) with 3SP to form 3SP-CoA. Afterwards, an elimination of sulfite via a putative desulfinase is expected. acnB encodes a putative 2-methylisocitrate dehydratase. Therefore, a new pathway is proposed for the catabolism of DTDP via 3MP, 3SP, and 3SP-CoA toward propionyl-CoA, which is then further catabolized via the 2-methylcitric acid cycle in T. mimigardefordensis.
The biotechnological relevance of 3,3′-dithiodipropionic acid (DTDP) is its application as a precursor substrate for microbially synthesized polythioesters (PTEs) (36). Furthermore, this organic sulfur compound (OSC) is employed in electrochemical and thermodynamic studies (49), for development of secondary batteries (58), in amino acid analysis (59), and for construction of self-assembly monolayers (15). The chemical structure of DTDP is very similar to that of the disulfide amino acid cystine. The absence of amino groups in DTDP is the only difference, yielding a higher melting point of cystine (247 to 249°C) than of DTDP (152 to 157°C). The occurrence of DTDP in natural habitats has not been described, to our knowledge, although this OSC may well be formed, because it is the disulfide of two molecules of 3-mercaptopropionic acid (3MP) and an oxidative disulfide formation is not unlikely. 3MP, along with cysteine and glutathione, belongs to the most frequently detected thiols in natural aquatic environments (1, 69). However, it occurs only in nanomolar concentrations (24). 3MP was found as a central intermediate during catabolism of the marine alga osmolyte dimethylsulfoniopropionate (11, 29, 57, 60, 61, 64) and also in freshwater habitats, as an anaerobic degradation product of sulfur-containing organic compounds, such as homocysteine and methionine (31, 43). Furthermore, it is generated by abiotic reactions of dissolved sulfide with dissolved organic matter in hypolimnetic waters (24). Therefore, 3MP is not a xenobiotic, though it is chemically produced at the scale of several thousand tons for applications as a bisphenol A cocatalyst. Moreover, it is used as a convulsant for studies of experimental epilepsy (16, 53) and for gold nanoparticle arrays to form three-dimensional monolayers (68). During application as a precursor substrate in PTE production, the microbial utilization of 3MP has been demonstrated clearly (35, 37, 38, 55). However, nothing is known about the pathway and the enzymes involved in the catabolism of 3MP or of its dimer, DTDP. Obviously, none of the PTE-producing microorganisms and only a few characterized strains utilize these two compounds as sole sources of carbon and energy (66). This is presumably due to the toxicity of these OSCs or of their intermediates during degradation (47).
Most information regarding the catabolism of naturally occurring OSCs is available for cysteine and methionine and also for dimethyl sulfoxide, dimethylsulfoniopropionate, and dimethylsulfide as intermediates of the sulfur cycle (28, 30, 33, 67). In addition, biodesulfurization of benzothiophenes (41) and of the fluorinated OSC bis-(3-pentafluorophenylpropyl)-sulfide (62) has been studied in detail. However, these OSCs are not related structurally to DTDP or 3MP.
It is presently not possible to synthesize PTEs from simple carbon sources and inorganic sulfur sources. To establish microbial synthesis independently of toxic and very expensive OSCs, the metabolism of a suitable microorganism has to be engineered. A first step in this direction was to obtain bacteria able to utilize one or more of the precursors as a carbon source. Because symmetric cleavage of DTDP via a putative disulfide reductase will yield two molecules of 3MP (36) and because DTDP is far less toxic to bacteria than 3MP is, we investigated the catabolism of DTDP. A proposal for the degradation pathway of this OSC in Tetrathiobacter mimigardefordensis strain DPN7T is presented in this report.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were cultivated aerobically in Luria-Bertani (LB) medium (48) at 37°C with the addition of applicable antibiotics, if necessary. E. coli XL1-Blue and the vector pBluescript SK (−) were used for DNA cloning and construction of genetic libraries. E. coli S17-1 harboring the suicide plasmid pSUP5011 was used for Tn5::mob mutagenesis of Tetrathiobacter mimigardefordensis strain DPN7. This betaproteobacterium was isolated from a matured compost heap and was found to grow on DTDP as the sole carbon source. Strain DPN7 was later characterized in more detail and allocated as the type strain in the new taxon T. mimigardefordensis (66). Deposition was accomplished at the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSM 17166T) and at the Laboratorium Microbiologie Universiteit Gent (LMG 22922T). Cells of T. mimigardefordensis were grown aerobically at 30°C in 0.8% (wt/vol) nutrient broth or in mineral salts medium (MSM) (50) containing the carbon source indicated in the text. Carbon sources were prepared as filter-sterilized 20% (wt/vol) stock solutions and adjusted to pH 7.0. Solid media contained 1.8% (wt/vol) purified agar-agar. Antibiotics were added to growth media at the following concentrations: ampicillin (Ap), 75 μg/ml; and kanamycin (Km), 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| S17-1 | thi-1 proA hsdR17 (rK− mK+) recA1; tra genes of plasmid RP4 integrated into the genome | 52 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 λ−lac [F′ proAB lacIqlacZΔM15 Tn10 (Tcr)] | 10 |
| Tetrathiobacter mimigardefordensis (DSM 17166T; LMG 22922T) | Wild type, DTDP-degrading organism | 66 |
| Plasmids | ||
| pSUP5011 | Apr Cmr Kmr Tn5::mob | 51 |
| pBluescript SK (−) | AprlacPOZ′ | Stratagene, San Diego, CA |
For abbreviations used in genotypes of E. coli, see reference 6.
Chemicals and synthesis of 3-sulfinopropionic acid.
Sulfur-containing substrates were purchased from Acros Organics (Geel, Belgium). Since 3-sulfinopropionate (3SP) could not be purchased, it was synthesized as a disodium salt according to the methods described by Jollés-Bergeret (26); the described procedure was slightly modified by one repetition of the alkaline cleavage of the intermediate bis-(2-carboxyethyl)sulfone. Starting from 111 g sodium formaldehyde sulfoxylate (purity, >98%) plus 108 ml acrylic acid (99.5%), 119 g of the intermediate bis-(2-carboxyethyl)sulfone was chemically synthesized. After alkaline scission, precipitation, and washing procedures, 99 g of the disodium salt of 3SP, with a purity of about 90%, was finally obtained. The success of synthesis and purity of the synthesized compound were confirmed by high-performance liquid chromatography (HPLC) and gas chromatography-mass spectrometry (GC-MS) analyses.
Tn5 mutagenesis.
