Abstract
Eukaryotes may influence pollutant degradation processes in groundwater ecosystems by activities such as predation on bacteria and recycling of nutrients. Culture-independent community profiling and phylogenetic analysis of 18S rRNA gene fragments, as well as culturing, were employed to obtain insight into the sediment-associated eukaryotic community composition in an anaerobic sandy aquifer polluted with landfill leachate (Banisveld, The Netherlands). The microeukaryotic community at a depth of 1 to 5 m below the surface along a transect downgradient (21 to 68 m) from the landfill and at a clean reference location was diverse. Fungal sequences dominated most clone libraries. The fungal diversity was high, and most sequences were sequences of yeasts of the Basidiomycota. Sequences of green algae (Chlorophyta) were detected in parts of the aquifer close (<30 m) to the landfill. The bacterium-predating nanoflagellate Heteromita globosa (Cercozoa) was retrieved in enrichments, and its sequences dominated the clone library derived from the polluted aquifer at a depth of 5 m at a location 21 m downgradient from the landfill. The number of culturable eukaryotes ranged from 102 to 103 cells/g sediment. Culture-independent quantification revealed slightly higher numbers. Groundwater mesofauna was not detected. We concluded that the food chain in this polluted aquifer is short and consists of prokaryotes and fungi as decomposers of organic matter and protists as primary consumers of the prokaryotes.
Food webs in aquifers may comprise eukaryotes in addition to bacteria and archaea. Groundwater eukaryotes range from single-celled heterotrophic nanoflagellates and fungi to amphipod crustaceans, each of which has important roles in the functioning of the groundwater ecosystem (8). The occurrence of fungi in low numbers has been described for a few pristine and polluted aquifers, but to our knowledge the identities and activities of these organisms have hardly been investigated (26, 27, 42). Protozoans, especially nanoflagellates, selectively graze on the biomass of the bacterial community (35) and recycle nutrients (36).
The abundance of protists, such as ciliates and flagellates, generally increases when there is pollution (27, 35). Biodegradation of organic pollutants often results in the development of anaerobic conditions. Under anoxic conditions, protists can survive and affect the abundance and diversity of bacteria (20). Protists can indirectly affect contaminant biodegradation. By feeding on bacteria, protists can reduce degrader populations, negatively influencing the process of biodegradation (21, 49). However, protists can also positively contribute to organic contaminant degradation by recycling limiting nutrients and making them available to pollutant-degrading bacteria (36), stimulating the activity of each bacterium (29, 30), or sustaining bioremediation by maintaining hydraulic conductivity of the aquifer as a result of reduced bacterial clogging (31, 43).
Landfill leachate is an important source of groundwater pollution. Studies on biodiversity in anaerobic landfill leachate-polluted groundwater, and polluted groundwater in general, have focused mainly on prokaryotes (7, 37). Despite the potential contribution of eukaryotes to ecosystem functioning and to biodegradation, the occurrence and diversity of eukaryotes in anaerobic aquifers have hardly been addressed. Phospholipid fatty acid (PLFA) biomarkers of fungi and algae were detected in an anaerobic aquifer polluted with leachate from a landfill in Denmark (24). The fungal and algal PLFA concentrations were highest near this landfill and decreased as the distance from it increased. PLFAs of protists were not detected, nor could protists be cultured (24).
Here, we determined the sediment-associated eukaryotic community structure in the anaerobic, sandy aquifer located downgradient from the Banisveld landfill (The Netherlands), using 18S rRNA gene-based denaturing gradient gel electrophoresis (DGGE). In order to obtain insight into the types of eukaryotes in anaerobic landfill leachate-polluted aquifers, culture-independent phylogenetic analysis was combined with sampling of groundwater metazoans and culturing of microeukaryotes. A phylogenetic analysis was performed with clone libraries derived from five sediment samples taken along a transect downgradient from the Banisveld landfill and one clean reference sample.
MATERIALS AND METHODS
Site description.
The Banisveld landfill is located 5 km southwest of Boxtel (The Netherlands) at 51°33′22"N, 05°16′55" E. This landfill has a surface area of approximately 6 ha, and the volume of waste dumped there is estimated to be 400,000 m3. The site functioned between 1965 and 1977 as a waste disposal site for household refuse and industrial wastes. After 1977 landfill operations were discontinued and the landfill was covered. Liners preventing migration of the leachate were not present, and a plume of leachate formed in the underlying aquifer. The lithology of this aquifer consists of a 7- to 9-m-thick layer of fine to coarse-grained unconsolidated clayey sands. Anaerobic iron reduction is the major process in the plume, while above and below the plume anaerobic conditions also prevail (51). The groundwater velocity is 4 to 10 m/year toward a nature reserve (51). More site characteristics have been described previously (51).
Groundwater mesofauna sampling.
Between December 2001 and November 2002, samples were obtained from 11 groundwater wells that were installed downgradient from the Banisveld landfill in June 1998 (51) and 28 groundwater monitoring wells in the surrounding Kampina nature reserve to determine the occurrence of groundwater fauna. Usually, each groundwater well downgradient from the Banisveld landfill contained three screens, one above the plume of pollution, one within the plume of pollution, and one below the plume of pollution. Two hundred to 3,000 liters of groundwater was pumped per screen using a Honda WX10 water pump and was filtered through 55-μm-mesh planktonic nets. The dross in the nets was collected in plastic jars, fixed in a 4% formaldehyde solution, and stained with Bengal Rose. These samples were analyzed to determine the presence of groundwater invertebrates using a stereomicroscope. The method used was successfully validated at a location (St. Agatha, Noord Brabant, The Netherlands) where groundwater fauna was previously encountered (41).
Sediment sampling.
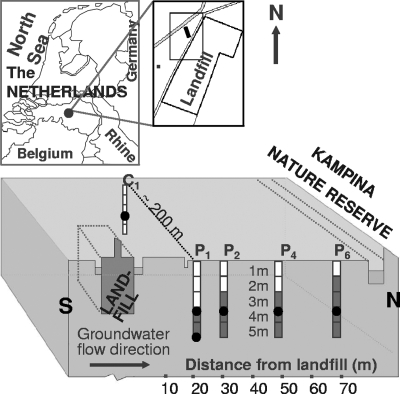
Sediment samples were obtained in June 2003 from five locations (Fig. 1). Four locations (P1, P2, P4, and P6) followed the main direction of the groundwater flow in the most polluted part of the aquifer (51) and were located 21, 30, 48, and 68 m downgradient from the landfill (Fig. 1). These locations were close to previously installed groundwater monitoring wells (51). The reference location chosen (C1) was in a clean part of the aquifer, approximately 200 m west of the polluted part of the aquifer. Sample cores were obtained at intervals of 1 m to a depth of 5.5 m. The sample designations indicate the depths; for example, “1m” indicates a sample obtained at a depth between 1.0 and 1.5 m, and “5m” indicates a sample obtained at a depth of 5.0 to 5.5 m.
