Abstract
Cellobiose utilization is a variable trait that is often used to differentiate members of the family Vibrionaceae. We investigated how Vibrio fischeri ES114 utilizes cellobiose and found a cluster of genes required for growth on this β-1,4-linked glucose disaccharide. This cluster includes genes annotated as a phosphotransferase system II (celA, celB, and celC), a glucokinase (celK), and a glucosidase (celG). Directly downstream of celCBGKA is celI, which encodes a LacI family regulator that represses cel transcription in the absence of cellobiose. When the celCBGKAI gene cluster was transferred to cellobiose-negative strains of Vibrio and Photobacterium, the cluster conferred the ability to utilize cellobiose. Genomic analyses of naturally cellobiose-positive Vibrio species revealed that V. salmonicida has a homolog of the celCBGKAI cluster, but V. vulnificus does not. Moreover, bioinformatic analyses revealed that CelG and CelK share the greatest homology with glucosidases and glucokinases in the phylum Firmicutes. These observations suggest that distinct genes for cellobiose utilization have been acquired by different lineages within the family Vibrionaceae. In addition, the loss of the celI regulator, but not the structural genes, attenuated the ability of V. fischeri to compete for colonization of its natural host, Euprymna scolopes, suggesting that repression of the cel gene cluster is important in this symbiosis. Finally, we show that the V. fischeri cellobioase (CelG) preferentially cleaves β-d-glucose linkages but also cleaves β-d-galactose-linked substrates such as 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal), a finding that has important implications for the use of lacZ as a marker or reporter gene in V. fischeri.
The family Vibrionaceae encompasses a diverse collection of marine bacteria that are studied both for their direct significance to humankind and as models for a variety of basic biological phenomena (54). For example, our research group and others study Vibrio fischeri as a model of bacterial bioluminescence and for its symbiotic host-bacteria interactions and pheromone-mediated gene regulation (4, 38, 53, 58, 68). Other Vibrio species are investigated because of their pathogenicity for humans or marine animals, their contributions to aquatic ecosystems and processes, or their genomic plasticity and remarkable adaptability (15, 18, 61, 63). Although there are exceptions, a hallmark of this family is the ability to grow rapidly in culture and to use a variety of nutrient sources. There is considerable interspecies variability in metabolic patterns, and such differences may reflect ecologically important traits for individual species. Nutrient utilization profiles have also been important in practice both for developing taxonomic schemes to identify Vibrio species and for growing and manipulating these bacteria in the laboratory.
Traditionally, carbon source utilization patterns have been among the criteria used to distinguish different species within the family Vibrionaceae (6, 18), and this information has also guided the development of semiselective media to enrich for specific species (14). For example, cellobiose utilization is among the variable traits used to describe and distinguish species within the family Vibrionaceae (6), and cellobiose-based media have been used to enrich for certain species, particularly Vibrio vulnificus (11, 33). Molecular markers and DNA sequence analyses are becoming more widespread taxonomic tools with great value, but carbon source utilization phenotypes are still useful discriminators and have been tested through decades of research (6, 18). In the future, once the genetic basis for the utilization of particular carbon sources is better understood, it should be possible to generate molecular DNA-based techniques that draw directly on the wealth of phenotypic information available for identifying Vibrio species.
Understanding the genetic basis for various metabolism patterns among different Vibrio species will also help elucidate the evolutionary history of the Vibrionaceae. Despite interest in both the metabolic variability of this family and the mechanisms underlying its evolution, much remains to be learned about these subjects. For example, it is not known whether cellobiose utilization was an ancestral trait lost by some members of this family or if it was a trait acquired by certain lineages more recently. Bioinformatic analyses of genome sequences in the family Vibrionaceae (12, 21, 32, 46, 52, 64) promise to help answer such questions, but gene and pathway annotations can be ambiguous or incorrect. Therefore, continued experimental determination of metabolic pathways will be necessary to connect genomic and phenotypic variability.
In this study, we describe a gene cluster that is both necessary for cellobiose utilization by V. fischeri and sufficient to confer cellobiose utilization on other Vibrio species. Based on our results and bioinformatic analyses, we propose a model for cellobiose utilization arising from the acquisition of distinct pathways by different lineages within the family Vibrionaceae. We also show that this cellobiose utilization cluster in V. fischeri is responsible for an unexpected cryptic β-galactosidase activity. This observation has immediate practical significance, because the β-galactosidase gene lacZ from Escherichia coli has been used as both a marker (17, 24) and a transcriptional reporter (30, 69, 72, 73) in V. fischeri.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains used in this study are described in Table 1. Plasmids were maintained in E. coli strain DH5α (20), except conjugative helper plasmid pEVS104 (57), which was maintained in CC118λpir (22); a pCR Blunt-TOPO-derivative, which was maintained in TOP-10 (Invitrogen, Carlsbad, CA); and other plasmids containing the R6Kγ replication origin, which were maintained in DH5αλpir (16). E. coli was incubated at 37°C in LB medium (34) or brain heart infusion (BHI) medium (Difco, Sparks, MD). For selection of E. coli, chloramphenicol, kanamycin, and trimethoprim were used at concentrations of 20, 40, and 10 μg ml−1, respectively. For the selection of E. coli with erythromycin (Em), 150 μg ml−1 was added to BHI medium. V. fischeri and all other Vibrionaceae strains were grown at 28°C in LBS medium (55) or by using a specific carbon source, as indicated, added to a minimal salts medium (0.340 mM NaPO4 [pH 7.5], 0.05 M Tris [pH 7.5], 0.3 M NaCl, 0.05 M MgSO4-7H2O, 0.01 M CaCl2-2H2O, 0.01 M NH4Cl, 0.01 M KCl, 0.01 mM FeSO4-7H2O, plus a carbon source). When chloramphenicol, kanamycin, Em, and trimethoprim were added to LBS or minimal medium for selection of V. fischeri or other Vibrionaceae strains, they were used at concentrations of 2, 100, 5, and 10 μg ml−1, respectively. d-Cellobiose (Acros Organics, Geel, Belgium) was added to solid and liquid media at concentrations of 5 mM and 10 mM, respectively. Glucose was added to media at a concentration of 20 mM. Bromocresol purple sodium salt (BCP; Eastman Kodak, Rochester, NY) and 5- bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal; Research Products International, Prospect, IL) were added to LBS medium at concentrations of 0.02 and 100 μg ml−1, respectively. Agar was added to a final concentration of 1.5% for solid media.
TABLE 1.
Bacterial strains, select plasmids, and oligonucleotides
| Bacteria, plasmid, or oligonucleotide | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| CC118-λpir | Δ(ara-leu) araD Δlac74 galE galK phoA20 thi-1 rpsE rpsB argE(Am) recA λpir | 22 |
| DH5α | F− Φ80dlacZΔ(lacZYA-argF) U169 deoR supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 20 |
| DH5α-λpir | DH5α lysogenized with λpir | 16 |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Str) endA1 nupG | Invitrogen |
| Vibrionaceae | ||
| ATCC 33653 | Vibrio mimicus | E. Lipp |
| ATCC 33564 | Vibrio hollisae | E. Lipp |
| ATCC 17803 | Vibrio parahaemolyticus | E. Lipp |
| DMA401 | ES114; celI::mini-Tn5-Em and 2-bp insertion in celG | This study |
| DMA420 | ES114; celI::mini-Tn5-Em (insertion at bp 914) | This study |
| DMA421 | ES114; celI::mini-Tn5-Em (insertion 53 bp before the predicted start of celI) | This study |
| DMA422 | ES114; celK::mini-Tn5-Em (insertion at bp 548) | This study |
| DMA423 | ES114; celA::mini-Tn5-Em (insertion at bp 20) | This study |
| DMA424 | ES114; ptsI::mini-Tn5-Em (insertion at bp 247) | This study |
| DMA425 | ES114; celG::mini-Tn5-Em (insertion at bp 502) | This study |
| DMA426 | ES114; celC::mini-Tn5-Em (insertion at bp 1166) | This study |
| DMA427 | ES114; celB::mini-Tn5-Em (insertion at bp 39) | This study |
| DMA428 | ES114; VF2408::mini-Tn5-Em (insertion at bp 614) and 11-bp deletion in celI (at bp 566) | This study |
| DMA429 | ES114; VF0170::mini-Tn5-Em (insertion at bp 314) and 11-bp deletion at in celI (at bp 566) | This study |
| ES114 | Wild-type isolate from E. scolopes | 7 |
| KNH6 | Photobacterium leiognathi | 57 |
| KV1319 | ES114; 2-bp insertion in celG | This study |
| KV2801 | ES114; ptsI::pTMO151 | 66 |
| VC4056 | Vibrio cholerae | R. Colwell |
| VC4103 | Vibrio cholerae | R. Colwell |
| Plasmids | ||
| pDMA171 | celI in pVSV107 | This study |
| pDMA193 | Pcel-gfp reporter in pVSV209 | This study |
| pEVS79 | oriVColE1oriTRP4 Cmr | 57 |
| pEVS170 | mini-Tn5-Em, oriVR6KγoriTRP4 Knr | N. Lyell |
| pKV151 | celCBGKAI plus 824 bp upstream of putative start in pVO8 | This study |
| pKV162 | 2-bp insertion in celG and flanking sequences in pEVS79; source of celG allele in KV1319 and DMA401 | This study |
| pVO8 | oriVP15AoriTRP4 Cmr EmrlacZα | 67 |
| pVSV107 | oriVR6KγoriTRP4oriVpES213 TprlacZα | 17 |
| pVSV209 | oriVR6KγoriTRP4oriVpES213rfp Knr promoterless Cmr-gfp | 17 |
| Oligonucleotides | ||
| dma91 | 5′CGGCGCTAGCGGTGCACGCCCAAGATCATATTATGAC 3′ | This study |
| dma92 | 5′CGCCGCTAGCCGCTGTAACAGCCAGAGCAACAGG 3′ | This study |
| dma93 | 5′GCCGCTAGCTGTGACTTCCTATATTTCAGCTTT 3′ | This study |
| dma94 | 5′GGCGCTAGCTGTTCACCCCTAATTAGAATTATAATTTA 3′ | This study |
Str, streptomycin resistance; Knr, kanamycin resistance; Emr, erythromycin resistance; Cmr, chloramphenicol resistance; Tpr, trimethoprim resistance.