For transposon mutagenesis of T. mimigardefordensis strain DPN7, the suicide plasmid technique described previously (52) was employed by transferring the vector pSUP5011 from E. coli S17-1 to the Km-susceptible strain DPN7 by conjugation, using the spot agar mating technique (21). Tn5-induced mutants were selected on MSM agar plates containing 50 μg Km ml−1 (MSMKm) and 0.5% (wt/vol) sodium gluconate, 0.2% (wt/vol) sodium propionate, or 0.2% (wt/vol) sulfinopropionate (master plates). Putative mutants were transferred in a coordinated pattern to MSMKm agar plates containing 0.4% (wt/vol) DTDP (selection plates) and to corresponding master plates for further analysis.
Isolation and manipulation of DNA.
Chromosomal DNAs of the Tn5-induced mutants of T. mimigardefordensis strain DPN7 and of the wild type were isolated as described by Marmur (39). Plasmid DNA was isolated by the method of Birnboim and Doly (7). Restriction enzymes and ligases were used according to the instructions of the manufacturers.
Transfer of DNA.
Competent cells of E. coli were prepared by the CaCl2 procedure and transformed with genomic DNA of Tn5-induced mutants of T. mimigardefordensis strain DPN7 ligated into plasmid pBluescript SK (−) (23).
Genotypic characterization of Tn5-induced mutants of T. mimigardefordensis strain DPN7.
Genomic DNAs of Tn5-induced mutants were digested with BamHI or SalI, and the resulting fragments were ligated into plasmid pBluescript SK (−). Recombinant E. coli clones were selected due to their Km resistance conferred by Tn5. The hybrid plasmids of the resulting clones harbored a BamHI or a SalI restriction fragment, respectively, which included the region of Tn5 located between the mob site and the IS50L element (including the Km resistance gene) plus genomic DNA adjacent to the Tn5 insertion locus. These recombinant plasmids were sequenced using an oligonucleotide (5′-GTTAGGAGGTCACATGG-3′) which hybridized specifically to the IS50L element of Tn5::mob and the oligonucleotide universal primers M13-forward (5′-GTAAAACGACGGCCAGT-3′) and M13-reverse (5′-CAGGAAACAGCTATGAC-3′).
PCR.
Amplifications of plasmid or genomic DNA were done as described previously (25). Inverse PCR (44) and direct genome walking using PCR (42) were done as specified to receive more information about the downstream and upstream regions of Tn5 insertion loci.
DNA-DNA hybridization.
Southern hybridizations to confirm Tn5 insertion were done by the method of Oelmüller et al. (45).
DNA sequencing and sequence analysis.
DNA sequencing was performed by applying a SequiTherm long-read cycle sequencing kit (Epicenter Technologies, WI) and IRD800-labeled oligonucleotides (MWG-Biotech, Ebersberg, Germany). Sequence reactions were accomplished by using a GeneReadIR 4200 DNA analyzer (LI-COR Inc., Biotechnology Division). Sequences were analyzed using the program BLAST (National Centre for Biotechnology Information [http://www.ncbi.nlm.nih.gov/BLAST/]) by searching the protein database using the translated nucleotide query (BlastX) (2, 3).
GC-MS analyses.
The compositions of the cell-free supernatants of cultures and of the obligate standards of the important OSCs were determined upon methanolysis after lyophilization in the presence of 15% (vol/vol) sulfuric acid (H2SO4) by GC analysis of the resulting methyl esters as described previously (8, 56).
HPLC analyses.
HPLC analysis was carried out with a LaChrom Elite HPLC apparatus (VWR-Hitachi International GmbH, Darmstadt, Germany) consisting of a Metacarb 67H advanced C column (Varian, Palo Alto, CA; Bio-Rad Aminex equivalent) and a 22350 VWR-Hitachi column oven. The column (300 mm by 6.5 mm) consisted of sulfonated polystyrene resin in the protonated form. The primary separation mechanism included ligand exchange, ion exclusion, and adsorption. A VWR-Hitachi refractive index detector (type 2490) with an active flow cell temperature control and automated reference flushing eliminating temperature effects on the refractive index baseline was used for detection. Aliquots of 20 μl were injected and eluted with 0.005 N sulfuric acid in double-distilled water at a flow rate of 0.8 ml/min. Online integration and analysis were done with EZ Chrome Elite software (VWR International GmbH, Darmstadt, Germany).
Nucleotide sequence accession numbers.
The complete DNA sequences and deduced amino acid sequences for lpdA (accession number EU423868), ddiox (accession number EU423869), and sucCD (accession number EU423870) and the partial DNA sequence for acnB (accession number EU423871) of T. mimigardefordensis strain DPN7T have been deposited in the GenBank database.
RESULTS
Biodegradation of DTDP by T. mimigardefordensis strain DPN7.
DTDP is utilized by T. mimigardefordensis strain DPN7 as the sole source of carbon and energy. Growth was initially observed on solid agar plates and then confirmed in liquid MSM, where an increase of turbidity was accompanied by a concomitant decrease of DTDP (Fig. 1). Controls (cultures without inoculum or without a carbon source) did not show any decrease of the DTDP concentration or increase of the optical density, respectively. A concentration of 60 μmol DTDP per mg protein was metabolized by cells of T. mimigardefordensis strain DPN7T. GC-MS analysis of the lyophilized cell-free supernatants identified, in addition to DTDP, two most likely intermediates of DTDP degradation (Fig. 2; see below), as they were absent in the sterile control.
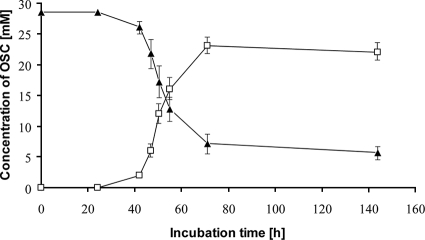
FIG. 1.
Degradation of DTDP by T. mimigardefordensis strain DPN7T and occurrence of 3MP. Cells were cultivated in a 250-ml Erlenmeyer flask without baffles on a rotary shaker at 30°C and 120 rpm in MSM containing 0.6% (wt/vol) or 28.6 mM DTDP as the sole carbon source. Analyses of DTDP and 3MP in the cell-free supernatants were done by GC. Data points presented are mean values for four replicates; error bars indicate the standard deviations. Symbols: ▴, DTDP; □, 3MP.
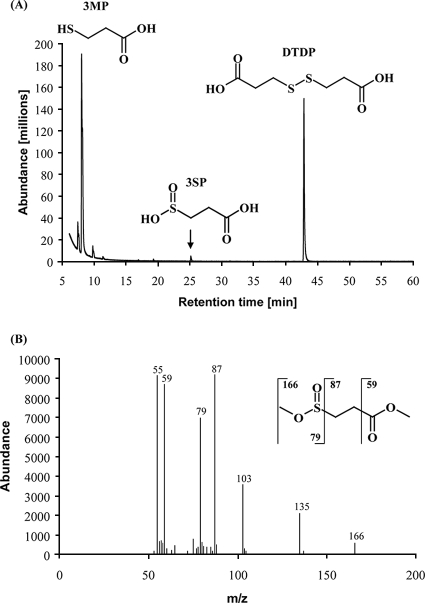
FIG. 2.