FIG. 1.
Map of the Banisveld landfill research area and a longitudinal section through the aquifer located downgradient from the landfill. Drilling locations (designated P1, P2, P4, P6, and C1) with 1-m sampling depth intervals are indicated; white intervals indicate clean sediment (EC, <750 μS cm−1), and dark intervals indicate polluted sediment (EC, >750 μS cm−1). Filled circles indicate the locations where sediment samples used for construction of clone libraries were obtained.
Sediments cores were obtained using a manual drilling device, which did not require the use of drilling fluid. The first 1.0 to 1.5 m was obtained by digging with an open clay auger (“ram horn”) made of steel in a form similar to a corkscrew. A polyvinyl chloride casing was then inserted into the borehole to prevent the hole from collapsing. A bailer drilling device (Eijkelkamp B.V., The Netherlands) was inserted through the casing and used for drilling in water-saturated sediments. When the desired depth was reached, an undisturbed sample was obtained by pushing an extensible core pusher equipped with a 50-cm-long polyvinyl chloride tube with an inside diameter of 6 cm through the sediment. After retrieval, the sediment cores were capped, tightly sealed with tape, kept at 4°C, and processed within 24 h. The outer ends of the cores (3 to 5 cm) were removed in the laboratory, as they were briefly exposed to oxygen and might have been contaminated on the outside. The remaining sediment was homogenized aseptically by manual mixing.
Three sediment samples (P1-3m, P4-3m, and C1-3m) were obtained anaerobically for subsequent anaerobic culturing. Immediately after collection, the sediment cores were capped in the field and transferred to cylindrical containers made of steel, which were tightly sealed and flushed with dinitrogen gas. In the laboratory, these samples were processed in an anaerobic glove box.
Physicochemical measurements and chemical analyses.
Pore water was extracted by centrifugation of 100 g (fresh weight) of sediment for 50 min at 4,500 rpm. Electrical conductivities (EC) and ammonium concentrations were determined to evaluate the landfill leachate contamination of the aquifer. EC was measured using a GMH 3410 CE digital conductivity meter (Greisinger Electronic, Regenstauf, Germany). Ammonium concentrations were determined by spectrophotometry using a Skalar SA-40 autoanalyzer (Skalar BV, Breda, The Netherlands) (22).
Community profiling.
Microorganisms were released from 50 g of fresh, homogenized sediment by extraction for 1 h in an orbital shaker at 150 rpm with 100 ml of 0.1% sodium pyrophosphate. The mixture was then allowed to stand for 60 s in order to allow heavy sediment particles to settle. The liquid phase containing microbial cells was filtered through 0.22-μm-mesh cellulose membrane filters (Sartorius AG, Göttingen, Germany) using a vacuum pump. The filters were frozen at −80°C until DNA was extracted. DNA was isolated using a FastDNA spin kit (BIO 101 Systems, Irvine, CA) according to the instructions of the manufacturer, after filters were aseptically cut into small pieces.
Eukarya 18S rRNA gene fragments were amplified using a 25-μl (total volume) mixture containing 0.4 μM forward primer Euk1A (11), 0.4 μM reverse primer Euk516r-GC (11), 0.4 mM of each deoxynucleoside triphosphate, 10 μg of bovine serum albumin, PCR buffer, 2 U of Taq polymerase (MRC Holland, Amsterdam, The Netherlands), and 1 μl of undiluted DNA template. The amplification protocol consisted of initial denaturation at 94°C for 130 s, followed by 40 cycles of 94°C for 30 s, 56°C for 45 s, and 72°C for 130 s and a final elongation at 72°C for 7 min.
DGGE was performed with a Bio-Rad DCode (Hercules, CA) system. PCR products were loaded onto 1-mm-thick 8% (wt/vol) polyacrylamide (ratio of acrylamide to bisacrylamide, 37.5:1) gels. A 20 to 35% linear denaturing gradient was used for Eukarya-specific PCR products; 100% denaturant was defined as 7 M urea and 40% (vol/vol) formamide. Electrophoresis was performed in 1× TAE buffer (40 mM Tris-acetate, 1 mM Na-EDTA; pH 8.0) at 200 V and 60°C for 4 h. The gels were stained in 1× TAE buffer containing 1 μg/ml ethidium bromide, and the results were recorded with a charge-coupled device camera system (The Imager; Appligen, Illkirch, France). To aid in later conversion and normalization of gels, a marker consisting of 12 clones was added to the sides of the gels, as well as after every four samples. The outer two lanes of each gel were not used.
DGGE images were converted, normalized, and analyzed with the GelCompar II software package (Applied Maths, Kortrijk, Belgium). Similarities between tracks were calculated with GelCompar II using the Pearson product-moment correlation coefficient (whole densitometric curve based) and were visualized using the unweighted pair group clustering method with arithmetic averages.
Culture-independent phylogenetic analysis of 18S rRNA gene fragments.
Fragments (1.4 kb) from the 18S rRNA gene were amplified using Eukarya-specific primers Euk1A and Euk1209R (11). PCR was performed using a 25-μl mixture containing 0.4 μM of each primer, 0.4 mM of each deoxynucleoside triphosphate, 10 μg of bovine serum albumin, PCR buffer, 2 U of Taq polymerase (MRC Holland, Amsterdam, The Netherlands), and 1 μl of undiluted DNA template. Amplification was performed with a Perkin-Elmer DNA ThermoCycler by using initial denaturation at 94°C for 130 s, followed by 40 cycles of 94°C for 30 s, 56°C for 45 s, and 72°C for 130 s and a final elongation at 72°C for 7 min. The PCR products were purified using a High Pure PCR product purification kit (Roche Diagnostics GmbH, Mannheim, Germany) and were cloned into competent Escherichia coli JM109 by using the pGEM-T vector system (Promega Corporation, Madison, WI). Transformants were checked for inserts that were the correct sizes by performing a PCR with pGEM-T vector-specific primers (T7 and Sp6). To classify clones, libraries were screened by amplified ribosomal DNA restriction analysis (ARDRA) with RsaI and HaeIII (5 U; Roche Diagnostics GmbH, Mannheim, Germany). Clones having identical ARDRA types were considered members of the same operational taxonomic unit. The percent coverage (Cx) was calculated to determine how much of the actual diversity was captured in the clone libraries, as follows: Cx = (1 − nx/N) × 100, where nx is the number of ARDRA types encountered only once in library x and N is the total number of clones analyzed in library x (17).
