Abstract
Vibrio tubiashii is a recently reemerging pathogen of larval bivalve mollusks, causing both toxigenic and invasive disease. Marine Vibrio spp. produce an array of extracellular products as potential pathogenicity factors. Culture supernatants of V. tubiashii have been shown to be toxic to oyster larvae and were reported to contain a metalloprotease and a cytolysin/hemolysin. However, the structural genes responsible for these proteins have yet to be identified, and it is uncertain which extracellular products play a role in pathogenicity. We investigated the effects of the metalloprotease and hemolysin secreted by V. tubiashii on its ability to kill Pacific oyster (Crassostrea gigas) larvae. While V. tubiashii supernatants treated with metalloprotease inhibitors severely reduced the toxicity to oyster larvae, inhibition of the hemolytic activity did not affect larval toxicity. We identified structural genes of V. tubiashii encoding a metalloprotease (vtpA) and a hemolysin (vthA). Sequence analyses revealed that VtpA shared high homology with metalloproteases from a variety of Vibrio species, while VthA showed high homology only to the cytolysin/hemolysin of Vibrio vulnificus. Compared to the wild-type strain, a VtpA mutant of V. tubiashii not only produced reduced amounts of protease but also showed decreased toxicity to C. gigas larvae. Vibrio cholerae strains carrying the vtpA or vthA gene successfully secreted the heterologous protein. Culture supernatants of V. cholerae carrying vtpA but not vthA were highly toxic to Pacific oyster larvae. Together, these results suggest that the V. tubiashii extracellular metalloprotease is important in its pathogenicity to C. gigas larvae.
Vibriosis caused by marine Vibrio species is considered one of the most serious diseases of hatchery-reared oyster larvae (10, 11, 17, 47, 52). The disease is characterized by a rapid and dramatic reduction in larval motility, detached vela, and necrotic soft tissue, which lead to high mortality rates, exceeding 90% within 1 day of infection (45). Pathogenic agents that cause larval bivalve vibriosis have intermittently and severely curtailed shellfish hatchery production on the Atlantic and Pacific coasts of the United States, causing substantial losses in the industry (3, 10, 13). Vibrio tubiashii, a bacterial species first reported by Tubiash et al. (51), was identified as a causative agent of vibriosis (originally referred to as bacillary necrosis) in larval and juvenile bivalves of the hard clam (Mercenaria mercenaria) and Eastern oyster (Crassostrea virginica). Estes et al. (14) characterized a number of pathogenic and nonpathogenic bacterial strains from diseased Pacific oysters (Crassostrea gigas) at shellfish hatcheries on the Pacific coast of North America and described some of the highly pathogenic bacterial isolates as V. tubiashii.
The genus Vibrio is the largest member of the family Vibrionaceae, which includes gram-negative and curved rod-shaped facultative anaerobes. The genus consists of at least 30 known species, which are widespread in the aquatic environment throughout the world. Vibrio species have a wide range of hosts, from humans to aquatic animals, including fish and shellfish. In shellfish diseases, larval and juvenile stages are particularly susceptible to bacterial infections (12, 35). The mortality caused by vibriosis at these early stages may occur suddenly and often results in severe losses of production in shellfish hatcheries (10, 11, 39, 40).