GC-MS analysis of DTDP and of intermediates occurring during degradation. Cultivation was accomplished under the conditions described in the legend to Fig. 1. (A) Besides DTDP, the intermediates 3MP and 3SP were separated in the chromatogram and identified. (B) Electron ionization mass spectrum of methylated 3SP as an example. Main fragmentations are indicated.
Identification of DTDP degradation metabolites in cell-free supernatants of cultures from T. mimigardefordensis strain DPN7T.
Degradation products of DTDP could be identified as 3MP and 3SP by GC-MS analysis when the data were compared with information provided by the National Institute of Standards and Technology library (Fig. 2). During cultivation of the cells in MSM at 30°C and at 120 rpm, with an incubation time of more than 20 hours, cleavage of DTDP resulted in a decrease of the DTDP concentration and an increase of the 3MP concentration in the supernatant, as revealed by GC analysis (Fig. 1). This was observed although 3MP was also spontaneously converted to DTDP, as revealed in control experiments using 3MP as the sole OSC in MSM. 3SP, in contrast, was detectable in the exponential growth phase; however, it occurred only at low concentrations in cultures of the wild type (Fig. 2).
Isolation and phenotypic characterization of auspicious Tn5::mob-induced mutants.
Transposon mutagenesis of T. mimigardefordensis strain DPN7 employing Tn5::mob was performed to generate mutants for investigation of DTDP catabolism and to identify genes coding for the involved enzymes in this bacterium. Insertion of Tn5::mob into the genomes of these mutants was confirmed by Southern hybridization using ApaI-digested genomic DNAs isolated from the mutants and the digoxigenin-labeled Km resistance gene of Tn5 as a probe. Extensive physiological characterizations preceded the mapping of the Tn5 insertion loci into the genome of each mutant relevant for this study. The genotypic characteristics of these mutants are summarized in Table 2. Eleven mutants (Jhw51c, Jhw90, Jhw101, KK14, KK15, Jhw17, Jhw103, JhwA8/121, Jhw13b, JhwI, and JhwV) exhibited fully impaired growth on MSM containing DTDP as the sole carbon source and were referred to as DTDP-negative mutants. Five mutants (Jhw38, JhwAA14, KK13, JhwIX, and JhwX) exhibited slower growth in the same medium and were therefore referred to as DTDP-leaky mutants. Most mutants grew like the wild type in MSM containing the following carbon sources at appropriate concentrations: acetate, gluconate, glucose, propionate, succinate, 3SP, sulfinoalanine, or taurine. Two exceptions were mutants JhwA8/121 and Jhw38, which exhibited diminished growth with 3SP (3SP-negative and -leaky, respectively). Mutant JhwA8/121 also showed partially impaired growth with sulfinoalanine as the sole carbon and energy source.
TABLE 2.
Genotypic characterization of Tn5::mob-induced mutants of T. mimigardefordensis defective in the utilization of DTDP
| Mutant | Phenotype regarding DTDP | Insertion locus of Tn5::mob (gene product)a | % Amino acid identity (strain) |
|---|---|---|---|
| Jhw51c | Negative | lpdA (dihydrolipoamide dehydrogenase) | 79 (Bordetella pertussis TohamaI) |
| Jhw90 | Negative | lpdA (dihydrolipoamide dehydrogenase) | 79 (Bordetella pertussis TohamaI) |
| Jhw101 | Negative | lpdA (dihydrolipoamide dehydrogenase) | 79 (Bordetella pertussis TohamaI) |
| KK14 | Negative | cdo (cysteine dioxygenase type I) | 65 (Verminephrobacter eiseniae EF01) |
| KK15 | Negative | cdo (cysteine dioxygenase type I) | 65 (Verminephrobacter eiseniae EF01) |
| Jhw17 | Negative | cdo (cysteine dioxygenase type I) | 65 (Verminephrobacter eiseniae EF01) |
| Jhw103 | Negative | cdo (cysteine dioxygenase type I) | 65 (Verminephrobacter eiseniae EF01) |
| JhwA8/121 | Negative | sucC (succinyl-CoA synthetase, beta chain) | 93 (Bordetella pertussis TohamaI) |
| Jhw38 | Leaky | sucC* (succinyl-CoA synthetase, beta chain) | 93 (Bordetella pertussis TohamaI) |
| JhwAA14 | Leaky | acnB (putative bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase) | 86 (Burkholderia sp. strain 383) |
| Jhw13b | Negative | acnB (putative bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase) | 86 (Burkholderia sp. strain 383) |
| KK13 | Leaky | lonA (ATP-dependent Lon protease) | 39 (Bacillus clausii KSM-K16) |
| JhwI | Negative | MOSC domain* (sulfur carrier) | 44(Bordetella bronchiseptica RB50) |
| JhwV | Negative | dnaK (chaperone protein) | 88 (Bordetella bronchiseptica RB50) |
| JhwIX | Leaky | dnaJ (molecular chaperone) | 80 (Bordetella avium 197N) |
| JhwX | Leaky | dnaJ (molecular chaperone) | 80 (Bordetella avium 197N) |
*, closest specified gene identified adjacent to the respective Tn5::mob insertion locus.
Genotypic characterization of Tn5::mob-induced mutants.
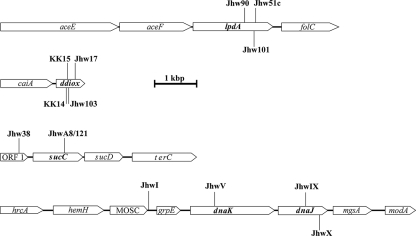
Total genomic DNA was isolated from all 16 transposon-induced DTDP-negative and -leaky mutants. The genomic regions comprising the Tn5::mob insertions were cloned into E. coli by selecting clones conferring Km resistance mediated by the transposon. Subsequently, the sequences directly adjacent to the Tn5::mob insertions were obtained by employing oligonucleotides hybridizing to Tn5. These sequences were then extended toward the upstream and downstream regions of the transposon insertions as described in Materials and Methods. By this method, in several cases identical, overlapping sequences were obtained, thus indicating that some transposon insertions had occurred in closely adjacent regions. The results of these analyses are summarized and presented graphically in Fig. 3, which provides the array of open reading frames (ORFs), including the positions of 13 transposon insertions in four different genomic regions relevant for DTDP catabolism in T. mimigardefordensis strain DPN7.
FIG. 3.