Representatives of ARDRA types that occurred more than once and all ARDRA types that occurred only once in a library were subjected to DGGE and sequencing. PCR with eukaryotic primers Euk1A and Euk516r-GC and subsequent DGGE were performed as described above. DGGE band positions were compared to the environmental DGGE profiles of the samples from which the clones had been derived. Both strands of the 18S rRNA gene (Saccharomyces cerevisiae 18S rRNA gene positions 4 to 1438) were sequenced. The sequences obtained were compared to sequences deposited in the GenBank DNA database by using the BLAST algorithm to obtain the most closely related sequences (3). Chimera checks of the 18S rRNA gene sequences were performed using the Chimera Check program from RDP (28), and chimeric sequences were removed from further phylogenetic analysis. Sequence alignment was performed by using ClustalW. Distance analysis of unambiguously aligned sequences using the correction of Jukes and Cantor (19) and bootstrap resampling (100 times) were done with the TREECON package (52). The distance matrix was used to construct a phylogenetic tree using the neighbor-joining method (39).
MPN-PCR quantification and profiling of eukaryotes.
Serial 10-fold dilutions of the DNA extracts were prepared using sterile water in triplicate and were used as templates in a PCR with the eukaryotic universal primers Euk1A and Euk516r (11). Dilutions that showed products were considered positive and used to calculate the most probable number (MPN). Positive PCR products were reamplified with the same primers, except that the Euk516r primer contained a GC clamp (Euk1A and Euk516r-GC). Products were run on DGGE gels to determine whether the dominant bands in the diluted DNA templates corresponded to the dominant bands in the undiluted DNA templates.
Enrichment and culture-based quantification of protists.
Protists were enriched from eight sediment samples (all six 1-m depth intervals at P1, P4-3m, and C1-3m). For each sample, 10 g (fresh weight) of sediment was transferred to a 100-ml glass bottle containing 50 ml of 0.03% tryptone soy broth (TSB). In addition, protists were aerobically and anaerobically enumerated for three of the sediment samples selected for cloning (P1-3m, P4-3m, and C1-3m) by using a microtiter plate-based 24-well MPN method as described by Darbyshire et al. (9) and modified by Rønn et al. (38). The medium used was 0.03% TSB. Four sets of 10-fold serial dilutions up to a dilution of 10−6 were prepared. Cultivation was initiated within 6 h after sampling. Bottles and microtiter plates were incubated in the dark at 20°C and were screened for the presence of protists with an inverted microscope at a magnification of ×600 after 1, 3, and 7 days of incubation. For anaerobic incubation, anoxic conditions were ensured by incubating preparations in an anaerobic atmosphere consisting of 5% CO2 and 95% dinitrogen gas, while aliquot samples for microscopic inspection were removed in an anaerobic glove box. Enrichments containing protists were maintained by successive transfers of 10 ml from a culture to 50 ml of fresh TSB in sterile 100-ml bottles.
Molecular screening of enrichments.
Isolation of DNA from concentrated 2-ml aliquots of the enrichments (5 min of centrifugation at 14,000 rpm) was performed with a FastDNA spin kit (BIO 101 Systems, Irvine, CA) used according to the instructions of the manufacturer. Each PCR was conducted with the eukaryotic primers Euk1A and Euk516rGC (11). The PCR products were loaded onto DGGE gels, and the profiles obtained for enrichments were related to the environmental DGGE profiles. In order to determine whether the enriched protists were related to the dominant sequences in sediment sample P1-5m, 560-bp PCR products were sequenced.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the GenBank DNA database under accession numbers EU091827 to EU091878.
RESULTS
Eukaryotic community structure.
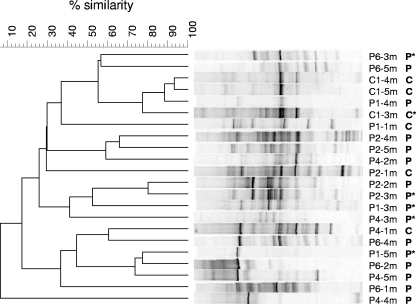
The eukaryotic community composition in subsurface sediment samples varied substantially over horizontal and vertical sections in the aquifer, and communities did not cluster visually according to depth, distance from the landfill, or the presence of pollution (Fig. 2). The presence of pollution was based on measurements of EC and the ammonium concentration, which are good indicators of the presence of landfill leachate at this research site (37, 51). An EC of 750 μS cm−1 marked the threshold between clean and polluted samples, as described previously (51). This threshold was also applicable in this study. The EC values at the clean reference location, C1, ranged from 60 to 578 μS cm−1, and the ammonia concentrations were between 2.6 and 3.6 mg/liter. Along the transect of drilling locations in the aquifer downgradient from the landfill, leachate was present 3 m below surface in June 2003 close to the landfill (P1) (Fig. 1). Pollution was detected only 1 to 2 m below the surface at distances more than 30 m from the landfill. The EC values ranged from 793 to 2,580 μS cm−1, while the ammonium concentrations were between 66 and 131 mg/liter in polluted sediment samples.
FIG. 2.
Clustering of 18S rRNA gene-based DGGE profiles of Eukarya (20 to 35% denaturant gradient) in sediment samples downgradient from the Banisveld landfill, determined using the unweighted pair group clustering method with arithmetic averages after Pearson correlation. For each sample, the location, depth (Fig. 1), and pollution level (P, polluted; C, clean) are indicated. Samples used for cloning are indicated by an asterisk.
Representation of eukaryotic diversity in 18S rRNA gene clone libraries.
Four sediment samples (P1-3m, P2-3m, P4-3m, and P6-3m) obtained from the plume of pollution at a depth of 3 m at different distances from the landfill (21, 30, 48, and 68 m, respectively) were selected for detailed phylogenetic analysis. For the location closest to the landfill (P1), an additional clone library was constructed for a sample taken at a depth of 5 m, because DGGE analysis revealed a dominant band that also occurred in other samples (Fig. 2). A reference clone library was constructed from a sample taken at a depth of 3 m in a part of the aquifer that was not influenced by the leachate (C1) (Fig. 1).