Marine Vibrio species are known to produce extracellular products, some of which are known pathogenicity factors. These toxic proteins include cytolysins, proteases, lipases, siderophores, exopolysaccharides, and effectors delivered via type III secretion systems (7, 38, 44, 46, 53, 54). In V. tubiashii, several secreted proteins, including a low-molecular-weight ciliostatic toxin, are suspected virulence factors in shellfish larval vibriosis (37). The east coast V. tubiashii strain ATCC 19105 has been described to produce a cytolysin/hemolysin (20) and an extracellular protease (9). Kothary et al. (20) showed that the hemolytic activity in culture supernatants could be detected with a variety of blood cells, including sheep erythrocytes. They further found that the N-terminal region (17 amino acids) shared significant similarity to VvhA, a hemolysin produced by Vibrio vulnificus that is known to function as a primary virulence factor (24). An extracellular protease of V. tubiashii was purified from ATCC 19105 (9) and was shown to be a zinc-containing metalloprotease, with the 20-amino-acid sequence of the N-terminal region being almost identical to those of other marine Vibrio metalloproteases. Although these previous studies showed that V. tubiashii is capable of producing several potential toxins, it is still unclear which role these potential pathogenicity factors play in V. tubiashii pathogenicity, if any. In this study, we have focused on the roles of an extracellular protease and a hemolysin in the toxicity of culture supernatants of V. tubiashii on Pacific oyster larvae.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Vibrio tubiashii strains RE22 and RE98 and an unknown bacterial isolate, RE15, isolated from a Pacific oyster (14), were grown on Luria-Bertani (LB) medium supplemented with 1% sodium chloride (LB-1% NaCl) at 30°C. For larval-toxicity assays, V. tubiashii strains were cultured overnight in 20 ml LB-1% NaCl at 25°C on a shaker. Vibrio cholerae strains used in this study were O1 classical biotype strain O395N1 (27) and V. cholerae 638, a HapA-deficient strain (41). V. cholerae strains were grown at 37°C on LB agar or in LB broth in a roller drum (New Brunswick Scientific, Edison, NJ). V. cholerae strains carrying V. tubiashii expression constructs or the empty vector were grown at 30°C in LB-1% NaCl, 100 μg ml−1 ampicillin, and 0.02% l-arabinose. Escherichia coli Top 10 cells (Invitrogen, Carlsbad, CA) were used for routine cloning and grown on LB agar supplemented with appropriate antibiotics. E. coli strain β2155 (thrB1004 pro thi strA hsdS lacZΔM15 [F9 lacZΔM15 laclq traD36 proA1 proB1] ΔdapA::erm [Ermr] pir::RP4 [kan {Kmr} from SM10]), which is auxotrophic for diaminopimelic acid (DAP) (8), was cultured in the presence of 1 mM DAP.
Larval-toxicity assay.
V. tubiashii strains cultured until the early stationary phase (optical density at 600 [OD600], ≅3.0) were centrifuged, and the supernatants were filter sterilized through a 0.22-μm polyethersulfone sterile filter. Ten-day-old oyster larvae (Coast Seafoods Company) in sterile seawater were aliquoted in a 96-well flat-bottomed plate (Nunc, Rochester, NY) at a density of approximately 20 larvae in 100 μl per well. Toxicity to larvae was assessed by adding various amounts of supernatants (0.5 to 2 μl) to the wells in triplicate, with various incubation times, to determine the threshold for toxicity. The same amount of growth medium was used as a control. Toxicity to larvae was determined by visualization with an inverted microscope. We considered oyster larvae dead when the larvae stopped moving, the velum was grossly damaged, and the larvae appeared to be darkened, similar to phenotypes described by Garland et al. (16). Concentrations of protease inhibitors were as follows: EDTA, 10 mM; 1,10-phenanthroline (PTL), 10 mM; tetraethylenepentamine (TEP), 10 mM; pepstatin A (PPA), 1 mM; phenylmethylsulfonyl fluoride (PMSF), 10 mM; and E-64, 1 mM. The concentration of the hemolysin inhibitor cholesterol was 50 ng/ml.
Enzyme assays.