Tn5::mob insertions in the genome of T. mimigardefordensis strain DPN7 and identification of genes adjacent to the insertion loci. The diagrams show the localization of Tn5::mob insertions in four regions of the T. mimigardefordensis strain DPN7 genome in the 13 independent mutants showing fully or partially impaired growth on DTDP. The positions of Tn5::mob insertions in the respective mutants (mutant codes are given in Table 2) are indicated. Lengths and directions of arrows showing the genes indicate the proportional lengths and directions of transcription, respectively, of the corresponding genes. Putative identities of the gene products suggested from the amino acid sequence identities to proteins in the GenBank database are as follows: aceE, pyruvate dehydrogenase; aceF, dihydrolipoamide acetyltransferase; lpdA, dihydrolipoamide dehydrogenase; folC, folylpolyglutamate synthase/dihydrofolate synthase; ddiox, thiol dioxygenase; caiA, acyl-CoA dehydrogenase; ORF1, no sequence similarities; sucC, succinyl-CoA synthetase, beta chain; sucD, succinyl-CoA synthetase, alpha chain; terC, tellurium resistance protein; hrcA, heat-inducible transcription repressor; hemH, ferrochelatase; MOSC, molybdenum cofactor sulfurase, C-terminal end; grpE, nucleotide exchange factor; dnaK, Hsp70-like chaperone; dnaJ, molecular chaperone; mgsA, methylglyoxal synthase; and modA, molybdate binding protein. Bar, 1,000 base pairs.
Mapping of the Tn5::mob insertion in three DTDP-negative mutants (Jhw51c, Jhw90, and Jhw101) revealed disruption of a dihydrolipoamide dehydrogenase gene (sequence similarity of 79% identical amino acids to the E3 component of the pyruvate dehydrogenase complex of Bordetella pertussis TohamaI [EC 1.8.1.4]). Dihydrolipoamide dehydrogenases are flavoproteins belonging to the family of disulfide-oxidoreductases (40). These enzymes consist of three domains, including a biotin/lipoyl attachment domain in the amino-terminal region; a pyridine nucleotide-disulfide oxidoreductase domain, which is actually a small NADH binding domain within a larger FAD binding domain (Rossmann fold superfamily); and a dimerization domain of the pyridine nucleotide-disulfide oxidoreductases, found in the carboxy-terminal region of the protein. The Tn5::mob insertions in the three mutants occurred distal to the NADH binding domain (mutant Jhw90) or between this domain and the dimerization domain (mutants Jhw101 and Jhw51c) (Fig. 3). Furthermore, these mutants grew normally on 3SP as the sole carbon and energy source.
In the genomes of four DTDP-negative mutants (KK14, KK15, Jhw17, and Jhw103), the transposon was localized in a 0.6-kbp ORF (Fig. 3) putatively coding for a type I cysteine dioxygenase (Cdo; EC 1.13.11.20). The translational product of this ORF showed 65% sequence similarity to the Cdo of Verminephrobacter eiseniae EF02. Those mutants were unable to utilize DTDP, but they grew on 3SP as the sole source of carbon and energy. Although they did not utilize DTDP, cleavage of this OSC was detected in cultivation experiments in MSM containing DTDP as a cosubstrate and gluconate or succinate as an alternative carbon source. In this experiment, accumulation of 3MP was detected, but at only slightly higher concentrations than those in the wild type. This is most likely due to the fact that under the cultivation conditions applied in this study, two molecules of 3MP are spontaneously oxidized to the dimer, thus yielding one molecule of DTDP. Therefore, accumulation of 3MP to high concentrations could not be detected in the supernatants. A negative effect on the utilization of cysteine or 3MP could not be demonstrated for any of these mutants because the wild type did not utilize either compound as the sole source of carbon and energy.
Amino acid sequences of the putative translational products deduced from the adjacent genomic DNA of the Tn5 insertion loci in the genomes of two mutants (JhwA8/121 and Jhw38) (Fig. 3) disclosed strong similarities to a gene coding for the β-chain of a succinyl-coenzyme A (succinyl-CoA) synthetase (EC 6.2.1.5). The direct vicinity of the Tn5::mob insertion in mutant Jhw38 revealed no sequence similarities; instead, the transposon insertion was mapped 298 base pairs upstream of sucC. To the corresponding protein of B. pertussis TohamaI, 93% amino acid identity was obtained. The mutants exhibited fully (JhwA8/121) or partially (Jhw38) impaired growth with DTDP as the sole carbon source and were also defective in the utilization of 3SP. If cultivated in MSM containing succinate or gluconate as a carbon source in addition to DTDP, mutant JhwA8/121 accumulated >5 mM 3SP in the supernatant, which is a significant amount in comparison to the wild type, which produces barely detectable amounts of this OSC (up to approximately 2 mM).
In one DTDP-negative (Jhw13b) and one DTDP-leaky (JhwAA14) mutant, Tn5::mob was mapped in a gene encoding a putative bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase (AcnB; EC 4.2.1.3). AcnB dehydrates 2-methylcitric acid to 2-methyl-cis-aconitic acid and subsequently hydrates it to 2-methylisocitric acid (5). Cultivation of mutant JhwAA14 in MSM containing 0.6% (wt/vol) succinate and 0.3% (wt/vol) DTDP revealed accumulation of 2-methylcitrate in the supernatant and therefore provided further evidence for an involvement of AcnB in DTDP catabolism.
Furthermore, for five mutants, Tn5::mob insertions were located in or adjacent to ORFs coding for various proteins involved in the heat shock response (32). In four of these mutants, the transposon was mapped in the chromosomal region between genes encoding the carboxy-terminal region of a molybdenum cofactor sulfurase (MOSC domain) (4) and a methylglyoxal synthase (mgsA) (Fig. 3). Two of those mutants exhibited a DTDP-negative phenotype (JhwI and JhwV). The Tn5 insertion in mutant JhwI was mapped in a noncoding region localized downstream of the MOSC-encoding gene and upstream of grpE, which encodes the nucleotide exchange factor GrpE (70). In mutant JhwV, Tn5 insertion occurred in the Hsp70-like chaperone-encoding gene dnaK. In two DTDP-leaky mutants (JhwIX and JhwX), insertions were located downstream of dnaK, inside dnaJ. Furthermore, in mutant KK13, the transposon disrupted an ORF coding for a putative LonA protein. The latter is an ATP-dependent protease (14) and is involved in intracellular protein degradation as a chaperone (22, 46).
DISCUSSION
Here we report on the first studies to unravel the biodegradation of DTDP by microorganisms. The gram-negative bacterium T. mimigardefordensis strain DPN7T was enriched for this purpose in a previous study (66) and was chosen for this investigation. Based on identified intermediates of degradation, in silico analyses of the Tn5 insertion loci, and the phenotypes of the mutants, we propose the pathway shown in Fig. 4.
FIG. 4.