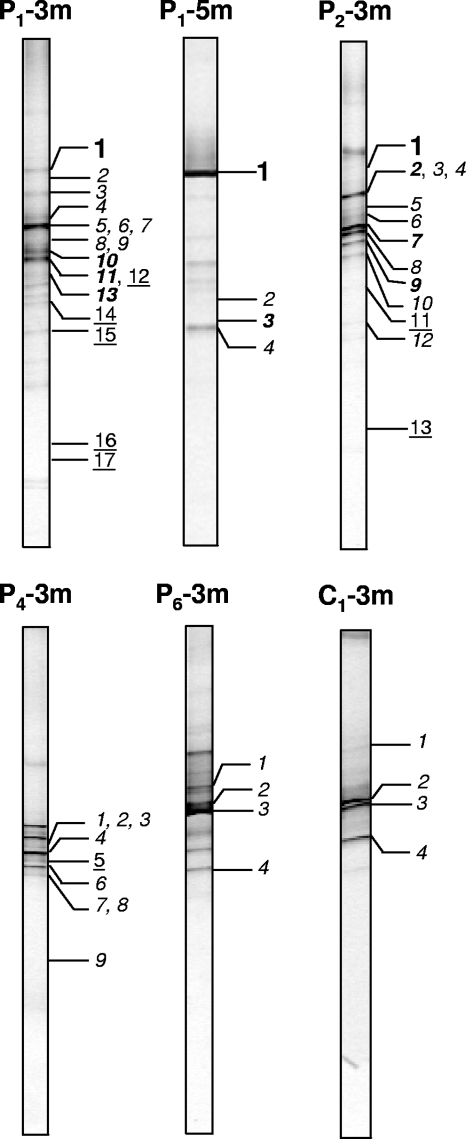
The levels of coverage, based on ARDRA screening of the clones, ranged from 66% (for P1-3m) to 98% (for P6-3m) (Table 1). DGGE screening of each unique ARDRA type revealed that, in general, the clone libraries represented the most intense DGGE bands in the environmental profiles (Fig. 3). The banding patterns of most clones corresponded to bands visible in DGGE profiles of the environmental samples. Thus, the diversity in the environmental samples was generally well covered by the relatively small clone libraries (32 to 43 clones per library).
TABLE 1.
Characteristics of eukaryotic communities in six sediment samples obtained from the aquifer polluted by the Banisveld landfilla
| Sample | Distance from landfill (m) | No. of clones analyzed | No. of ARDRA types | Coverage (%) | Aerobic MPN | Anaerobic MPN | MPN-PCR |
|---|---|---|---|---|---|---|---|
| P1-3m | 21 | 33 | 17 | 66 | 450 | 558 | 1,860 |
| P1-5m | 21 | 36 | 4 | 97 | NDb | ND | 760 |
| P2-3m | 30 | 43 | 13 | 81 | ND | ND | 3,000 |
| P4-3m | 48 | 37 | 9 | 95 | 1,260 | 558 | 86 |
| P6-3m | 68 | 42 | 4 | 98 | ND | ND | 1,860 |
| C1-3m | Nonpolluted | 32 | 4 | 97 | 138 | 138 | 860 |
See Fig. 1 for locations. The numbers of clones and unique ARDRA types and the coverage were derived from analysis of clone libraries. MPN estimates were obtained by culturing (eukaryotic cells/g sediment) and by PCR-based, culture-independent MPN analysis (18S rRNA gene copies/g sediment).
ND, not determined.
FIG. 3.
Linking eukaryotic clone identities to environmental DGGE profiles (20 to 35% denaturant gradient). The designations above the DGGE tracks indicate the sampling location and depth (Fig. 1). The identities of the numbered bands are shown in Fig. 5. Bold roman numbers indicate Cercozoa-like clones, bold italic numbers indicate Chlorophyta-like clones, light italic numbers indicate funguslike clones, and underlined numbers indicate other eukaryotes.
Phylogenetic diversity in microeukaryotes.
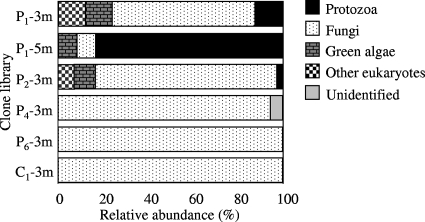
Gene fragments corresponding to positions 4 to 1438 in the S. cerevisiae 18S rRNA gene were sequenced for representatives of 51 clone types with unique ARDRA and DGGE profiles. Fungi were the most abundant group encountered (Fig. 4) and dominated the clone libraries generated for samples from the 3-m depth. The percentages of fungi were lower at locations closer to the landfill (P1-3m, P1-5m, and P2-3m; 8 to 81% of the total clones) than at locations farther away and in the clean reference sample (95 to 100% of the clones in the clone libraries) (Fig. 4). Sequences most closely related to protozoans (the cercozoan flagellate H. globosa) (Fig. 5) were identified only in polluted sediments obtained near the landfill (P1 and P2) and dominated (83% of the clones) the clone library generated for the sample obtained from a depth of 5 m at the location 21 m from the landfill (Fig. 4). Likewise, sequences most closely related to Chlorophyta (green algae) were also found only near the landfill (P1 and P2) and constituted 8 to 12% of the clones in the clone libraries (Fig. 4).
FIG. 4.
Relative abundance of various phylogenetic groups of eukaryotes in clone libraries generated from aquifer sediment samples.
FIG. 5.