DNA and plasmid manipulations.
Standard methods were used to generate plasmids and to clone DNA fragments. Plasmids were mobilized from E. coli into recipients by triparental mating, using pEVS104 as a conjugative helper, as described previously (57). Restriction enzymes, DNA ligase, and Klenow fragment were obtained from New England Biolabs (Ipswich, MA). Chromosomal DNA was purified using an Easy-DNA kit (Invitrogen, Carlsbad, CA). Plasmids were isolated using a QIAprep spin miniprep kit (Qiagen, Valencia, CA) or a GenElute plasmid miniprep kit (Sigma, St. Louis, MO). The Zero Blunt TOPO PCR cloning kit (Invitrogen, Carlsbad, CA) was used to clone PCR products into pCR-BluntII-TOPO. DNA fragments were purified using a DNA Clean-up & Concentrator-5 kit (Zymo Research, Orange, CA). PCR was performed using KOD HiFi DNA polymerase (Novagen, Madison, WI), following the manufacturer's recommendations for cycle programs based on predicted DNA product size. Oligonucleotides (Table 1) were obtained from Integrated DNA Technologies (Coralville, IA). The annealing temperature for primer pair dma91 and dma92 was 55°C, and for dma93 and dma94, it was 47°C. PCR was performed by using an iCycler unit (Bio-Rad Laboratories, Hercules, CA). DNA sequencing was conducted on an ABI automated DNA sequencer at the University of Michigan DNA sequencing core, and sequences were analyzed using Sequencher version 4.6 software (Gene Codes, Ann Arbor, MI).
Construction of mutants and complementation plasmids.
Descriptions of select plasmids and the primers used in their construction are provided in Table 1. Details of plasmid construction are as follows. To generate a 2-bp insertion in celG, we first screened an existing library of XbaI-digested ES114 DNA cloned into pBluescript (Stratagene, La Jolla, CA) to isolate pKV150, which contains the cel gene cluster. pKV150 was digested with BglII, and the cel gene cluster was subcloned into BamHI-digested pEVS79, yielding pKV153. pKV153 was digested with SpeI and self-ligated to roughly center celG within the insert, making pKV156. pKV156 was then digested with ClaI, the overhangs were filled in with Klenow fragment, and the plasmid was recircularized by self-ligation, yielding pKV162, which has a frame-shifting 2-bp insertion in celG. This mutation was placed on the chromosomes of ES114 and DMA420 by allelic exchange, yielding strains KV1319 and DMA401, respectively.
The celI complementation plasmid pDMA171 was generated by first amplifying celI with ∼500 bp of upstream sequence, thereby incorporating NheI sites that were engineered near the 5′ end of each primer (dma91 and dma92). This PCR product was cleaned and digested with NheI before being cloned directly into AvrII-digested pVSV107. Plasmid pKV151 contains the active cel cluster and was isolated from a library of BglII-digested ES114 DNA cloned into pV08 (2).
To construct pDMA193, the Pcel-gfp reporter, the intergenic DNA upstream of celC was PCR amplified with NheI sites incorporated near the 5′ ends of the primers (dma93 and dma94) and cloned into pCR-BluntII-TOPO (Invitrogen, Carlsbad, CA), yielding pDMA181. pDMA181 was digested with SpeI and XhoI, and the promoter fragment was subcloned into pEVS79, digested with the same enzymes, making pDMA184. pDMA184 was digested with NheI, and the promoter was subcloned into AvrII-digested pVSV209, which contains a promoterless gfp gene, completing the construction of pDMA193.
Transposon mutagenesis was performed by conjugating the mini-Tn5 delivery plasmid pEVS170 (N. Lyell and E. Stabb, unpublished results) into wild-type V. fischeri strain ES114. After conjugation proceeded for more than 8 h, the conjugation mixture was diluted and plated onto selective media. The mini-Tn5 mutagenesis was performed in three independent experiments for each screen, encompassing ∼10,000 colonies per screen. In one screen, mutant colonies were isolated based on their blue color on LBS-X-gal supplemented with Em. In the other screen, mutant colonies were examined for yellowish-white color on LBS medium supplemented with cellobiose, Em, and X-gal. The site of transposon insertion in each mutant strain was determined by cloning the transposon and flanking DNA and then sequencing across the transposon::chromosome junction, using the M13 Forward primer. Insertions were cloned by digesting chromosomal DNA with HhaI, self-ligating the fragments, and recovering the transposon and flanking DNA as a plasmid, taking advantage of the origin of replication and Em resistance gene contained within the transposon.
Carbon utilization assays.
The ability to grow on glucose or on cellobiose as the sole carbon source was tested by adding these sugars to a minimal medium and then streaking single colonies of each strain onto plates, which were incubated at 28°C for ∼48 h and assessed for growth. To test for acid production by strains in the presence of glucose or cellobiose, single colonies were used to inoculate test tubes containing LBS medium with BCP and either cellobiose, glucose, or no sugar added. Cultures were incubated at 28°C with shaking (200 rpm) for 24 h, and acidification was scored as a change in the BCP from purple to yellow.
cel induction measurements in culture.
Overnight cultures of V. fischeri carrying pDMA193(Pcel-gfp) were grown in LBS with appropriate antibiotics and diluted 1:500 into 30 ml of antibiotic-free LBS medium, with or without cellobiose or glucose, in 125-ml baffled flasks and were then incubated at 24°C with shaking (200 rpm). The reporter and control plasmids used are derived from a vector that is stable in V. fischeri and does not require selection for maintenance (17). Samples (500-μl) were removed at intervals and the culture optical density at 595 nm (OD595) was determined by using a BioPhotometer unit (Brinkman Instruments, Westbury, NY). Fluorescence was measured using a TD-700 fluorometer (Turner Designs, Sunnyvale, CA), using excitation and emission filters of 486 nm and >510 nm, respectively. The fluorescence reported is the average of measurements taken when the OD595 readings were approximately 2.5. The fluorescence of strains carrying the promoterless gfp construct in pVSV209 was subtracted as background.
To examine the ability of various carbon sources to induce the cel operon, 15 μl of 100 mM stocks of cellobiose, raffinose, sucrose, maltose, lactose, N-acetyl-glucosamine, fructose, mannose, ribose, galactose, xylose, arabinose, and glucose were spotted onto filter disks placed on LBS-X-gal plates and spread-plated with ES114. After 24 h of incubation, plates were examined for the induction of the cel operon, which was scored as rings of blue in the lawn surrounding the sugar-impregnated disk. Parallel plates with the celG mutant KV1319 served as negative controls and did not develop blue color.
Enzyme assays using pNP-conjugated substrates.