V. tubiashii supernatants were assayed for proteolytic and hemolytic activity as previously described by Halpern et al. (18) and Chan and Foster (4), respectively. Proteolytic activity of the sterile filtered V. tubiashii supernatants was assessed by using azocasein. Briefly, 100 μl of supernatant was incubated with 400 μl of 1% azocasein for 30 min at 37°C. The reaction was stopped by the addition of 600 μl of 10% trichloroacetic acid and incubated on ice for 30 min before being centrifuged at about 16,000 × g for 5 min. Eight hundred microliters of supernatant from the centrifuged reaction was added to 200 μl of 1.8 N sodium hydroxide, and the absorbance at 420 nm was measured in a Bio-Rad SmartSpec Plus spectrophotometer. Hemolytic activity was determined by incubating 50 μl of 3.5% sheep blood (Colorado Serum Co.) in phosphate-buffered saline with 450 μl of either undiluted supernatant or a 10-fold dilution at 30°C for 1 hour. The reaction mixtures were centrifuged at 4,000 rpm for 10 min, and the absorbances at 405 nm were measured.
Zymography analyses.
Proteolytic activity was also assessed using zymogram gel electrophoresis. V. tubiashii filtered supernatants were resolved in 10% gelatin zymogram gels (Bio-Rad Laboratories) for 2 hours at 90 V. Gels were incubated in zymogram renaturing buffer (Bio-Rad) for 1 hour, followed by incubation in zymogram development buffer (Bio-Rad) for an additional hour. Gels were stained with Coomassie brilliant blue for 24 h at 37°C and visualized using a BioDocIt imaging station.
V. tubiashii genes.
All PCR and cloning reactions were conducted using standard procedures (1). Sequences for the vtpA (GenBank accession no. 1087431) and vthAB (GenBank accession no. 1087428) open reading frames (ORFs) were obtained by PCR, using genomic DNA of V. tubiashii strain RE22 with primer pairs designed based on homologous genes from various Vibrio species. Several primer pairs per gene were designed, and PCRs with all primer pair combinations were performed under low-stringency conditions to find a pair that successfully amplified a segment of the V. tubiashii genome (Table 1). These PCR products were then cloned into pCR2.1-TOPO by use of a TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Resulting clones were sequenced at the Oregon State University Center for Genome Research and Biocomputing core lab facility, using M13 forward and reverse primers. Sequences were verified by BLAST searches. To obtain the entire ORFs for these genes, inverse PCR was performed as previously described (42), using primers designed from the V. tubiashii sequences. In brief, genomic DNA of V. tubiashii was digested in 12 reactions with the following restriction enzymes: BamHI, BanII, PstI, EcoRV, SalI, BglII, XhoI, SacI, EcoRI, SmaI, SpeI, and PvuII. Following digestion and subsequent enzyme inactivation, intramolecular ligation reactions were performed with T4 DNA ligase (Invitrogen, Carlsbad, CA) at 16°C overnight. The ligation reaction product was used as the template for inverse PCR, using the primers shown in Table 1. Products obtained from inverse PCR were TA cloned into pCR 2.1-TOPO (Invitrogen, Carlsbad, CA) and sequenced using the M13 forward and reverse primers. The Oregon State University Center for Genomic Research and Biocomputing website was used for bioinformatic tools (http://bioinfo.cgrb.oregonstate.edu/).
TABLE 1.
Primers used in this study
| Primer and use | Sequencea (5′ to 3′) |
|---|---|
| Initial identification of vtpA and vthA | |
| HA5′deg1 | GGNCCNGGNGGNAAYCARAARAC |
| HA3′4 | GCCCGCTTTGACATACAGATCCGCATCGCCC |
| Cy5′2 | CGCGTCAATGTGGCACAAATGCGCTCGGTACAATC |
| Cy3′2 | TAGGCCCCAAACTTGGTTCCAACTGCCGTGACAGC |
| Inverse PCR | |
| 5HapOut | GTCCGTACCGAAGTTGTATTG |
| 3HapOut | GACAGCAAACAGCACGTTTGA |
| 5CytOut | GCCAATGTACGTGCGGCGGTTCGCCCAGCTTTGG |
| 3CytOut | GCAACGACAGGATGGTCAATTCCACGTACGGTTCG |
| pBAD expression cloning | |
| 5hapexp | GAGGAATAATAAATGAAACAACGTCAAATGCTTTG |
| 3hapexp | TTAGTCTAATCTTAGTGTCACGC |
| 5cytexp | GAGGAATAATAAATGAAAACTTCTACAATTTTCAC |
| 3cytexp | TTAAAGTTTGATTTGCTGTAGTG |
| vtpA internal deletion | |
| 5HapDel | GGGGGGCGGCCGCCAATACAACTTCGGTACGGAC |
| 3HapDel | GGGGGACTAGTTCAAACGTGCTGTTTGCTGTC |
N = A, G, C, or T; Y = C or T; R = A or G.