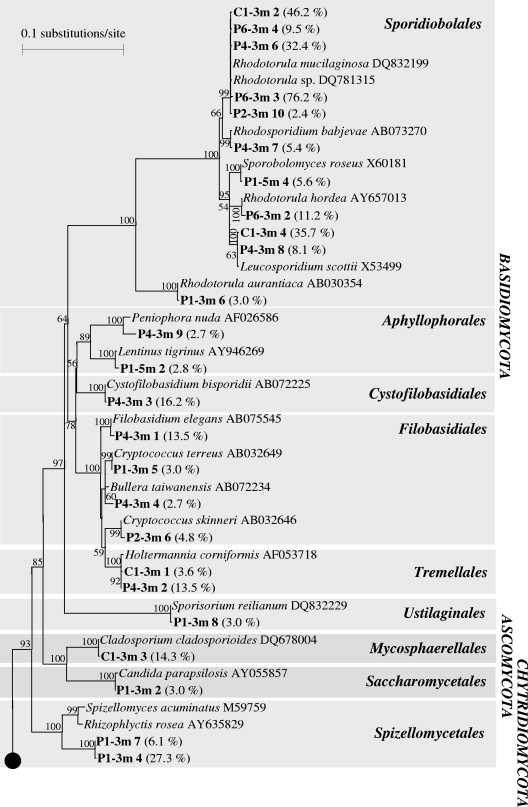
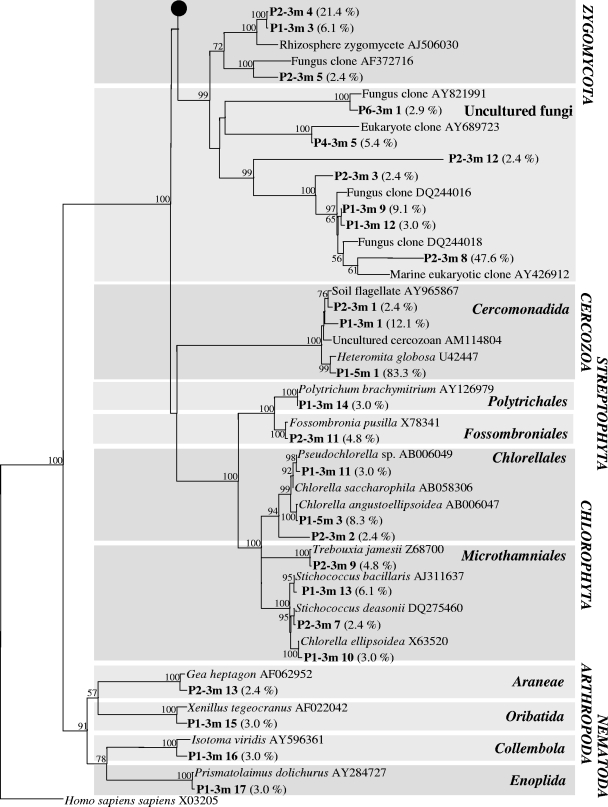
Phylogenetic tree based on almost complete 18S rRNA sequences for clones (bold type) in six libraries derived from the aquifer downgradient from the Banisveld landfill (The Netherlands). Each clone designation includes the library designation (e.g., P4-3m), which indicates the location and depth from which the clone was retrieved (Fig. 1), followed by a number corresponding to a numbered band in Fig. 3; the frequency at which the clone type occurred in the clone library is indicated in parentheses. Only bootstrap values greater than 50% are shown.
The most abundant group among the fungi was the Basidiomycota (50% of all analyzed clones). Sequences most closely related to the basidiomycete Rhodotorula mucilaginosa dominated the clone library generated for the clean reference sample C1-3m. Other fungal sequences detected in the C1-3m sample were most closely related to Tremellales and Leucosporidiales of the basidiomycetes and Mycosphaerellales of the ascomycotes (Fig. 5). Only basidiomycote sequences were identified in samples obtained 48 to 68 m downgradient from the landfill (P4-3m and P6-3m clone libraries) (Fig. 5). Sequences most closely related to R. mucilaginosa (Sporidiobolales) were also the most abundant basidiomycote sequences in these clone libraries. Closer to the landfill (P1-3m, P1-5m, and P2-3m), Rhodotorula-like sequences were less frequently encountered, and greater fungal diversity was observed (Fig. 5). In addition to basidiomycetes, members of the Ascomycota (Saccharomycetales and Mycosphaerellales) and Chytridiomycota (Spizellomycetales) were present.
A number of clones (P1-3m clones 3, 9, and 12; P2-3m clones 3, 4, 5, 8, and 12, P4-3m clone 5, and P6-3m clone 3) (Fig. 5) fell into a cluster for which cultured representatives are not yet known. These sequences were related to uncultured fungi that were previously encountered in lakes (23, 44, 53) and anoxic sediments (10).
Green algae (Chlorophyta) were represented in the clone libraries by sequences most closely related to members of the Chlorellales and Microthamniales (P1-3m clone 10, P1-3m clone 11, P1-5m clone 3, P2-3m clone 2, P2-3m clone 7, and P2-3m clone 9 in Fig. 5). All clones that were most closely related to the cercozoan flagellate H. globosa (P1-3m clone 1, P1-5m clone 1, and P2-3m clone 1 in Fig. 5), whose sequence was the only protozoan-like sequence encountered in the six clone libraries, exhibited the same migration in DGGE (Fig. 3, bands 1 in tracks P1-3m, P1-5m, and P2-3m).
Quantification of eukaryotes by MPN-PCR and culture-based MPN methods.
A culture-independent PCR-MPN approach revealed that the number of copies of eukaryotic 18S rRNA genes ranged from 86 copies per g of sediment (for P4-3m) to 3,000 copies per g of sediment (for P2-3m). In general, these numbers were higher than the numbers obtained by culturing (Table 1). DGGE screening revealed that, in general, the bands observed in DGGE profiles derived from the diluted DNA templates corresponded to the most intense bands observed for the undiluted DNA extracts (data not shown). For the location nearest the landfill (P1) at a depth of 5 m, a DGGE band corresponding to that of cloned H. globosa sequences was recognized for all DNA dilutions that showed positive amplification with eukaryotic primers, suggesting that the nanoflagellate H. globosa was dominant at this location.
The number of culturable microeukaryotes ranged from 138 to 1,260 cells/g sediment. Anaerobic cultivation revealed numbers of eukaryotes comparable to the numbers obtained by aerobic cultivation (Table 1), with the exception of the P4-3m sample (48 m from landfill), in which the number of protozoans obtained by aerobic culturing was twice the number obtained by anaerobic culturing. No obvious relationship between the number of microeukaryotes and the distance from the landfill or degree of pollution (clean versus polluted) was observed.
Culturable protists.