The strains tested were grown to an OD595 of ∼2.0, pelleted, and lysed by freezing at −80°C for 20 min, and the pellets were resuspended in the original volume of a 500 mM sodium phosphate buffer (pH 7.0). One hundred microliters of this lysate was added to 400 μl of a 10 mM p-nitrophenol (pNP)-conjugated substrate dissolved in 50 mM sodium phosphate buffer (pH 7.0). Parallel reaction mixtures were incubated at 28°C and 37°C until a yellow color was observed or for a maximum of 24 h. The assay was stopped by adding 2 ml of 1 M Na2CO3 (final concentration of 800 mM). A 1-ml sample from the reaction mixture was centrifuged for 5 min to pellet cell debris. The absorbance was read at 410 nm (A410) to determine the amount of pNP generated from enzymatic cleavage, and at A550 to determine light scattering from residual cell debris. To calculate pmol of pNP generated min−1 ml−1, the A410 reading from each sample minus the A550 reading for each sample was compared to a linear standard of pNP, and this was divided by the incubation time, and the 0.1 ml of lysate was added to the reaction mixture.
Squid colonization assays.
E. scolopes host animals were maintained in Instant Ocean (Aquarium Systems, Mantor, OH) mixed to ∼36 ppt. To determine whether a mutant strain had a competitive disadvantage in the symbiosis relative to the wild type, cultures for inoculation were grown as previously described (17), and juvenile squid were exposed to a ∼1:1 mixture of the wild-type and mutant strains for 14 h and then moved to V. fischeri-free Instant Ocean. Squid were homogenized after 48 h to determine the ratio of wild-type to mutant strains. The relative competitive index (RCI) was determined by dividing the mutant-to-wild type ratio for each individual squid by the ratio for the inoculum. Log-transformed data were used to calculate the average RCI and to determine statistical significance.
Bioinformatic analyses.
Protein sequence comparisons to GenBank entries were generated using BLASTp (3). V. salmonicida LFI1238 sequence was obtained from the Sanger Institute (http://www.sanger.ac.uk/Projects/V_salmonicida/) as a shotgun database, and homologs of specific V. fischeri genes were determined by using Artemis software (48). Genomes with similar regions surrounding the CelC open reading frame (ORF) were found by using the SEED pinned region search (40). The similarities reported between homologs were determined by MatGAT software using the default settings (10). Phylogenetic and molecular evolutionary analyses were conducted with MEGA software version 4.0, using the default settings (60). Using the MEGA program, consensus neighbor-joining phylogenetic trees were constructed by using the amino-Poisson correction. The unweighted-pair group method with arithmetic mean (UPGMA) and minimum evolution trees were also constructed with similar results (data not shown). Bootstrap values for the trees were obtained from a consensus tree based on 1,000 randomly generated trees, using MEGA 4.0 software (60).
RESULTS
Mutations in celI reveal cryptic β-galactosidase activity in V. fischeri.
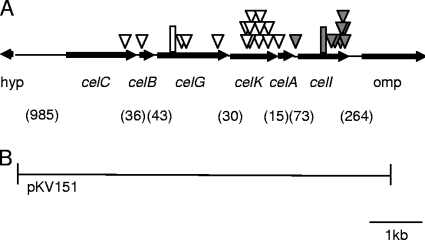
This study was initiated by the serendipitous observation that when V. fischeri ES114 was mutagenized with mini-Tn5 and plated on selective medium with X-gal, approximately 1 in 1,000 colonies was dark blue, suggesting that some mutations could reveal a cryptic β-galactosidase activity in this strain. We isolated 10 such blue mutants and determined the location of the transposon insertion in these strains. Eight of the mutants had insertions in or directly upstream of VF0608, a gene that encodes a putative LacI family transcriptional regulator. We have now designated VF0608 as celI. The other two blue mutants, DMA428 and DMA429, had insertions in VF2408, a putative LuxR family transcriptional regulator, and VF0170, a putative O antigen flippase. However, these mutants, like the other eight, could be complemented to the parental yellowish-white colony color on LBS medium supplemented with X-gal (LBS-X-gal) by the reintroduction of celI on pDMA171 but not by the introduction of the parent vector pVSV107. Subsequent analyses showed that DMA428 and DMA429 each also had an identical 11-bp deletion in celI, which could account for their blue phenotype on LBS-X-gal plates (Fig. 1).
FIG. 1.
Schematic representation of the genetic organization around the cel gene cluster in V. fischeri ES114. (A) Arrows represent ORFs and indicate the direction of gene transcription, as well as the gene size, which is presented relative to the scale bar. Shaded triangles represent transposon insertions that resulted in a blue colony phenotype on LBS-X-gal plates. Open triangles represent transposon insertions that resulted in a yellowish-white colony phenotype on LBS-X-gal cellobiose plates. A hypothetical gene is designated “hyp.” The shaded rectangle denotes the 11-bp deletion detected in mutants DMA428 and DMA429. The open rectangle denotes the 2-bp insertion in strain KV1319. Numbers in parentheses represent the base pairs between ORFs. (B) The region cloned into plasmid pKV151.
Identification of the cellobiose utilization gene cluster.
Analysis of the genome near celI revealed that it was downstream of a gene cluster annotated as functioning in disaccharide uptake and catabolism, including a putative glucosidase (cellobioase) gene (Fig. 1). We hypothesized that the glucosidase gene encoded the enzyme responsible for the cleavage of X-gal and that celI repressed this operon in the absence of cellobiose. Consistent with this hypothesis, when wild-type ES114 was plated on LBS-X-gal and 5 mM cellobiose, all the colonies were blue. We then repeated the mini-Tn5 mutagenesis but plated the mutants on LBS-X-gal and 5 mM cellobiose and screened for the loss of blue colony color, predicting that the inability to cleave X-gal would correlate with the loss of the ability to grow on cellobiose as the sole carbon source. After more than 10,000 colonies were screened, 16 transposon mutants were isolated based on their yellowish-white color on LBS-X-gal cellobiose plates. All but one insertion was localized to the predicted cellobiose utilization gene cluster upstream of celI (Fig. 1), and all of the mutants with insertions in this cluster grew poorly or did not grow with cellobiose as the sole carbon source. Moreover, in BCP assays, none of these mutants acidified the medium in the presence of cellobiose as the wild type did (Table 2). Each of the mutants with an insertion in the genes upstream of celI could be complemented to a wild-type-like phenotype upon reintroduction of this gene cluster on pKV151 (Fig. 1) but not by introduction of the parent vector pVO8. In light of these data, the genes in the cluster were named based on their predicted function: phosphotransferase system (PTS) IIA component, celA (VF0607); PTS IIB component, celB (VF0604); PTS IIC component, celC (VF0603); 6-phospho-β-glucosidase, celG (VF0605); glucokinase, celK (VF0606) and, as mentioned above, LacI-like family transcriptional regulator, celI (VF0608) (Fig. 1).
TABLE 2.
Growth and acid production of Vibrio strains on glucose and cellobiose
| Strain | Growth on minimal medium plus cellobiose
|
Acidification of LBS-BCP medium in the presence of:
|
||||
|---|---|---|---|---|---|---|
| Glucose
|
Cellobiose
|
|||||
| pVO8 | pKV151 | pVO8 | pKV151 | pVO8 | pKV151 | |
| ES114 | + | + | + | + | + | + |
| KV1319 | − | + | + | + | − | + |
| Photobacterium leiognathi | − | + | + | + | − | − |
| Vibrio cholerae VC4103 | − | + | + | + | − | + |
| Vibrio cholerae VC4056 | − | + | + | + | + | + |
| Vibrio parahaemolyticus | − | + | + | + | − | + |
| Vibrio hollisae | − | + | + | + | − | − |
| Vibrio mimicus | − | + | + | + | − | + |
The single mutant with an insertion outside this gene cluster, DMA424, had an insertion in VF1896, which is annotated as a PTS system enzyme. Visick et al. (66) previously characterized a mutation in this gene and determined that the VF1895 and VF1896 ORFs were actually one gene, an ortholog to E. coli ptsI. Similarly to the ptsI mutant isolated by Visick et al. (66), we found that our ptsI mutant grew poorly in minimal medium with cellobiose or with glucose as the sole carbon source and had a slower growth rate than the wild type in LBS medium (data not shown). The ptsI gene encodes the E1 component of the PTS system, one of two proteins with essential roles as general components for all PTS systems. Because there is no other E1 encoded in V. fischeri ES114, the PTS II system comprised by CelA, CelB, and CelC should be severely attenuated or nonfunctional in the DMA424 mutant.
celG encodes a β-glucosidase with lower β-galactosidase activity.