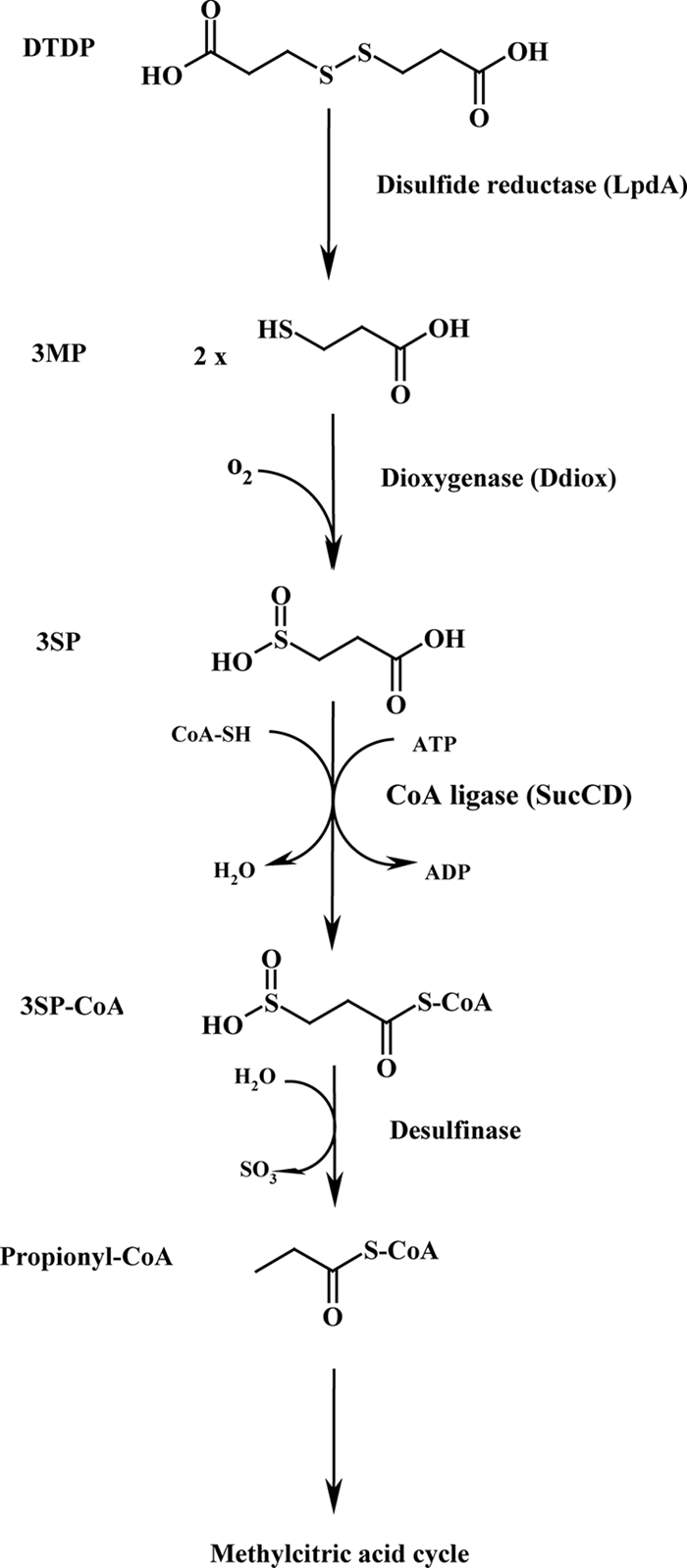
Putative pathway for degradation of DTDP in T. mimigardefordensis. Initially, DTDP is cleaved into two molecules of 3MP, which are then oxygenated by a dioxygenase, thereby yielding 3SP. After linkage to CoA by a thiokinase, the sulfur moiety is putatively removed by a desulfinase, resulting in propionyl-CoA, which is then metabolized via the methylcitric acid cycle toward intermediates of the central metabolism. Whereas most enzyme reactions are based on experimental data and on general predictions of the respective enzymes, the desulfination reaction is hypothetical and is based on theoretical considerations and indirect experimental evidence.
The initial step in the biodegradation of DTDP was predicted to yield two molecules of 3MP (36). Further evidence for this was obtained in this study. First, 3MP formation always accompanied degradation of DTDP (Fig. 1). Second, disruption of an lpdA homologue in three independent mutants (Table 2; Fig. 3) resulted in a DTDP-negative phenotype, whereas growth of these mutants on 3SP was not affected. Flavoprotein disulfide reductases exhibited high sequence and structural similarities. These enzymes catalyze the reduction of compounds which are linked by disulfide bonds (65). Therefore, the predicted function of the LpdA homologue in T. mimigardefordensis is the symmetric cleavage of DTDP into two molecules of 3MP (Fig. 4).
In four other independent mutants (Table 2), Tn5::mob was mapped in a cdo-homologous putative thiol dioxygenase-encoding gene (ddiox) (Fig. 3). Cdos from eukaryotes are well characterized (34), whereas Cdos were detected only recently in prokaryotes (19). The gene product of ddiox and homologous enzymes belong to the nonheme Fe2+-dioxygenases and the cupin superfamily. An occurrence of these enzymes was shown for mammalian cells, some yeast species, and a few bacteria (27). Already described thiol dioxygenases catalyze the irreversible oxidation of the OSC sulfhydryl group to a sulfinic acid, i.e., cysteine is oxidized to sulfinoalanine (19) (Fig. 5). Oxidation of cysteamine to hypotaurine by a Cdo homologue was also shown (12, 18), but only in eukaryotic cells until now. 3MP is a hitherto atypical substrate for a Cdo and was described only as an inhibitor of Cdo activity without the detection of any oxidation product of 3MP (13, 19). Furthermore, recent studies on the biodegradation of 3,3′-thiodipropionic acid in a different bacterium provide strong evidence for the involvement of a Cdo in the catabolism of 3MP (N. Bruland, J. H. Wübbeler, and A. Steinbüchel, unpublished data). Therefore, conversion of 3MP into 3SP as shown in Fig. 5 is most likely catalyzed by the Cdo-homologous enzyme in strain DPN7T.
FIG. 5.