For heterologous expression of V. tubiashii genes vthA and vtpA in V. cholerae, the V. tubiashii ORFs were PCR amplified using primers designed based on the start and stop codons, with the addition of the Shine-Dalgarno ribosome binding site sequence incorporated into the 5′ primer. The PCR product was cloned into pBAD-TOPO using a pBAD TOPO TA expression kit (Invitrogen). Plasmids carrying the inserts in the desired orientation (i.e., placing the gene under the control of the arabinose-inducible promoter) were identified and confirmed by sequencing. V. cholerae strains O395N1 and 638 were transformed with the V. tubiashii genes via electroporation.
V. tubiashii mutants.
A vtpA V. tubiashii mutant was constructed by insertional mutagenesis. Briefly, a 773-bp internal DNA fragment of the gene located 627 bases from the start and 424 bases from the stop was amplified by PCR (Table 1) and cloned into the vector pWM91 (29). Since V. tubiashii strain RE22 is naturally ampicillin resistant, a kanamycin resistance cassette was also cloned into this vector. The resulting construct was transformed into chemically competent E. coli β2155 cells. Transformed cells were selected on LB agar with kanamycin and DAP and were subsequently conjugated with V. tubiashii RE22 on LB-1% NaCl agar plus DAP and incubated at 30°C overnight. Conjugates were harvested by suspending the biomass from the plate in LB-1% NaCl broth and were then plated on LB-1% NaCl agar supplemented with kanamycin but without DAP and incubated at 30°C.
RESULTS
Extracellular protease and hemolysin production by oyster larva-pathogenic and -nonpathogenic bacterial isolates.
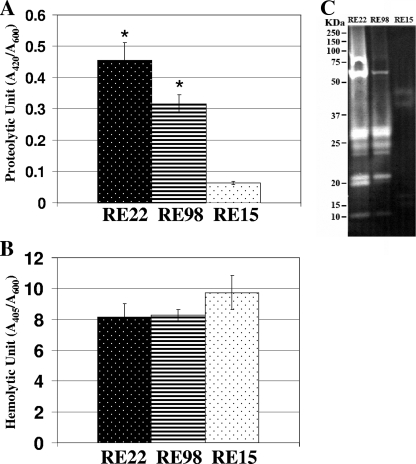
Previous studies identified extracellular products, such as a metalloprotease and a cytolysin/hemolysin, from culture supernatants of V. tubiashii strain ATCC 19105 (9, 20). To determine if bacterial strains isolated from diseased Pacific oyster larvae also produce these secreted proteins, we grew V. tubiashii strains RE22 and RE98, which were previously described as pathogenic strains (14), and RE15, which was categorized as a nonpathogenic bacterial isolate. Harvested culture supernatants were filter sterilized and assayed for proteolytic and hemolytic activities. Figure 1A shows that culture supernatants of RE22 and RE98 contained approximately sevenfold and fivefold higher levels of protease, respectively, than that of RE15 in the quantitative azocasein assay. Zymography is a sensitive and functional assay for detecting proteolytic activity. As shown in Fig. 1C, culture supernatants of RE22 and RE98 showed similar profiles of multiple proteolytic bands of different sizes on the zymogram gel. In contrast, the intensities of proteolytic bands from supernatants of RE15 were much lower, and fewer bands were apparent in the zymographic gel. On the other hand, the hemolysin quantitative assay with sheep blood revealed that culture supernatants of RE15 contained approximately 1.2 times more hemolytic activity than did those of RE22 and RE98 (Fig. 1B).