To investigate whether the putative 6-phospho-β-glucosidase encoded by celG was responsible for the cleavage of cellobiose and X-gal, we generated mutant KV1319, which contains a 2-bp insertion in celG. KV1319 was unable to cleave X-gal and unable to utilize cellobiose as a sole carbon source and did not acidify cellobiose-containing media in BCP assays. This provides further evidence that CelG is responsible for the cleavage of both cellobiose and X-gal, a supposition that is also supported by the enzymatic assays described below. To test the prediction that the celI mutant colony's blue phenotype on LBS-X-gal was due to the loss of CelI-mediated repression of celG, we incorporated the 2-bp frameshifting mutation in celG into DMA420, a celI transposon mutant. As predicted, the resulting strain (DMA401 celG; mutant, celI::mini-Tn5-Em) frameshift was yellowish-white in contrast to the blue color of the celI mutant on LBS-X-gal plates (data not shown).
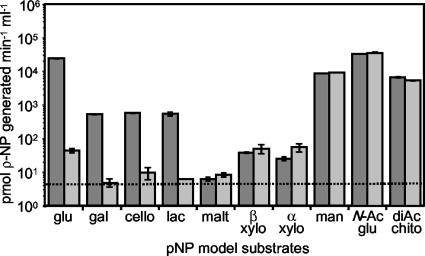
To further test the substrates targeted by CelG, we assayed enzymatic activity in cell lysates by using sugar substrates para-linked to a nitrophenol group. We examined lysates of the celI mutant DMA420, which should enhance CelG activity by allowing the derepression of celG. To determine whether the utilization of a particular substrate was specific to CelG and not to some other enzyme in the whole-cell lysate, we compared the activity of celI mutant lysates (Fig. 2, dark gray bars) to that of lysates of the celI celG double mutant, DMA401 (Fig. 2, light gray bars). Thus, Fig. 2 shows both CelG-dependent activity, which is the difference between the activities in strains DMA420 and DMA401, and CelG-independent activity, which is the activity in strain DMA401.
FIG. 2.
celG-dependent enzymatic activity. Activities in lysates from DMA420 (celI::mini-Tn5-Em) are shown as dark gray bars, and activities from DMA401 lysates (celG frameshift, celI::mini-Tn5-Em) are shown as light gray bars. Activities represent the ability to cleave pNP from the substrate. Error bars, some too small to visualize, indicate standard error (n = 3). The dashed line represents the limit of detection. Results shown are representative of two experiments at 37°C and are also similar to data obtained from two experiments at 28°C. Substrate abbreviations: glu, pNP-β-d-glucopyranoside (model substrate for cellobiose); gal, pNP-β-d-galactopyranoside; cello, pNP-β-d-cellobioside; lac, pNP-β-d-lactopyranoside; malt, pNP-β-d-maltoside; β xylo, pNP-β-d-xylopyranoside; α xylo, pNP-α-d-xylopyranoside; man, pNP-β-d-mannopyranoside; N-Ac glu, pNP-N-acetyl-β-d-glucosaminide; and diAC chito, pNP-N,N′-diacetyl-β-d-chitobioside.
Figure 2 shows that the celG mutant loses the ability to cleave the model substrate for cellobiose cleavage, pNPβ-d-glucopyranoside (Fig. 2, glu). Lower but significant CelG-dependent activity levels were observed with pNP-β-d-galactopyranoside (Fig. 2, gal), pNPβ-d-cellobioside (Fig. 2, cello), and pNP-β-d-lactopyranoside (Fig. 2, lac). We also observed modest (0.25%) residual activity toward pNP-β-d-glucopyranoside (Fig. 2, glu) even in the celG mutant DMA401, which could indicate that the frameshift in celG does not completely eliminate CelG function or that other enzymes in the lysate catalyze a relatively minor amount of hydrolysis of the substrate. To differentiate between these possibilities, we tested the celG::mini-Tn5-Em mutant, DMA425, and found it had activity toward pNP-β-d-glucopyranoside (Fig. 2, glu) similar to that of the celG frameshift mutant, supporting the idea that the residual activity is due to an unidentified enzyme rather than partial activity from the frameshift allele (data not shown).
Although each of the other substrates cleaved by CelG contains a β-1,4 linkage, the promiscuity of the enzyme was evident in its activity as both a β-glucosidase and a β-galactosidase. The latter may reflect a coincidental and physiologically irrelevant activity. Although CelG is apparently able to cleave pNP-β-d-lactopyranoside (Fig. 2, lac), V. fischeri is unable to utilize lactose as a carbon source. Moreover, the data below indicate that CelG is induced by the presence of cellobiose but not by lactose (Fig. 3). Given the ability of CelG to direct X-gal cleavage, the β-galactosidase activity attributed to CelG in this assay was expected; however, it is worth noting that this activity was ∼50-fold lower than the β-glucosidase activity (Fig. 2).
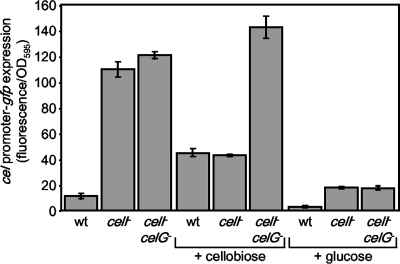
FIG. 3.
Specific fluorescence generated from the Pcel-gfp reporter in ES114 (wild type [wt]), the mutant DMA420 (celI mutant), or DMA401 (celI celG double mutant). Cultures of strains carrying pDMA193 were grown in LBS with and without 10 mM cellobiose or 20 mM glucose. Data represent the average specific fluorescence with standard error (n = 3).
The assays with other pNP-linked substrates indicate that enzymes other than CelG in the lysates direct cleavage of pNP-β-d-mannopyranoside (Fig. 2, man), pNP-N-acetyl-β-d-glucosaminide (Fig. 2, N-Ac glu), pNP-N,N′-diacetyl-β-d-chitobioside (Fig. 2, diAC chito), pNP-β-d-xylopyranoside (Fig. 2, β xylo), and pNP-α-d-xylopyranoside (Fig. 2, α xylo). We saw no enzymatic activity toward pNP-β-d-maltoside (Fig. 2, malt).
celI-cellobiose- and glucose-mediated control of cel expression.
To determine when the cel genes are induced, we developed a transcriptional reporter plasmid, pDMA193, containing the region immediately upstream of the cel gene cluster, driving the expression of gfp. Without cellobiose, the reporter's fluorescence in ES114 was slightly elevated above that of the background, but with growth in cellobiose, there was an increase of almost 5-fold in fluorescence (Fig. 3). In the celI::mini-Tn5-Em mutant DMA420, the reporter was expressed with and without added cellobiose, further supporting our prediction that CelI represses the cel gene cluster when cellobiose is not present.
Interestingly, in the presence of cellobiose, fluorescence of the reporter in the celI mutant DMA420 decreases compared to when it is growing without cellobiose. We hypothesized that the generation of glucose due to the cleavage of cellobiose might cause catabolite repression of the gene cluster, resulting in this inhibitory effect of cellobiose on cel expression. To examine this, both the wild type ES114 and the celI mutant DMA420 containing the reporter plasmid were grown in the presence of glucose. In both strains, fluorescence from the reporter plasmid decreased substantially when cells were grown in LBS medium supplemented with glucose (Fig. 3). Furthermore, we moved the reporter plasmid into DMA401, the celI celG double mutant, to assess whether the loss of CelG, which should reduce the breakdown of cellobiose to glucose, would allow induction of the gene cluster in the presence of cellobiose. In media with glucose, reporter expression in DMA401 was reduced, as it was in the other strains (Fig. 3). However, supplementation with cellobiose did not affect reporter expression in DMA401 (Fig. 3). Thus, it appears that cel expression is repressed by glucose that is either added exogenously or generated by CelG-dependent cleavage of cellobiose.
Taking advantage of the promiscuity of CelG and its ability to degrade X-gal, we next determined whether carbon sources other than cellobiose could induce the expression of the cel gene cluster, using a disc assay with ES114 or the celG mutant KV1319 plated on LBS-X-gal plates. We tested glucose, galactose, lactose, chitin-hexamers, cellulose, N-acetyl-glucosamine, and cellobiose and found that only cellobiose was able to induce the expression of the cel gene cluster, resulting in CelG-dependent cleavage of X-gal and development of blue color in the growth of ES114 cells around the disc (data not shown). Thus, even if CelG is able to cleave substrates other than cellobiose (as described above), the substrates above do not induce celG and are therefore unlikely to be physiologically relevant targets for the cel gene cluster.