Protists were observed by microscopy in aerobic and anaerobic enrichments derived from eight sediment samples (P1-0m to P1-5m, P4-3m, and C1-3m). For samples P1-3m, P4-3m, and C1-3m, DGGE screening revealed that the enrichments obtained for the sediments were more complex than those obtained for sample P1-5m (data not shown). DGGE profiling revealed that a single dominant band was present for P1-5m. The migration behavior of this band corresponded to that of the H. globosa-like sequence present in the clone library obtained from the same location. Sequencing revealed a level of similarity for the cultured flagellate and the cloned H. globosa-like sequences of 100%. This enrichment was transferred 16 times successively over a 14-month period, and H. globosa was the only eukaryote observed by microscopy as well as by DGGE screening and sequencing. For location P1 (nearest the landfill), where aerobic enrichment of protists was performed in a vertical transect from the surface to a depth of 5 m, ciliates were detected in sediments from the surface and a depth of 1 m, while only flagellates were detected in sediments from depths of 1 to 5 m (data not shown).
Groundwater mesofauna.
Multicellular groundwater inhabitants (stygobiontic organisms) were not detected in groundwater samples from the polluted aquifer located downgradient from the Banisveld landfill or at clean reference locations surrounding it. Only very low numbers of nonindigenous (stygoxenic) soil-like invertebrates, such as mites, collembolans, oligochaetes, tardigrades, nematodes, and stygoxenic crustaceans, were observed (range, 0.03 to 2 individuals per 100 liters). Of the 218 cloned 18S rRNA genes, a few sequences were most closely related to multicellular eukaryotes, but they were related only to nonindigenous mosses (Bryophyta) and soil invertebrate metazoans (Arachnida, Acarina, Collembola, and Nematoda) (Fig. 5).
DISCUSSION
Food web structure in the aquifer polluted by the Banisveld landfill.
The food chains in the anaerobic aquifer downgradient from the Banisveld landfill are short and consist of prokaryotes and of nanoflagellates as the top predators. True groundwater invertebrates (stygofauna) were not detected in approximately 150 m3 of groundwater pumped and filtered from the monitoring wells. The absence of groundwater fauna is probably a consequence of the lack of oxygen and the presence of fine-grained sediments (50) that leave insufficient living space for groundwater invertebrates.
Both culture-dependent and culture-independent analyses revealed the presence of a cercozoan flagellate most closely related to H. globosa in landfill leachate-affected sediment. Members of the genus Heteromita are common and abundant heterotrophic soil flagellates (16). Our study is the first study to show that H. globosa can be a dominant member of eukaryotic communities in anaerobic aquifers. The recovery of H. globosa in aerobic and anaerobic enrichments showed that viable cells are present and that culture-independent detection of this organism is not due to amplification of extracellular DNA. A Heteromita sp. has previously been encountered in landfills (15), but its ability to grow anaerobically (30, 31) suggests that Heteromita spp. might also be active in the aquifer itself and that their presence is not simply due to transport of cysts out of the landfill. H. globosa was relatively abundant in the proximity of the landfill. Here, the rate of dissolved organic carbon degradation is highest (50), suggesting that the associated, relatively high bacterial production is capable of supporting an additional trophic level consisting of bacterium-predating H. globosa.
The presence of Heteromita spp. has significance for anaerobic biodegradation; laboratory studies have indicated that H. globosa influences toluene and alkylbenzene biodegradation (29, 30) by enhancing the activity per bacterium. These studies were done under aerobic conditions, but similar stimulation may occur under anaerobic conditions. H. globosa was able to reduce bacterial clogging under denitrifying conditions (31), which should contribute to maintaining the hydraulic conductivity of aquifers and sustaining biodegradation (43).
Also, sequences most closely related to green algae (Chlorophyta) were encountered close to the landfill. Green algae have previously been encountered in aquifers (5, 40, 42), but their function in the subsurface is still unclear. Algae are most likely introduced into aquifers from the surface by percolating rainwater or from surface water recharge (e.g., lakes or streams) (42). Mixotrophic algae can survive by feeding on bacteria in the absence of light (18). However, the ability to grow mixotrophically has not been described yet for the algal species that we encountered. Contamination of samples with algae during drilling seems unlikely as the same drilling water-free technology and sampling procedures were used at the other locations, and analysis of the clone libraries based on these samples did not reveal the presence of green algae.
The numbers of protists (86 to 3,000 organisms per g of sediment) observed in the present study was intermediate between the numbers observed at other contaminated anaerobic aquifers; 100 to 101 eukaryotes per g (dry weight) of sediment were obtained for hydrocarbon-contaminated aquifers in South Dakota (United States) and Germany (48, 55), while 104 to 105 protists per g (dry weight) of sediment were identified in an aquifer polluted by wastewater in Massachusetts (United States) (20).
Fungal communities.
A group of little-studied eukaryotic organisms that occur in (contaminated) aquifers is the fungi. Unlike the few studies in which fungi in groundwater were just enumerated (26, 27, 42) or in which fungal PLFAs were detected (24), in the present study we identified various types of fungi. To our knowledge, the study of Luo and coworkers (25) is the only other DNA-based culture-independent study of an anaerobic aquifer, and in that case the aquifer was polluted with hydrocarbons. These authors identified mainly ascomycetous and basidiomycetous fungal sequences in a clone library. While Pezizomycetes ascomycotes were the dominant group in the study of Luo et al. (25), in our study Sporidiobolales basidiomycotes were the most abundant organisms.
Sequences most closely related to the yeasts Rhodotorula, Cryptococcus, and Leucosporidium were dominant members of the Banisveld aquifer clone libraries. Strains belonging to these genera are able to catabolize benzene compounds and to assimilate lignin monomeric degradation products as sole carbon sources under aerobic conditions (32, 34). Cryptococcus species are capable of utilizing humic acids as sole carbon and nitrogen sources (14). The ascomycetous yeast Candida parapsilosis, to which a clone obtained from the P1-3m sample 21 m from the landfill was most closely related, degrades phenols and hydroxybenzoates aerobically (33). Members of the genera Rhodotorula, Cryptococcus, and Candida can grow anaerobically (13, 54) by fermentation and were also encountered in another anaerobic aquifer (25). Thus, the fungal species identified in our study could be quite versatile in terms of their physiological abilities. Fungi may contribute to the decomposition of organic matter, either within the landfill (from which the fungi leached out to the groundwater) or in the leachate plume. Lignin and humic acids, compounds that are degraded by some of the cultured relatives most closely related to our sequences, are abundant in landfills and landfill leachate (6). On the downside, Rhodotorula, Cryptococcus, and Candida species have also been implicated in various infectious diseases (1, 45) and mycoses (2, 4).
Fungi have also been encountered in other anaerobic environmental settings, such as anaerobic marine environments (12, 47) and anaerobic marine intertidal (10) and marsh (46) sediments, where they constituted just a small fraction of the total eukaryote community. However, fungal sequences dominated clone libraries generated from anaerobic lake freshwater and brackish anoxic sediments (10), as well as from anaerobic sediments in a freshwater pond and an anaerobic sewage digestor (25).