FIG. 1.
Proteolytic and hemolytic analyses of bacterial culture supernatants. Quantitative data are shown for proteolytic (A) and hemolytic (B) activities of culture supernatants of V. tubiashii pathogenic strains RE22 and RE98 and the nonpathogenic bacterial isolate RE15. (C) Zymography analysis of these supernatants. Bacteria were grown in LB medium supplemented with 1% NaCl at 25°C and were harvested at an OD600 of approximately 3.0. Proteolytic and hemolytic activities were determined using azocasein and sheep blood, respectively, as described in Materials and Methods. The error bars indicate standard deviations (n = 3). Data for proteolytic and hemolytic activities were evaluated by Student's t test (*, P < 0.01 compared with RE15). Bands of proteolytic activity in the zymogram gel are shown as clear protein bands in a dark background. The molecular masses (kDa) on the left indicate the positions of molecular size markers.
Growth-dependent production of extracellular protease and hemolysin in V. tubiashii.
For further analyses of the extracellular proteins produced by V. tubiashii, we used strain RE22 because it has been described as one of the most pathogenic isolates (14). Similar proteins produced by other Vibrio species are known to be regulated by the growth phase of the bacteria (30). To determine if the growth phase had any effect on the production of secreted protease and hemolysin in V. tubiashii, we assayed for the production of these proteins at multiple points during bacterial growth. As shown in Fig. 2B, V. tubiashii began to produce detectable levels of hemolysin at the early log phase (OD600, ≅1.8), with the production reaching the highest level during the late log phase and then declining shortly after the mid-stationary phase. In contrast, protease production was gradually increased during the late log phase (OD600, ≅3.2), and the proteolytic activity was the highest at the mid-stationary phase (Fig. 2A). Thus, while V. tubiashii hemolysin is highly produced until early stationary phase, protease production reached its highest levels at the late stationary phase.
FIG. 2.
Relationship between growth and proteolytic (A) and hemolytic (B) activities of culture supernatants of V. tubiashii strain RE22. Bacteria were grown in LB medium supplemented with 1% NaCl at 25°C, and samples were harvested at different times during bacterial growth. The error bars indicate standard deviations (n = 3).
Effects of inhibitors on protease and hemolysin functions and toxicity to Pacific oyster larvae.
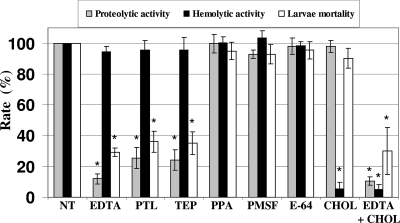
We hypothesized that extracellular metalloprotease and/or hemolysin produced by V. tubiashii may be required for toxicity to Pacific oyster larvae. To determine the effects of proteolytic and hemolytic enzymes secreted by V. tubiashii on the mortality of the larvae, we treated culture supernatants with various protease and hemolysin inhibitors. The data in Fig. 3 show that the proteolytic activity was severely reduced by metalloprotease inhibitors, including EDTA, TEP, and PTL, whereas the activity was not dramatically affected by treatment with the aspartic protease inhibitor PPA, the serine protease inhibitor PMSF, and the cysteine protease inhibitor E-64. None of these protease inhibitors showed any notable effects on hemolytic activity (Fig. 3). Cholesterol has been described as a strong hemolysin inhibitor in V. tubiashii (20). Consistent with this finding, the hemolytic activity was severely decreased by treatment of culture supernatants with cholesterol (Fig. 3). Interestingly, the mortality of Pacific oyster larvae was not affected by the loss of hemolytic activity but was strongly inhibited by the presence of the metalloprotease inhibitors EDTA, TEP, and PTL and not by other classes of protease inhibitors (PPA, PMSF, and the cysteine protease inhibitor E-64) or the hemolysin inhibitor cholesterol (Fig. 3).
FIG. 3.