The celCBGKAI gene cluster on pKV151 confers cellobiose utilization on six cellobiose-negative Vibrio strains.
To determine whether this cellobiose-utilizing gene cluster in V. fischeri was sufficient to confer cellobiose utilization to other Vibrio strains, pKV151 (Fig. 1) was moved into six different Vibrio or Photobacterium strains that are negative for cellobiose utilization (Table 2). Transconjugants were tested for growth on solid medium with cellobiose as the sole carbon source and were also tested for growth in the presence of glucose (the breakdown product of cellobiose) as a control. Both ES114 and the celG frameshift mutant were also included as positive and negative controls, respectively. All of the strains grew regardless of the plasmid when glucose was the sole carbon source (data not shown); however, of the strains carrying the insertless vector pVO8, only ES114 grew on cellobiose (Table 2). Thus, for each of the other Vibrio or Photobacterium strains, the V. fischeri cel gene cluster on pKV151 conferred the ability to grow on cellobiose (Table 2).
Substrate utilization is often tested indirectly based on the production of fermentation acids in the presence of a particular sugar, resulting in a pH shift that can be detected by the colorimetric change in the dye BCP. We therefore also tested strains carrying pKV151 in BCP assays. All strains with either the control vector or pKV151 acidified glucose-containing LBS medium (Table 2). For Vibrio cholerae VC4103, V. mimicus, and V. parahaemolyticus, pKV151 conferred not only the ability to grow on cellobiose but also the production of acid in LBS supplemented with cellobiose. For the other strains, the ability to grow on cellobiose did not correlate with acid production in the presence of cellobiose. Photobacterium leiognathi and V. hollisae were unable to acidify the cellobiose-containing medium regardless of whether they carried the control vector or pKV151, whereas V. cholerae VC4056 acidified the medium regardless of whether it contained pKV151 or the control vector. Thus, although acid production is often used as an indirect indicator of sugar catabolism by Vibrio species (6, 18), direct testing for growth on cellobiose was a more reliable measure of this metabolic capability.
Bioinformatic analyses of the cel gene cluster.
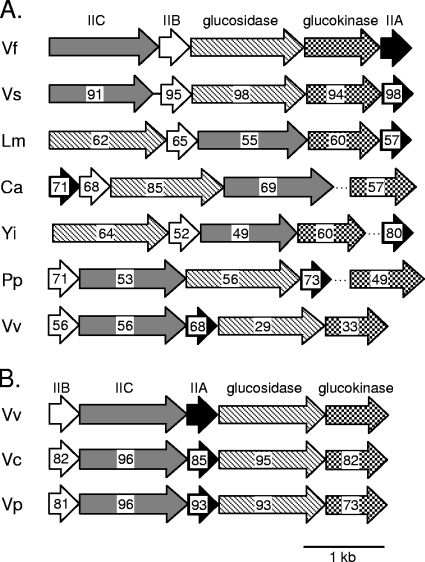
Using a combination of bioinformatic programs and databases (see Materials and Methods), we sought to determine whether celCBGKAI was an ancestral locus present in all cellobiose-utilizing members of the family Vibrionaceae and, if this was not the case, to determine the likely origin(s) of these genes. Comparisons of nucleotide or encoded protein sequences yielded similar results, and for the most part, only the latter are reported here, with nucleotide sequences used for reporting gene arrangement. We found that V. salmonicida strain LFI1238, which utilizes cellobiose (N.-P. Willassen, personal communication), has a homologous celCBGKAI cluster. The cel gene order is conserved in V. salmonicida, the encoded proteins were >90% similar to the respective homologs in V. fischeri, and the V. salmonicida cel gene cluster included both the genes for cellobiose utilization (Fig. 4A) and the regulator celI (not shown).
FIG. 4.
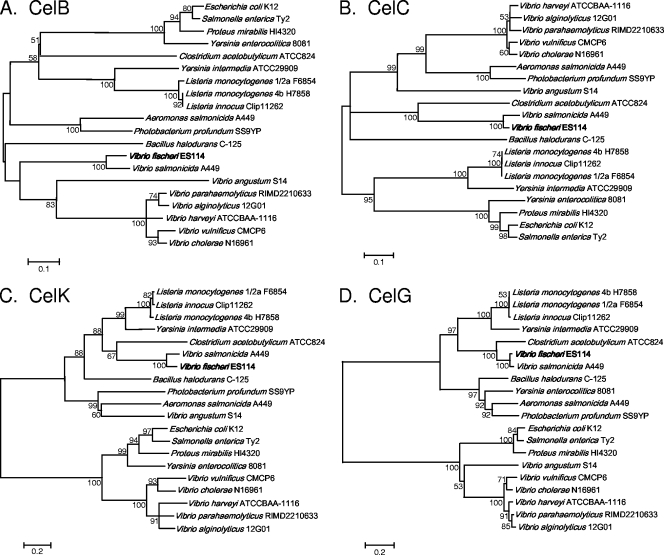
Homologs of the V. fischeri cellobiose-utilizing genes. Gene arrangement and orientation for each particular strain are indicated with arrows. Arrows with the same shading or pattern are putative homologs. Genes encoding PTS components A, B, and C are shaded black, white, or gray, respectively. Genes encoding glucokinases or glucosidases are filled with checkerboard or crosshatch patterns, respectively. Dotted lines indicate that glucokinase or PTS IIA genes in these bacteria are not genetically linked to the other genes shown. Numbers within the arrows represent the overall similarity to the respective protein from V. fischeri (panel A) or V. vulnificus (panel B) as determined by MatGAT software (10). Species abbreviations are: Vf, Vibrio fischeri; Vs, Vibrio salmonicida; Lm, Listeria monocytogenes; Ca, Clostridium acetobutylicum; Yi, Yersinia intermedia; Pp, Photobacterium profundum; Vv, Vibrio vulnificus; Vc, Vibrio cholerae; Vp, Vibrio parahaemolyticus.
Photobacterium profundum SS9 and V. vulnificus, which are cellobiose-positive bacteria (18, 37), also had gene clusters that included PTS II system genes, a glucosidase gene, and, in the latter bacterium, a glucokinase gene as well; however, neither gene had impressive similarity to the V. fischeri cel cluster. For example, the putative glucosidase and glucokinase encoded by this V. vulnificus cluster shared only 29% and 33% similarity to CelG and CelK, respectively (Fig. 4A). Although V. vulnificus is able to grow on cellobiose, we speculate that this might not be the gene cluster responsible for cellobiose catabolism. In support of this idea, V. cholerae and V. parahaemolyticus are both cellobiose-negative bacteria, yet both have a gene cluster that is highly homologous to that described above in V. vulnificus (Fig. 4B). Partial genomic sequences are available for other cellobiose-utilizing Vibrio species, but additional clusters similar to celCBGKAI were not found. Indeed, homologs of the individual V. fischeri CelG and CelK proteins were notably absent. Taken together, our analyses suggest that celCBGKAI underpins cellobiose utilization in the V. fischeri/V. salmonicida clade but that distinct pathways may direct cellobiose catabolism in other members of the family Vibrionaceae.
Despite further analyses, the ancestry of the celCBGKAI genes in V. fischeri and V. salmonicida remains uncertain, and these genes may have multiple origins. It seems likely that celI, which encodes the LacI family regulator, originated within the family Vibrionaceae, as it shares high similarity with many regulators in this bacterial family (data not shown). Interestingly, however, CelG and CelK clustered most closely with ORFs found in the phylum Firmicutes, particularly in the Clostridium, Bacillus, and Listeria species. This is illustrated both in the comparisons of similar gene clusters (Fig. 4) and in the neighbor-joining trees that compare the specific proteins encoded by these gene clusters (Fig. 5). Gene clusters in Clostridium acetobutylicum and Listeria monocytogenes were more similar to the V. fischeri cel cluster than were the gene clusters from other Vibrio species (Fig. 4A). The closest homolog of CelG was found in C. acetobutylicum (Fig. 4A and 5D), while close homologs to CelK were found in L. monocytogenes and Yersinia intermedia (Fig. 4A and 5C). Although Y. intermedia is a gammaproteobacterium, it appeared to be exceptional in this group in that the proteins encoded by the Y. intermedia cluster group more closely with homologs in Listeria than with proteins in other proteobacteria. The gene cluster in L. monocytogenes is also similar to the V. fischeri cel cluster, and the gene order is conserved with that in V. fischeri, except that the PTS IIC and glucosidase genes are switched (Fig. 4A). Moreover, codon usage by Listeria is so similar to that in V. fischeri that genes transferred between the two cannot be recognized as foreign by using this criterion (data not shown). The origin of the celA, celB, and celC genes is especially difficult to infer, as the PTS IIC component tends to group with homologs in the phylum Firmicutes (Fig. 5B), whereas the PTS IIB and IIA components tend to group more closely with homologs within the family Vibrionaceae (Fig. 5A and data not shown). The PTS IIB and IIA components must interface with other endogenous proteins, and the respective genes might be expected to face greater selective pressure to adapt to a new host and therefore appear less foreign. Overall, it seems plausible that at least some of the genes for cellobiose uptake and catabolism may have been transferred horizontally from a marine Firmicutes organism to an ancestor of the V. fischeri/V. salmonicida clade, although other scenarios remain possible.