In conclusion, the food web in the polluted Banisveld aquifer is simple, consisting of bacteria and fungi as potential decomposers of organic matter and flagellate protozoans as grazers. Further information on the activity of eukaryotes in polluted aquifers with respect to their direct and indirect contributions to contaminant biodegradation is important for assessing, monitoring, and predicting natural attenuation.
Acknowledgments
The TRIAS program, a joint venture of The Netherlands Organization of Scientific Research (NWO), The Netherlands Center for Soil Quality Management and Knowledge Transfer (SKB), and Delft Cluster, and The Netherlands' BSIK Ecogenomics program supported this project financially.
We are grateful to Jos Notenboom for fruitful discussions regarding the groundwater fauna and for his enthusiasm in joining our sampling field trips. We also thank Frank Fiers for his kind assistance in identifying the copepods and cladocerans.
Footnotes
Published ahead of print on 9 May 2008.
REFERENCES
- 1.Abi-Said, D., E. Anaissie, O. Uzun, I. Raad, H. Pinzcowski, and S. Vartivarian. 1997. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin. Infect. Dis. 24:1122-1128. [DOI] [PubMed] [Google Scholar]
- 2.Alliot, C., B. Desablens, R. Garidi, and S. Tabuteau. 2000. Opportunistic infection with Rhodotorula in cancer patients treated by chemotherapy: Two case reports. Clin. Oncol. 12:115-117. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Anaissie, E., G. P. Bodey, H. Kantarjian, J. Ro, S. E. Vartivarian, R. Hopfer, J. Hoy, and K. Rolston. 1989. New spectrum of fungal infections in patients with cancer. Rev. Infect. Dis. 11:369-378. [DOI] [PubMed] [Google Scholar]
- 5.Beloin, R. M., J. L. Sinclair, and W. C. Ghiorse. 1988. Distribution and activity of microorganisms in subsurface sediments of a pristine study site in Oklahoma. Microb. Ecol. 16:85-95. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, J. B., D. L. Jensen, C. Gron, Z. Filip, and T. H. Christensen. 1998. Characterization of the dissolved organic carbon in landfill leachate-polluted groundwater. Water Res. 32:125-135. [Google Scholar]
- 7.Christensen, T. H., P. Kjeldsen, P. L. Bjerg, D. L. Jensen, J. B. Christensen, A. Baun, H. J. Albrechtsen, and C. Heron. 2001. Biogeochemistry of landfill leachate plumes. Appl. Geochem. 16:659-718. [Google Scholar]
- 8.Danielopol, D. L., C. Griebler, A. Gunatilaka, and J. Notenboom. 2003. Present state and future prospects for groundwater ecosystems. Environ. Conserv. 30:104-130. [Google Scholar]
- 9.Darbyshire, J. F., R. E. Whitley, M. P. Greaves, and R. H. E. Inkson. 1974. A rapid micromethod for estimating bacterial and protozoan populations in soil. Rev. Ecol. Biol. Sol. 11:465-475. [Google Scholar]
- 10.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Díez, B., C. Pedrós-Alió, T. L. Marsh, and R. Massana. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgcomb, V. P., D. T. Kysela, A. Teske, A. De Vera Gomez, and M. L. Sogin. 2002. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. USA 99:7658-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekendahl, S., A. H. O'Neill, E. Thomsson, and K. Pedersen. 2003. Characterisation of yeasts isolated from deep igneous rock aquifers of the Fennoscandian shield. Microb. Ecol. 46:416-428. [DOI] [PubMed] [Google Scholar]
- 14.Filip, Z., and P. Bielek. 2002. Susceptibility of humic acids from soils with various contents of metals to microbial utilization and transformation. Biol. Fertil. Soils 36:426-433. [Google Scholar]
- 15.Finlay, B. J., and T. Fenchel. 1991. An anaerobic protozoon, with symbiotic methanogens, living in municipal landfill material. FEMS Microbiol. Ecol. 85:169-180. [Google Scholar]
- 16.Fredslund, L., F. Ekelund, K. S. Jacobsen, and K. Johnsen. 2001. Development and application of a most-probable-number-PCR assay to quantify flagellate populations in soil samples. Appl. Environ. Microbiol. 67:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Good, I. J. 1953. The population frequencies of species and the estimation of the population parameters. Biometrika 40:237-264. [Google Scholar]
- 18.Jones, R. I. 2000. Mixotrophy in planktonic protists: an overview. Freshwater Biol. 45:219-226. [Google Scholar]
- 19.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, New York, NY. [Google Scholar]
- 20.Kinner, N., R. W. Harvey, D. M. Shay, D. W. Metge, and A. Warren. 2002. Field evidence for a protistan role in a organically-contaminated aquifer. Environ. Sci. Technol. 36:4312-4318. [DOI] [PubMed] [Google Scholar]
- 21.Kota, S., R. C. Borden, and M. A. Barlaz. 1999. Influence of protozoan grazing on contaminant biodegradation. FEMS Microbiol. Ecol. 29:179-189. [Google Scholar]
- 22.Krom, N. 1980. Spectrophotometric determination of ammonia: a study of a modified Berthelot reaction using salicylate and dichloro-iso-cyanurate. Analyst 105:305-316. [Google Scholar]
- 23.Lefranc, M., A. Thenot, C. Lepere, and D. Debroas. 2005. Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 71:5935-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludvigsen, L., H. J. Albrechtsen, D. B. Ringelberg, F. Ekelund, and T. H. Christensen. 1999. Distribution and composition of microbial populations in a landfill leachate contaminated aquifer (Grindsted, Denmark). Microb. Ecol. 37:197-207. [DOI] [PubMed] [Google Scholar]
- 25.Luo, Q., L. R. Krumholz, F. Z. Najar, A. D. Peacock, B. A. Roe, D. C. White, C. David, and M. S. Elshahed. 2005. Diversity of the microeukaryotic community in sulfide-rich Zodletone Spring (Oklahoma). Appl. Environ. Microbiol. 71:6175-6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madsen, E. L., and W. C. Ghiorse. 1993. Groundwater microbiology: subsurface ecosystem processes, p. 167-213. In T. E. Ford (ed.), Aquatic microbiology: an ecological approach. Blackwell Scientific Publications, Boston, MA.