Effects of protease and hemolysin inhibitors on proteolytic and hemolytic activities and on toxicity to Pacific oyster larvae. Enzymatic activities and toxicity levels are shown as percentages of those for nontreated samples. For the toxicity assay, filter-sterilized supernatants were added to a final concentration of 1%. The error bars indicate standard deviations (n = 3). Data for proteolytic and hemolytic activities were evaluated by Student's t test (*, P < 0.01 compared with the nontreated control). Data for larval mortality were evaluated by chi-square test (*, P < 0.05 compared with nontreated control). NT, nontreated; TEP, tetraethylene pentamine; OPA, 1,10-phenanthroline; PMSF, phenylmethylsulfonyl fluoride; PPA, pepstatin A; CHOL, cholesterol.
Cloning of metalloprotease gene and sequence analyses.
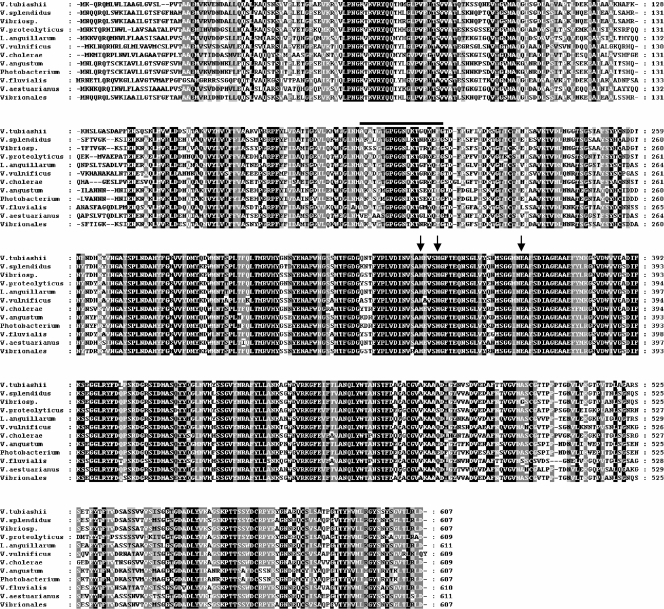
We successfully obtained DNA fragments containing the entire ORF of the metalloprotease gene (vtpA) (GenBank accession no. EU675309). The coding region of vtpA is 1,824 bp, sufficient to encode a putative polypeptide of 607 amino acid residues (Fig. 4). The predicted size of the protein is approximately 65.5 kDa, based on the deduced amino acid sequence. Frequently, these types of metalloproteases are preceded by a signal peptide, an N-terminal propeptide followed by a C-terminal propeptide, which is removed from the mature protease after autoprocessing (32, 49). The apparent molecular mass (35 kDa) of the metalloprotease from V. tubiashii strain ATCC 19105, which was previously described by Delston et al. (9), matched well with the theoretical molecular mass of mature VtpA, assuming that C-terminal processing removes a 10-kDa segment. Amino acid sequence alignment of VtpA with proteins produced by other bacterial species revealed that VtpA shared the highest homology (76% identity and 88% similarity) with zinc metalloproteases of a Vibrio sp. and a Vibrio splendidus strain (Fig. 4). VtpA also shared 81 to 87% similarity with proteases from Vibrio proteolyticus, a Vibrionales bacterium, Vibrio (Listonella) anguillarum, V. vulnificus, V. cholerae, Vibrio aestuarianus, Vibrio angustum, Photobacterium sp., and Vibrio fluvialis. These data show that this type of protease is widespread among Vibrio and other marine bacterial species. Previous studies have categorized microbial zinc metalloproteases into several groups, based on critical and functional motifs and key residues (23, 31). VtpA possesses a zinc binding motif (HEXXH) at residues 341H to 345H as well as another critical ligand, 365E, located 18 amino acid residues from the HEXXH motif (Fig. 4, arrows). A previous study by Delston et al. (9) determined the N-terminal amino acid sequence (AQATGTGPGGNQKTGQYNFG) of a mature metalloprotease secreted by V. tubiashii strain ATCC 19105. Our results showed that these 20 amino acid residues were 100% conserved in the V. tubiashii VtpA sequence (Fig. 4).