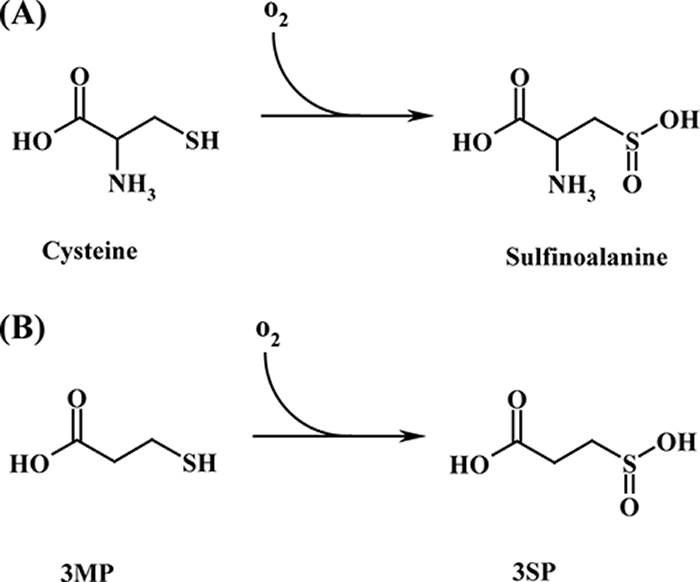
Reaction schemes of the enzymatic conversions of cysteine into sulfinoalanine by cysteine dioxygenases (EC 1.13.20.11) (A) and of 3MP into 3SP by the 3MP dioxygenase encoded by ddiox from T. mimigardefordensis (B).
The third proposed step in the catabolism of DTDP is the activation of the resulting 3SP and its ligation to CoA, yielding 3SP-CoA (Fig. 4). In gram-negative bacteria, the holoenzyme of SucCD is a tetramer consisting of two αβ dimers, which are encoded by sucC (β-chain) and sucD (α-chain) (9). The bacterial enzyme catalyzes the phosphorylation of ADP to ATP during aerobic metabolism in the citric acid cycle, coupled to cleavage of succinyl-CoA to succinate plus CoA. However, the thiokinase activity in the reverse direction is also relevant, e.g., in anabolism. Therefore, during catabolism of DTDP, the SucCD homologue of T. mimigardefordensis strain DPN7T should exhibit thiokinase activity. This is not unlikely, because 3SP is structurally analogous to succinate; the only difference in the structures between succinate and 3SP is the exchange of the carboxyl group with a sulfinic group.
Further degradation of the resulting 3SP-CoA and the fourth step of DTDP catabolism must be catalyzed by a desulfinase-like enzyme. This step should yield propionyl-CoA (Fig. 4) but is most unknown for T. mimigardefordensis because none of the DTDP-negative or -leaky mutants pointed to a gene encoding such an enzyme. The reasons for this could by manifold, and further investigations are necessary to identify the desulfinating enzyme. It is most certain that propionyl-CoA is actually being formed, because in two of the Tn5::mob-induced mutants the transposon was mapped in a gene coding for AcnB (Table 2). This aconitase is one of the typical enzymes of the 2-methylcitric acid cycle, which catalyzes in many bacteria the conversion of propionyl-CoA to pyruvate (54). Accumulation of significant amounts of 2-methylcitrate in the medium during cultivation of these mutants in MSM containing DTDP plus succinate confirms this conclusion. This also means that the fifth step of DTDP degradation is catalyzed by the 2-methylcitric acid cycle in T. mimigardefordensis.
It can be concluded that the organic sulfur compound DTDP is biodegradable and is used as the sole carbon and energy source for growth by a few soil microorganisms. It has to be investigated whether the utilization of this OSC depends on rare functions and low substrate specificities of the described well-known enzymes or whether these enzymes are specifically synthesized for degradation of DTDP. Detailed biochemical studies on the enzymes are currently being done. Unusual sulfur-containing metabolites occurred as intermediates during degradation. The potential inhibitory effects exerted by some of these compounds cause stress to the cells and probably require the presence of various heat shock proteins. Hints for involvement of these chaperones were obtained not only in this study (Table 2; Fig. 3) but also during identification of putative dimethylsulfoniopropionate degradation genes (11). Metabolites of 3MP were identified as potent inhibitors of β-oxidation (17, 47), but activation is important and necessary for the development of toxicity. In contrast to 3MP, 3SP is not known to occur naturally in the environment, and biochemical or microbial studies including this sulfinic acid are very rare. It was used as an analogous substance to succinate and sulfinoalanine in kinetic and substrate specificity studies of enzymes (20) and in studies about the radioprotective activity of various sulfinic acids, which revealed a certain radioprotective activity property but also toxicity of 3SP (63). Therefore, it is not surprising that chaperones are essentially required for the functionality of some enzymes catalyzing DTDP degradation.
Acknowledgments
We thank Heinrich Luftmann (Organisch-Chemisches Institut, Westfälische Wilhelms-Universität Münster) for analyzing the synthesized 3-sulfinopropionic acid.
Footnotes
Published ahead of print on 2 May 2008.
REFERENCES
- 1.Al-Farawati, R., and C. M. G. van den Berg. 2001. Thiols in coastal waters of the North Sea and English Channel. Environ. Sci. Technol. 35:1902-1911. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Ghish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Anatharam, V., and L. Aravind. 2002. MOSC domains: ancient, predicted sulfur-carrier domains, present in diverse metal-sulfur cluster biosynthesis proteins including molybdenum cofactor sulfurases. FEMS Microbiol. Lett. 207:55-61. [DOI] [PubMed] [Google Scholar]
- 5.Aoki, H., H. Uchiyama, H. Umetsu, and T. Tabuchi. 1995. Isolation of 2-methylisocitrate dehydratase, a new enzyme serving in the methylcitric acid cycle for propionate metabolism, from Yallowia lipolytica. Biosci. Biotechnol. Biochem. 59:1825-1828. [Google Scholar]
- 6.Bachmann, B. J. 1987. Linkage map of Escherichia coli K-12, p. 807-876. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 7th ed., vol. 2. American Society for Microbiology, Washington, DC. [Google Scholar]
- 7.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandl, H., A. Gross, R. W. Lenz, and R. C. Fuller. 1988. Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 54:1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buck, D., M. E. Spencer, and J. R. Guest. 1985. Primary structure of the succinyl-CoA synthetase of Escherichia coli. Biochemistry 24:6245-6252. [DOI] [PubMed] [Google Scholar]
- 10.Bullock, W. O., J. M. Fernandez, and J. M. Stuart. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 11.Bürgmann, H., E. C. Howard, W. Ye, F. Sun, S. Sun, S. Napierala, and M. A. Moran. 2007. Transcriptional response of Silicibacter pomeroyi DSS-3 to dimethylsulfoniopropionate (DMSP). Environ. Microbiol. 9:2742-2755. [DOI] [PubMed] [Google Scholar]
- 12.Cavallini, D., R. Scandurra, and C. De Marco. 1963. The enzymatic oxidation of cysteamine to hypotaurine in the presence of sulfide. J. Biol. Chem. 238:2999-3005. [PubMed] [Google Scholar]