FIG. 5.
Comparison of the V. fischeri CelB, CelC, CelK, and CelG proteins to proteins encoded by other bacteria with similar gene clusters. Consensus neighbor-joining trees constructed using MEGA 4.0 software (60) are shown. Trees constructed using both UPGMA and minimum evolution algorithms were similar to those shown in the figure (data not shown). Bootstrap values of >50% are indicated at the respective nodes. The scale bar represents a corrected sequence divergence of 0.1 or 0.2 as indicated. Trees in panels A, B, C, and D include V. fischeri proteins CelB, CelC, CelK, and CelG, respectively.
Symbiotic colonization of E. scolopes.
Our laboratories and others study the symbiotic interaction between V. fischeri ES114 and the Hawaiian bobtail squid E. scolopes, and we therefore investigated the symbiotic phenotype of the cel mutants. If cellobiose utilization contributed to colonization proficiency, this would provide insight into the symbiotic nutritional environment of the host light organ. On the other hand, if mutants had no symbiotic attenuation, then mutant cel alleles could be useful as neutral markers, with a readily scored phenotype on X-gal.
Mutants with insertions in each of the cel genes were checked for their ability to compete with the wild type for colonization in E. scolopes. Mutants with an insertion in celI or in the noncoding region upstream of celI were consistently outcompeted by ∼2.2-fold as indicated by RCI values of ∼0.45 (Table 3). In contrast, strains with mutations in genes responsible for the transport and degradation of cellobiose, celA, celB, celC, celG, and celK, had no significant competitive defect relative to that of the wild type (Table 3). We competed DMA401 (celG mutant, celI::mini-Tn5-Em) to see if the competitive defect of a mutation in celI was dependent on the overexpression of a functional cel gene cluster and cellobioase activity. This competition yielded an RCI that was essentially the same as the single celI mutant alone, indicating that the negative effect of knocking out celI on symbiotic colonization is independent of cellobioase activity. Instead, this attenuation of competitiveness may simply be from the overexpression of the Cel proteins. Not surprisingly, the ptsI mutant DMA424 was outcompeted by the wild type; however, this mutant's slower growth in culture indicates that its defect in colonizing the host cannot be considered symbiosis specific.
TABLE 3.
Colonization competitiveness of V. fischeri cel mutants relative to that of ES114
| Strain | Genotype | No. of animalsa | RCIb |
|---|---|---|---|
| DMA420 | celI::mini-Tn5-Em | 42 | 0.40c |
| 41 | 0.48c | ||
| 44 | 0.40c | ||
| DMA421 | celI upstream region::Tn5 | 44 | 0.44c |
| DMA401 | celI::mini-Tn5-Em, celG mutant (frameshift) | 32 | 0.39c |
| DMA425 | celG::Tn5-Em | 32 | 0.90 |
| KV1319 | celG mutant (frameshift) | 32 | 1.21 |
| DMA422 | celK::mini-Tn5-Em | 26 | 0.68 |
| DMA423 | celA::mini-Tn5-Em | 29 | 0.95 |
| DMA427 | celB::mini-Tn5-Em | 32 | 1.04 |
| DMA426 | celC::mini-Tn5-Em | 32 | 0.91 |
| DMA424 | ptsI::mini-Tn5-Em | 32 | 0.02c |
Number of animals used in an individual experiment.
The RCI was derived by the ratio of the mutant strain to the wild type in the light organ at 48 h postinoculation divided by the ratio in the inoculum.
The mutant was significantly outcompeted by ES114; P value of <0.01.
DISCUSSION
In this study, we describe a gene cluster that is required for the utilization of cellobiose by V. fischeri and is sufficient to confer this property onto cellobiose-negative Vibrio species. The genes in this cluster encode a PTS transport system (celA, celB, and celC), a glucokinase (celK), a glucosidase (celG), and a LacI-like transcriptional regulator (celI) that inhibits expression of the cluster when cellobiose is not present (Fig. 1 and 3). Mutational analyses and enzymatic assays show that the cel cluster is responsible for cleavage of both cellobiose, which is a β-1,4-linked glucose disaccharide, and X-gal, which is a β-1,4 linked galactoside. Although CelG is promiscuous with respect to the β-1,4-linked substrates it cleaves and, theoretically, it could direct catabolism of a β-1,4-galactoside such as lactose, we propose that its activity as a β-1,4-glucosidase is physiologically relevant, whereas its galactosidase activity is coincidental. In support of this, CelG appeared to have greater activity as a glucosidase, and it was required for growth on cellobiose, while lactose is not even utilized by V. fischeri as a carbon source. Moreover, the glucoside cellobiose (or a mutation in celI) induced the cel cluster, whereas several other sugars including lactose did not.
Cellobiose is a disaccharide-breakdown product of cellulose, but V. fischeri apparently lacks cellulose-degrading capacity, and the importance of its cel gene cluster is unclear. There is no evidence that cellobiose is released by E. scolopes to symbiotic V. fischeri, and it seems unlikely that this predatory invertebrate would produce or accumulate cellulose or cellobiose. Moreover, losing the ability to utilize cellobiose did not result in symbiotic attenuation (Table 3). V. fischeri has been isolated from the water column, even in areas where symbiotic hosts are not found (27, 45), and it has also been isolated from the bacterial consortium in the guts of herbivorous marine fishes, where it can survive and persist (42, 59). These observations suggest a niche where cellobioase activity may be important for V. fischeri. Many herbivorous fish partially digest cellulose by acid hydrolysis in their stomachs (74), and it is believed that bacterial cellulases contribute to the digestion of the cellulose in the gut, as with termites and ruminants (47, 70). Although V. fischeri does not contain a cellulase, it is possible that either the acidity of the fish's stomach or other bacteria in a fish's intestine could break down cellulose into cellobiose, allowing V. fischeri to consume it.
Interestingly, other cellobiose-utilizing Vibrio species, notably V. vulnificus, lack the celCBGKAI gene cluster, and we speculate that cellobiose catabolism may have arisen in members of the family Vibrionaceae by multiple distinct events. Based on our bioinformatic data, we speculate that a Firmicutes species horizontally transferred the cellobiose degradation glucokinase and glucosidase genes, and possibly the PTS genes, to an ancestor of the V. fischeri and V. salmonicida lineages. Firmicutes species have been found in the guts of marine fishes (23, 36) and in the marine environment (9, 44), so it seems plausible for the ancestor of V. fischeri to have acquired the gene cluster from a member of the phylum Firmicutes by horizontal gene transfer. Moreover, codon usage patterns of V. fischeri and Listeria species isolated from marine environments are not distinguishably different, suggesting that the expression of genes transferred between these species may be readily possible. As genome sequences become available for additional marine species, particularly those of Firmicutes, the origin of the V. fischeri gene cluster may become more apparent.
Our laboratory and those of others use the β-galactosidase gene lacZ as a transcriptional reporter (30, 69, 72, 73) in V. fischeri, and the discovery of a cryptic β-galactosidase activity in V. fischeri strikes a cautionary note for such applications. For example, when lacZ is used as a transcriptional reporter and transposon mutants are screened to find regulators of these lacZ fusions, knockouts of celI will also result in blue colonies on media containing X-gal. It may be useful in such situations to use the celG mutant allele in KV1319 in the reporter strain to prevent celG expression from confounding screens for lacZ activity. Alternatively, celI could be introduced on the pDMA171 plasmid into strains with apparent increases in lacZ activity to eliminate the possibility that a celI mutation and concomitant celG expression are responsible for β-galactosidase activity. Whatever the experimental setup, appropriate controls and careful interpretations are warranted whenever lacZ is used in a celG+ background.
lacZ has also been used as a marker in V. fischeri, so that the ratio of two strains in a mixed inoculum or infection can be determined by blue/white screen plating on media with X-gal. Determining strain ratios underlies competition experiments, which enable researchers to detect even subtle differences in symbiotic fitness (8, 24, 26, 29, 31, 35, 39, 56, 65, 66, 71, 72). Recently, V. fischeri strains have been marked for competition assays with the introduction of a stable plasmid containing lacZ (1, 17, 24). However, the lacZ-carrying plasmids can be lost, albeit at a low rate, and their use is inconsistent with other plasmids (e.g., for complementation). Our data suggest a fresh approach that does not rely on a plasmid-borne lacZ gene but retains the convenience of blue/white scoring to determine strain ratios. In this approach, one strain could be marked with the celG mutant allele present in KV1319. We have shown that this mutation has no effect on colonization competitiveness (Table 3), yet it results in the loss of blue color when it is plated on medium containing cellobiose and X-gal.