- 27.Madsen, E. L., J. L. Sinclair, and W. C. Ghiorse. 1991. In situ biodegradation: microbiological patterns in a contaminated aquifer. Science 252:830-833. [DOI] [PubMed] [Google Scholar]
- 28.Maidak, B. L., J. R. Cole, J. Charles, T. Parker, G. M. Garrity, N. Larsen, B. Li, T. J. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattison, R. G., and S. Harayama. 2001. The predatory soil flagellate Heteromita globosa stimulates toluene biodegradation by a Pseudomonas sp. FEMS Microbiol. Lett. 194:39-45. [DOI] [PubMed] [Google Scholar]
- 30.Mattison, R. G., and S. Harayama. 2005. The soil flagellate Heteromita globosa accelerates bacterial degradation of alkylbenzenes through grazing and acetate excretion in batch culture. Microb. Ecol. 49:142-150. [DOI] [PubMed] [Google Scholar]
- 31.Mattison, R. G., H. Taki, and S. Harayama. 2002. The bacterivorous soil flagellate Heteromita globosa reduces bacterial clogging under denitrifying conditions in sand-filled aquifer columns. Appl. Environ. Microbiol. 68:4539-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Middelhoven, W. J. 1993. Catabolism of benzene compounds by ascomycetous and basidiomycetous yeasts and yeastlike fungi. A literature review and an experimental approach. Antonie van Leeuwenhoek 63:125-144. [DOI] [PubMed] [Google Scholar]
- 33.Middelhoven, W. J., A. Coenen, B. Kraakman, and M. D. Sollewijn Gelpke. 1992. Degradation of some phenols and hydroxybenzoates by the imperfect ascomycetous yeasts Candida parapsilosis and Arxula adeninivorans: evidence for an operative gentisate pathway. Antonie van Leeuwenhoek 62:181-187. [DOI] [PubMed] [Google Scholar]
- 34.Middelhoven, W. J., M. Koorevaar, and G. W. Schuur. 1992. Degradation of benzene compounds by yeasts in acidic soils. Plant Soil 45:37-43. [Google Scholar]
- 35.Novarino, G., A. Warren, H. Butler, G. Lambourne, A. Boxshall, J. Bateman, N. E. Kinner, R. W. Harvey, R. A. Mosse, and B. Teltsch. 1997. Protistan communities in aquifers. A review. FEMS Microbiol. Rev. 20:261-276. [DOI] [PubMed] [Google Scholar]
- 36.Ratsak, C. H., K. A. Maarsen, and S. A. L. M. Kooijman. 1996. Effects of protozoa on carbon mineralization in activated sludge. Water Res. 30:1-12. [Google Scholar]
- 37.Röling, W. F. M., B. M. Van Breukelen, M. Braster, B. Lin, and H. W. Van Verseveld. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 67:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rønn, R., F. Ekelund, and S. Christensen. 1995. Optimizing soil extract and broth media for MPN-enumeration of naked amoebae and heterotrophic flagellates in soil. Pedobiologia 39:10-19. [Google Scholar]
- 39.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 40.Sargent, K. A., and C. B. Fliermans. 1989. Geology and hydrology of the deep subsurface microbiology sampling sites at the Savannah River Plant, South Carolina. Geomicrobiol. J. 7:3-13. [Google Scholar]
- 41.Schminke, H. K., and J. Notenboom. 1990. Parastenocarididae (Copepoda, Harpacticoida) from the Netherlands. Bijdr. Dierkd. 60:299-304. [Google Scholar]
- 42.Sinclair, J. L., and W. C. Ghiorse. 1989. Distribution of aerobic bacteria, protozoa, algae, and fungi in deep subsurface sediments. Geomicrobiol. J. 7:15-31. [Google Scholar]
- 43.Sinclair, J. L., D. H. Kampbell, M. L. Cook, and J. T. Wilson. 1993. Protozoa in subsurface sediments from sites contaminated with aviation gasoline or jet fuel. Appl. Environ. Microbiol. 59:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slapeta, J., D. Moreira, and P. López-García. 2005. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc. R. Soc. Lond. Ser. B 272:2073-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sobel, J. D., and J. A. Vazquez. 1999. Fungal infections of the urinary tract. World J. Urol. 17:410-414. [DOI] [PubMed] [Google Scholar]
- 46.Stoeck, T., and S. Epstein. 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 69:2657-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoeck, T., G. T. Taylor, and S. Epstein. 2003. Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean Sea). Appl. Environ. Microbiol. 69:5656-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, J. M., C. L. Bruce, V. R. Gordy, K. L. Duston, S. R. Hutchins, J. L. Sinclair, and C. H. Ward. 1997. Assessment of the microbial potential for nitrate-enhanced bioremediation of a JP-4 fuel-contaminated aquifer. J. Ind. Microbiol. Biotechnol. 18:213-221. [Google Scholar]
- 49.Travis, B. J., and N. D. Rosenberg. 1997. Modeling in situ bioremediation of TCE at Savannah River: effects of product toxicity and microbial interactions on TCE degradation. Environ. Sci. Technol. 31:3093-3102. [Google Scholar]
- 50.Van Breukelen, B. M., J. Griffioen, W. F. M. Röling, and H. W. Van Verseveld. 2004. Reactive transport modeling of biogeochemical processes and carbon isotope geochemistry inside a landfill leachate plume. J. Contam. Hydrol. 70:249-269. [DOI] [PubMed] [Google Scholar]
- 51.Van Breukelen, B. M., W. F. M. Röling, J. Groen, J. Griffioen, and H. W. Van Verseveld. 2003. Biogeochemistry and isotope geochemistry of a landfill leachate plume. J. Contam. Hydrol. 65:245-268. [DOI] [PubMed] [Google Scholar]
- 52.Van de Peer, Y., and Y. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of phylogenetic trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 53.Van Hannen, E. J., W. Mooij, M. P. Van Agterveld, H. J. Gons, and H. J. Laanbroek. 1999. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visser, W., W. A. Scheffers, W. H. Batenburg-van der Vegte, and J. P. Van Dijken. 1990. Oxygen requirements of yeast. Appl. Environ. Microbiol. 56:3785-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zarda, B., G. Mattison, A. Hess, D. Hahn, P. Höhener, and J. Zeyer. 1998. Analysis of bacterial and protozoan communities in an aquifer contaminated with monoaromatic hydrocarbons. FEMS Microbiol. Ecol. 27:141-152. [Google Scholar]