- 13.Chai, S. C., A. A. Jerkins, J. J. Banik, I. Shalev, J. L. Pinkham, P. C. Uden, and M. J. Maroney. 2004. Heterologous expression, purification, and characterization of recombinant rat cysteine dioxygenase. J. Biol. Chem. 280:9865-9869. [DOI] [PubMed] [Google Scholar]
- 14.Charette, M. F., G. W. Henderson, and A. Markovitz. 1981. ATP hydrolysis-dependent protease activity of the Lon (CapR) protein of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 78:4728-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Codognoto, L., E. Winter, J. A. R. Paschoal, H. B. Suffredini, M. F. Cabral, S. A. S. Machado, and S. Rath. 2007. Electrochemical behavior of dopamine at 3,3′-dithiodipropionic acid self-assembled monolayers. Talanta 72:427-433. [DOI] [PubMed] [Google Scholar]
- 16.Crick, E. W., I. Osorio, N. C. Bhavaraju, T. H. Linz, and C. E. Lunte. 2007. An investigation into the pharmacokinetics of 3-mercaptopropionic acid and development of a steady-state chemical seizure model using in vivo microdialysis and electrophysiological monitoring. Epilepsy Res. 74:116-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuebas, D., J. D. Beckmann, F. E. Frerman, and H. Schulz. 1985. Mitochondrial metabolism of 3-mercaptopropionic acid. J. Biol. Chem. 260:7330-7336. [PubMed] [Google Scholar]
- 18.Dominy, J. E., Jr., C. R. Simmons, L. L. Hirschberger, J. Hwang, R. M. Coloso, and M. H. Stipanuk. 2007. Discovery and characterization of a second mammalian thiol dioxygenase, cysteamine dioxygenase. J. Biol. Chem. 282:25189-25198. [DOI] [PubMed] [Google Scholar]
- 19.Dominy, J. E., Jr., C. R. Simmons, P. A. Karplus, A. M. Gehring, and M. H. Stipanuk. 2006. Identification and characterization of a bacterial cysteine dioxygenase: a new route of cysteine degradation for eubacteria. J. Bacteriol. 188:5561-5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foote, J., A. M. Lauritzen, and W. N. Lipscomb. 1985. Substrate specificity of aspartate transcarbamylase. Interaction of the enzyme with analogs of aspartate and succinate. J. Biol. Chem. 260:9624-9629. [PubMed] [Google Scholar]
- 21.Friedrich, B., C. Hogrefe, and H. G. Schlegel. 1981. Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J. Bacteriol. 147:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goff, S. A., L. P. Casson, and A. L. Goldberg. 1984. Heat shock regulatory gene htpR influences rates of protein degradation and expression of the lon gene in Escherichia coli. Proc. Natl. Acad. Sci. USA 81:6647-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 136:557-580. [DOI] [PubMed] [Google Scholar]
- 24.Hu, H., G. Benoit, and S. E. Mylon. 2006. Distribution of the thiols glutathione and 3-mercaptopropionic acid in Connecticut lakes. Limnol. Oceanogr. 51:2763-2774. [Google Scholar]
- 25.Innis, M. A., D. H. Gelfaud, J. J. Suinsky, and T. J. White. 1990. PCR-protocol: a guide to methods and applications. Academic Press Inc., San Diego, CA.
- 26.Jollés-Bergeret, B. 1974. Enzymatic and chemical synthesis of 3-sulfinopropionic acid an analog of succinic acid. Eur. J. Biochem. 42:349-353. [DOI] [PubMed] [Google Scholar]
- 27.Joseph, C. A., and M. J. Maroney. 2007. Cysteine dioxygenase: structure and mechanism. Chem. Commun. 2007:3338-3349. [DOI] [PubMed] [Google Scholar]
- 28.Kertesz, M. A. 1999. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol. Rev. 24:135-175. [DOI] [PubMed] [Google Scholar]
- 29.Kiene, R. P., and B. F. Taylor. 1988. Biotransformation of organosulphur compounds in sediments via 3-mercaptopropionate. Nature 332:148-150. [Google Scholar]
- 30.Kiene, R. P., L. J. Linn, and J. A. Bruton. 2000. New and important roles for DMSP in marine microbial communities. J. Sea Res. 43:209-224. [Google Scholar]
- 31.Kiene, R. P., K. D. Malloy, and F. B. Taylor. 1990. Sulfur-containing amino acids as precursors of thiols in anoxic coastal sediments. Appl. Environ. Microbiol. 56:156-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindquist, S. 1986. The heat shock response. Annu. Rev. Biochem. 55:1151-1191. [DOI] [PubMed] [Google Scholar]
- 33.Lomans, B. P., C. van der Drift, and H. J. M. Op den Camp. 2002. Microbial cycling of volatile organic sulfur compounds. Cell. Mol. Life Sci. 59:575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lombardini, J. B., T. P. Singer, and P. D. Boyer. 1969. Cystein oxygenase. II. Studies on the mechanism of the reaction with 18oxygen. J. Biol. Chem. 244:1172-1175. [PubMed] [Google Scholar]
- 35.Lütke-Eversloh, T., A. Fischer, U. Remminghorst, J. Kawada, R. H. Marchessault, A. Bögershausen, M. Kalwei, H. Eckert, R. Reichelt, S.-J. Liu, and A. Steinbüchel. 2002. Biosynthesis of novel thermoplastic polythioesters by engineered Escherichia coli. Nat. Mater. 1:236-240. [DOI] [PubMed] [Google Scholar]
- 36.Lütke-Eversloh, T., and A. Steinbüchel. 2003. Novel precursor substrates for polythioesters (PTE) and limits of PTE biosynthesis in Ralstonia eutropha. FEMS Microbiol. Lett. 221:191-196. [DOI] [PubMed] [Google Scholar]
- 37.Lütke-Eversloh, T., J. Kawada, R. H. Marchessault, and A. Steinbüchel. 2002. Characterization of biological polythioesters: physical properties of novel copolymers synthesized by Ralstonia eutropha. Biomacromolecules 3:159-166. [DOI] [PubMed] [Google Scholar]
- 38.Lütke-Eversloh, T., K. Bergander, H. Luftmann, and A. Steinbüchel. 2001. Identification of a new class of biopolymer: bacterial synthesis of sulfur-containing polymer with thioester linkages. Microbiology 147:11-19. [DOI] [PubMed] [Google Scholar]
- 39.Marmur, J. 1961. A procedure for the isolation of desoxyribonucleic acid from microorganisms. J. Mol. Biol. 1:208-218. [Google Scholar]
- 40.Massey, V. 1963. Lipoyldehydrogenase, p. 275-306. In P. D. Boyer, H. Lardy, and K. Myrback (ed.), The enzymes, 2nd ed., vol. 7. Academic Press, New York, NY. [Google Scholar]
- 41.McFarland, B. L. 1999. Biodesulfurization. Curr. Opin. Microbiol. 2:257-264. [DOI] [PubMed] [Google Scholar]
- 42.Mishra, R. N., S. J. Singla-Pareek, S. Nair, S. K. Sopory, and M. K. Reddy. 2002. Directional genome walking using PCR. BioTechniques 33:830-834. [DOI] [PubMed] [Google Scholar]
- 43.Mopper, K., and B. F. Taylor. 1986. Biogeochemical cycling of sulfur-thiols in coastal marine sediments, p. 324-339. In M. Sohn (ed.), Organic marine geochemistry. American Chemical Society, Washington, DC.