The family Vibrionaceae is an important and diverse family of bacteria in which species are continually being discovered (5, 25, 28, 41, 43, 50, 62) and with an apparent capacity for rapid evolution, given the periodic emergence of new pathogenic biotypes (13, 19, 49, 51). Traditionally, phenotypic markers such as strains' catabolic capacities have been used to help define Vibrio species. However, as more genomes are sequenced for important Vibrio species, molecular probes and DNA-based techniques will likely play an ever-larger role in identifying and defining important species or emergent biotypes. Our bioinformatic and phenotypic analyses suggest, not surprisingly, that caution is warranted when automated genome annotations are viewed. For example, an automated annotation of celA indicated that it directed “diacetylchitobiose-specific” transport, which seems, clearly, not to be the case given its importance in cellobiose catabolism. Similarly, PTS gene clusters were annotated as cellobiose transport systems in V. parahaemolyticus and V. cholerae, two species that are cellobiose negative. Experimental studies linking genes with taxonomically useful phenotypes, such as our dissection of the cel gene cluster reported here, will be useful in the future to improve Vibrio genome annotations and to connect molecular and phenotypic identification techniques.
Acknowledgments
We thank Jeff Turner and Erin Lipp for providing strains, Larry Shimkets and Jan Mrazek for helpful discussions, and Eduardo de Aquino Ximenes, Emily DeCrescenzo Henriksen, James Henriksen, Dana Cook, Nathan Montgomery, and Line Skoufos for technical assistance. We also thank Nils-Peder Willassen and the Sanger Institute for allowing us access to the V. salmonicida genome before publication.
This work was supported by NIH grant GM59690 to K.L.V., National Science Foundation CAREER grant MCB-0347317 to E.V.S., and NIH grant A150661 to M. McFall-Ngai.
Footnotes
Published ahead of print on 16 May 2008.
REFERENCES
- 1.Adin, D. M., N. J. Phillips, B. W. Gibson, M. A. Apicella, E. G. Ruby, M. J. McFall-Ngai, D. B. Hall, and E. V. Stabb. 2008. Characterization of htrB and msbB mutants of the light organ symbiont Vibrio fischeri. Appl. Environ. Microbiol. 74:633-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aeckersberg, F., C. Lupp, B. Feliciano, and E. G. Ruby. 2001. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J. Bacteriol. 183:6590-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Antunes, L. C., A. L. Schaefer, R. B. Ferreira, N. Qin, A. M. Stevens, E. G. Ruby, and E. P. Greenberg. 2007. Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J. Bacteriol. 189:8387-8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ast, J. C., I. Cleenwerck, K. Engelbeen, H. Urbanczyk, F. L. Thompson, P. De Vos, and P. V. Dunlap. 2007. Photobacterium kishitanii sp. nov., a luminous marine bacterium symbiotic with deep-sea fishes. Int. J. Syst. Evol. Microbiol. 57:2073-2078. [DOI] [PubMed] [Google Scholar]
- 6.Baumann, P., A. L. Furniss, and J. V. Lee. 1984. Genus I. Vibrio Pacini 1854, p. 518-538. In N. R. Kreig and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 7.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bose, J. L., U. Kim, W. Bartkowski, R. P. Gunsalus, A. M. Overley, N. L. Lyell, K. L. Visick, and E. V. Stabb. 2007. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 65:538-553. [DOI] [PubMed] [Google Scholar]
- 9.Bou-M'handi, N., C. Jacquet, A. El Marrakchi, and P. Martin. 2007. Phenotypic and molecular characterization of Listeria monocytogenes strains isolated from a marine environment in Morocco. Foodborne Pathog. Dis. 4:409-417. [DOI] [PubMed] [Google Scholar]
- 10.Campanella, J. J., L. Bitincka, and J. Smalley. 2003. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerda-Cuellar, M., J. Jofre, and A. R. Blanch. 2000. A selective medium and a specific probe for detection of Vibrio vulnificus. Appl. Environ. Microbiol. 66:855-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, A. L., J. D. Oliver, A. DePaola, E. J. Feil, and E. F. Boyd. 2007. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl. Environ. Microbiol. 73:5553-5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donovan, T. J., and P. van Netten. 1995. Culture media for the isolation and enumeration of pathogenic Vibrio species in foods and environmental samples. Int. J. Food Microbiol. 26:77-91. [DOI] [PubMed] [Google Scholar]
- 15.Dryselius, R., K. Kurokawa, and T. Iida. 2007. Vibrionaceae, a versatile bacterial family with evolutionarily conserved variability. Res. Microbiol. 158:479-486. [DOI] [PubMed] [Google Scholar]
- 16.Dunn, A. K., M. O. Martin, and E. V. Stabb. 2005. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54:114-134. [DOI] [PubMed] [Google Scholar]
- 17.Dunn, A. K., D. S. Millikan, D. M. Adin, J. L. Bose, and E. V. Stabb. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farmer, J. J. I., and F. W. Hickman-Brenner. 2006. The genera Vibrio and Photobacterium, p. 508-563. In M. Dworkin, S. Falkow, E. Rosenberg, K. Schleifer, and E. Stackenbrandt (ed.), The prokaryotes, 3rd ed., vol. 6. Springer, New York, NY. [Google Scholar]
- 19.Fouz, B., F. J. Roig, and C. Amaro. 2007. Phenotypic and genotypic characterization of a new fish-virulent Vibrio vulnificus serovar that lacks potential to infect humans. Microbiology 153:1926-1934. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 21.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holben, W. E., P. Williams, M. A. Gilbert, M. Saarinen, L. K. Sarkilahti, and J. H. Apajalahti. 2002. Phylogenetic analysis of intestinal microflora indicates a novel Mycoplasma phylotype in farmed and wild salmon. Microb. Ecol. 44:175-185. [DOI] [PubMed] [Google Scholar]
- 24.Hussa, E. A., T. M. O'Shea, C. L. Darnell, E. G. Ruby, and K. L. Visick. 2007. Two-component response regulators of Vibrio fischeri: identification, mutagenesis, and characterization. J. Bacteriol. 189:5825-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung, S. Y., Y. T. Jung, T. K. Oh, and J. H. Yoon. 2007. Photobacterium lutimaris sp. nov., isolated from a tidal flat sediment in Korea. Int. J. Syst. Evol. Microbiol. 57:332-336. [DOI] [PubMed] [Google Scholar]
- 26.Lee, K.-H., and E. G. Ruby. 1994. Competition between Vibrio fischeri strains during initiation and maintenance of a light organ symbiosis. J. Bacteriol. 176:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, K.-H., and E. G. Ruby. 1994. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 60:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Roux, F., A. Goubet, F. L. Thompson, N. Faury, M. Gay, J. Swings, and D. Saulnier. 2005. Vibrio gigantis sp. nov., isolated from the haemolymph of cultured oysters (Crassostrea gigas). Int. J. Syst. Evol. Microbiol. 55:2251-2255. [DOI] [PubMed] [Google Scholar]
- 29.Lupp, C., R. E. Hancock, and E. G. Ruby. 2002. The Vibrio fischeri sapABCDF locus is required for normal growth, both in culture and in symbiosis. Arch. Microbiol. 179:57-65. [DOI] [PubMed] [Google Scholar]
- 30.Lupp, C., and E. G. Ruby. 2005. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 187:3620-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lupp, C., M. Urbanowski, E. P. Greenberg, and E. G. Ruby. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50:319-331. [DOI] [PubMed] [Google Scholar]
- 32.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of Vibrio cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 33.Massad, G., and J. D. Oliver. 1987. New selective and differential medium for Vibrio cholerae and Vibrio vulnificus. Appl. Environ. Microbiol. 53:2262-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Millikan, D. S., and E. G. Ruby. 2004. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J. Bacteriol. 186:4315-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moran, D., S. J. Turner, and K. D. Clements. 2005. Ontogenetic development of the gastrointestinal microbiota in the marine herbivorous fish Kyphosus sydneyanus. Microb. Ecol. 49:590-597. [DOI] [PubMed] [Google Scholar]