- 44.Ochman, H., A. L. Gerber, and D. L. Hartl. 1988. Genetic application of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oelmüller, U., N. Krüger, A. Steinbüchel, and C. G. Friedrich. 1990. Isolation of prokaryotic RNA and detection of specific mRNA with biotinylated probes. J. Microbiol. Methods 11:73-84. [Google Scholar]
- 46.Phillips, T. A., R. A. VanBogelen, and F. C. Neidhardt. 1984. lon gene product of Escherichia coli is a heat shock protein. J. Bacteriol. 159:283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabbagh, E., D. Cuebas, and H. Schulz. 1985. 3-Mercaptopropionic acid, a potent inhibitor of fatty acid oxidation in rat heart mitochondria. J. Biol. Chem. 260:7337-7342. [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 49.Saxena, R. S., and A. Gupta. 1984. Electrochemical studies on the composition, stability-constants and thermodynamics of Ti (I) complexes with dithiodipropionic acid. Monatsschr. Chem. 115:1293-1298. [Google Scholar]
- 50.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Mikrobiol. 38:209-222.13747777 [Google Scholar]
- 51.Simon, R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-mob transposon. Mol. Gen. Genet. 196:413-420. [DOI] [PubMed] [Google Scholar]
- 52.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 53.Sprince, H., C. M. Parker, and G. G. Smith. 1970. 3-Mercaptopropionic acid: convulsant and lethal properties compared with other sulfur-convulsants; protection therefrom. Agents Actions 1:231-233. [DOI] [PubMed] [Google Scholar]
- 54.Textor, S., V. F. Wendisch, A. A. Graf, U. Müller, M. I. Linder, D. Linder, and W. Buckel. 1997. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch. Microbiol. 168:428-436. [DOI] [PubMed] [Google Scholar]
- 55.Thakor, N., T. Lütke-Eversloh, and A. Steinbüchel. 2005. Application of the BPEC pathway for large-scale biotechnological production of poly(3-mercaptopropionate) by recombinant Escherichia coli including a novel in situ isolation method. Appl. Environ. Microbiol. 71:835-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Timm, A., D. Byrom, and A. Steinbüchel. 1990. Formation of blends of various poly(3-hydroxyalkanoic acids) by a recombinant strain of Pseudomonas oleovorans. Appl. Microbiol. Biotechnol. 33:296-301. [Google Scholar]
- 57.Todd, J. D., R. Rogers, Y. G. Li, M. Wexler, P. L. Bond, L. Sun, A. R. J. Curson, G. Malin, M. Steinke, and A. W. B. Johnston. 2007. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315:666-669. [DOI] [PubMed] [Google Scholar]
- 58.Tsutsumi, H., S. Okada, and T. Oishi. 1998. A potentially biodegradable polyamide containing disulfide bonds as a positive material for secondary batteries. Electrochim. Acta 43:427-429. [Google Scholar]
- 59.Tuan, Y.-H., and R. D. Phillips. 1997. Optimized determination of cystine/cysteine and acid-stable amino acids from a single hydrolysate of casein- and sorghum-based diet and digesta samples. J. Agric. Food Chem. 45:3535-3540. [Google Scholar]
- 60.van der Maarel, M. J. E. C., and T. A. Hansen. 1996. Anaerobic microorganisms involved in the degradation of DMS(P), p. 351-360. In R. P. Kiene, P. T. Visscher, M. D. Keller, and G. O. Kirst (ed.), Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum Press, New York, NY.
- 61.van der Maarel, M. J. E. C., M. Jansen, and T. A. Hansen. 1995. Methanogenic conversion of 3-S-methylmercaptopropionate to 3-mercaptopropionate. Appl. Environ. Microbiol. 61:48-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Hamme, J. D., P. M. Fedorak, J. M. Foght, M. R. Gray, and H. D. Dettman. 2004. Use of a novel fluorinated organosulfur compound to isolate bacteria capable of carbon-sulfur bond cleavage. Appl. Environ. Microbiol. 70:1487-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vasil'eva, T. P., and V. V. Znamenskii. 1994. Synthesis and radioprotective properties of 2-arylethylammonium salts of sulfinic acids. Pharmaceut. Chem. J. 28:47-50. [Google Scholar]
- 64.Visscher, P. T., and B. F. Taylor. 1994. Demethylation of dimethylsulfoniopropionate to 3-mercaptopropionate by an aerobic marine bacterium. Appl. Environ. Microbiol. 60:4617-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams, C. H., Jr. 1992. Lipoamide dehydrogenase, glutathione reductase, thioredoxin reductase, and mercuric ion reductase—a family of flavoenzyme transhydrogenases, p. 121-211. In F. Muller (ed.), Chemistry and biochemistry of flavoenzymes, vol. III. CRC Press, Boca Raton, FL. [Google Scholar]
- 66.Wübbeler, J. H., T. Lütke-Eversloh, P. Vandamme, S. Van Trappen, and A. Steinbüchel. 2006. Tetrathiobacter mimigardefordensis sp. nov., isolated from compost, a betaproteobacterium capable of utilizing the organic disulfide 3,3′-dithiodipropionic acid. Int. J. Syst. Evol. Microbiol. 56:1305-1310. [DOI] [PubMed] [Google Scholar]
- 67.Yoch, D. C. 2002. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68:5804-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, J., and M. Oyama. 2007. Electrocatalytic activity of three-dimensional monolayer of 3-mercaptopropionic acid assembled on gold nanoparticle arrays. Electrochem. Commun. 9:459-464. [Google Scholar]
- 69.Zhang, J., F. Wang, J. D. House, and B. Page. 2004. Thiols in wetland interstitial waters and their role in mercury and methylmercury speciation. Limnol. Oceanogr. 49:2276-2286. [Google Scholar]
- 70.Ziemienowicz, A., D. Skowyra, J. Zeilstra-Ryalls, O. Fayet, C. Georgopoulos, and M. Zylicz. 1993. Both the Escherichia coli chaperone systems, GroEL/GroES and DnaK/DnaJ/GrpE, can reactivate heat-treated RNA polymerase. Different mechanisms for the same activity J. Biol. Chem. 268:25425-25431. [PubMed] [Google Scholar]