- 37.Nogi, Y., N. Masui, and C. Kato. 1998. Photobacterium profundum sp. nov., a new, moderately barophilic bacterial species isolated from a deep-sea sediment. Extremophiles 2:1-7. [DOI] [PubMed] [Google Scholar]
- 38.Nyholm, S. V., and M. J. McFall-Ngai. 2004. The winnowing: establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol. 2:632-642. [DOI] [PubMed] [Google Scholar]
- 39.Nyholm, S. V., E. V. Stabb, E. G. Ruby, and M. J. McFall-Ngai. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA 97:10231-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crecy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park, Y. D., K. S. Baik, C. N. Seong, K. S. Bae, S. Kim, and J. Chun. 2006. Photobacterium ganghwense sp. nov., a halophilic bacterium isolated from sea water. Int. J. Syst. Evol. Microbiol. 56:745-749. [DOI] [PubMed] [Google Scholar]
- 42.Ramesh, A., and V. K. Venugopalan. 1988. Cellulolytic activity of luminous bacteria. World J. Microbiol. Biotechnol. 4:227-230. [Google Scholar]
- 43.Rivas, R., P. Garcia-Fraile, P. F. Mateos, E. Martinez-Molina, and E. Velazquez. 2006. Photobacterium halotolerans sp. nov., isolated from Lake Martel in Spain. Int. J. Syst. Evol. Microbiol. 56:1067-1071. [DOI] [PubMed] [Google Scholar]
- 44.Rodas-Suárez, O. R., J. F. Flores-Pedroche, J. M. Betancourt-Rule, E. I. Quiñones-Ramirez, and C. Vázquez-Salinas. 2006. Occurrence and antibiotic sensitivity of Listeria monocytogenes strains isolated from oysters, fish, and estuarine water. Appl. Environ. Microbiol. 72:7410-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruby, E. G., E. P. Greenberg, and J. W. Hastings. 1980. Planktonic marine luminous bacteria: species distribution in the water column. Appl. Environ. Microbiol. 39:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell, J. B., and J. L. Rychlik. 2001. Factors that alter rumen microbial ecology. Science 292:1119-1122. [DOI] [PubMed] [Google Scholar]
- 48.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 49.Safa, A., N. A. Bhuyian, S. Nusrin, M. Ansaruzzaman, M. Alam, T. Hamabata, Y. Takeda, D. A. Sack, and G. B. Nair. 2006. Genetic characteristics of Matlab variants of Vibrio cholerae O1 that are hybrids between classical and El Tor biotypes. J. Med. Microbiol. 55:1563-1569. [DOI] [PubMed] [Google Scholar]
- 50.Sawabe, T., Y. Fujimura, K. Niwa, and H. Aono. 2007. Vibrio comitans sp. nov., Vibrio rarus sp. nov. and Vibrio inusitatus sp. nov., from the gut of the abalones Haliotis discus discus, H. gigantea, H. madaka and H. rufescens. Int. J. Syst. Evol. Microbiol. 57:916-922. [DOI] [PubMed] [Google Scholar]
- 51.Serichantalergs, O., N. A. Bhuiyan, G. B. Nair, O. Chivaratanond, A. Srijan, L. Bodhidatta, S. Anuras, and C. J. Mason. 2007. The dominance of pandemic serovars of Vibrio parahaemolyticus in expatriates and sporadic cases of diarrhoea in Thailand, and a new emergent serovar (O3 : K46) with pandemic traits. J. Med. Microbiol. 56:608-613. [DOI] [PubMed] [Google Scholar]
- 52.Seshadri, R., S. W. Joseph, A. K. Chopra, J. Sha, J. Shaw, J. Graf, D. Haft, M. Wu, Q. Ren, M. J. Rosovitz, R. Madupu, L. Tallon, M. Kim, S. Jin, H. Vuong, O. C. Stine, A. Ali, A. J. Horneman, and J. F. Heidelberg. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188:8272-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stabb, E. V. 2005. Shedding light on the bioluminescence “paradox.” ”>ASM News 71:223-229. [Google Scholar]
- 54.Stabb, E. V. 2006. The Vibrio fischeri-Euprymna scolopes light organ symbiosis, p. 204-218. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington DC.
- 55.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stabb, E. V., and E. G. Ruby. 2003. Contribution of pilA to competitive colonization of the squid Euprymna scolopes by Vibrio fischeri. Appl. Environ. Microbiol. 69:820-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 58.Stabb, E. V., K. L. Visick, D. S. Millikan, A. A. Corcoran, L. Gibson, S. V. Nyholm, M. McFall-Ngai, and E. G. Ruby. 2001. The Vibrio fischeri-Euprymna scolopes symbiosis: a model marine animal-bacteria interaction, p. 269-277. In N. K. Saxena (ed.), Recent advances in marine science and technology. PACON International, Honolulu, HI.
- 59.Sugita, H., and Y. Ito. 2006. Identification of intestinal bacteria from Japanese flounder (Paralichthys olivaceus) and their ability to digest chitin. Lett. Appl. Microbiol. 43:336-342. [DOI] [PubMed] [Google Scholar]
- 60.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 61.Thompson, F. L., B. Austin, and J. G. Swings. 2006. The biology of vibrios. Blackwell Publishing, Boston, MA.
- 62.Thompson, F. L., B. Hoste, C. C. Thompson, J. Goris, B. Gomez-Gil, L. Huys, P. De Vos, and J. Swings. 2002. Enterovibrio norvegicus gen. nov., sp. nov., isolated from the gut of turbot (Scophthalmus maximus) larvae: a new member of the family Vibrionaceae. Int. J. Syst. Evol. Microbiol. 52:2015-2022. [DOI] [PubMed] [Google Scholar]
- 63.Thompson, F. L., and K. E. Klose. 2006. Vibrio2005: the first international conference on the biology of vibrios. J. Bacteriol. 188:4592-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vezzi, A., S. Campanaro, M. D'Angelo, F. Simonato, N. Vitulo, F. M. Lauro, A. Cestaro, G. Malacrida, B. Simionati, N. Cannata, C. Romualdi, D. H. Bartlett, and G. Valle. 2005. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459-1461. [DOI] [PubMed] [Google Scholar]
- 65.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Visick, K. L., T. M. O'Shea, A. H. Klein, K. Geszvain, and A. J. Wolfe. 2007. The sugar phosphotransferase system of Vibrio fischeri inhibits both motility and bioluminescence. J. Bacteriol. 189:2571-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Visick, K. L., and E. G. Ruby. 1998. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J. Bacteriol. 180:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Visick, K. L., and E. G. Ruby. 2006. Vibrio fischeri and its host: it takes two to tango. Curr. Opin. Microbiol. 9:632-638. [DOI] [PubMed] [Google Scholar]
- 69.Visick, K. L., and L. M. Skoufos. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Warnecke, F., P. Luginbuhl, N. Ivanova, M. Ghassemian, T. H. Richardson, J. T. Stege, M. Cayouette, A. C. McHardy, G. Djordjevic, N. Aboushadi, R. Sorek, S. G. Tringe, M. Podar, H. G. Martin, V. Kunin, D. Dalevi, J. Madejska, E. Kirton, D. Platt, E. Szeto, A. Salamov, K. Barry, N. Mikhailova, N. C. Kyrpides, E. G. Matson, E. A. Ottesen, X. Zhang, M. Hernandez, C. Murillo, L. G. Acosta, I. Rigoutsos, G. Tamayo, B. D. Green, C. Chang, E. M. Rubin, E. J. Mathur, D. E. Robertson, P. Hugenholtz, and J. R. Leadbetter. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560-565. [DOI] [PubMed] [Google Scholar]
- 71.Whistler, C. A., and E. G. Ruby. 2003. GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association with an animal host. J. Bacteriol. 185:7202-7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yip, E. S., K. Geszvain, C. R. DeLoney-Marino, and K. L. Visick. 2006. The symbiosis regulator rscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 62:1586-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yip, E. S., B. T. Grublesky, E. A. Hussa, and K. L. Visick. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol. Microbiol. 57:1485-1498. [DOI] [PubMed] [Google Scholar]
- 74.Zemke-White, W. L., K. D. Clements, and P. J. Harris. 1999. Acid lysis of macroalgae by marine herbivorous fishes: myth or digestive mechanism? J. Exp. Mar. Biol. Ecol. 233:95-113. [Google Scholar]