FIG. 4.
Alignment of deduced V. tubiashii metalloprotease amino acid sequence with those of various bacterial species. Numbers on the right refer to the positions of the amino acid residues. The black bar indicates the previously identified region by Delston et al. (9). Arrows indicate critical residues for zinc binding. Black shaded areas indicate identical amino acids in all strains, and gray shaded areas indicate identical or similar amino acids in eight or more strains at any position. The following sequences were aligned using ClustalW: zinc metalloproteases of Vibrio sp. strain MED222 (GenBank accession no. NZ_AAND01000005), V. splendidus strain 12B01 (accession no. ZP_00990032), V. proteolyticus (accession no. AAA27548), Vibrionales bacterium strain SWAT-3 (ZP_01816166), Vibrio (Listonella) anguillarum strain M93Sm (accession no. AAR88093), Vibrio vulnificus strain YJ016 (accession no. NP_937521), V. cholerae strain 623-39 (accession no. ZP_01980763), V. aestuarianus strain 01/32 (accession no. AAU04777), V. angustum strain S14 (accession no. ZP_01236251), Photobacterium sp. strain SKA34 (accession no. ZP_01158654), and V. fluvialis strain AQ0005 (accession no. BAB86344).
Cloning of hemolysin genes and sequence analyses.
The coding region of the V. tubiashii hemolysin gene, vthA (GenBank accession no. EU675308), from strain RE22 was 1,416 bp, encoding a putative polypeptide of 471 amino acids (Fig. 5C). The predicted molecular mass of the protein was approximately 53 kDa, which matched well with that of the previously described hemolysin produced by V. tubiashii strain ATCC 19105 (20). VthA showed significant homology only with the V. vulnificus hemolysin, VvhA (76% identity and 86% similarity). vvhA has been reported to be cotranscribed with the upstream gene, vvhB, as an operon (6). Our sequence data revealed that a putative vvhB homolog (vthB) (GenBank accession no. 1087428) is located upstream of vthA in V. tubiashii strain RE22 (Fig. 5A). VthB shares high homology with VvhB (60% identity and 77% similarity) (Fig. 5B). In addition, the genetic arrangement of vthA and vthB in V. tubiashii is similar to that in V. vulnificus, with only one nucleotide gap between these genes (data not shown). The sequence of the first 17 amino acids (DDYVPVVEKVYYITSSK) of the hemolysin secreted by V. tubiashii strain ATCC 19105 was previously identified by Korthary et al. (20). We also observed the identical sequence in VthA of V. tubiashii strain RE22, after a putative signal sequence (Fig. 5C).
FIG. 5.
Alignments of deduced amino acid sequences of VthA and VthB with V. vulnificus VvhA and VvhB. (A) Genetic organization of the vthA and vthB genes in V. tubiashii strain RE22. ClustalW alignments were done with VthB (B) and VthA (C) and their V. vulnificus homologs (GenBank accession no. AB124803). The black bar indicates the region previously identified by Kothary et al. (20).
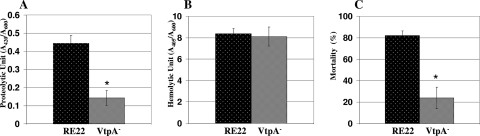
Effects of loss of VtpA on pathogenicity of V. tubiashii for Pacific oyster larvae.
To see the effects of VtpA on protease production and mortality of Pacific oyster larvae, we isolated a VtpA-deficient mutant of V. tubiashii strain RE22. Culture supernatants derived from the mutant were tested for toxicity to Pacific oyster larvae. Figure 6A and B show that compared to the wild-type strain, the mutant produced markedly reduced levels of protease but comparable levels of hemolysin. These culture supernatants were also assayed for lethality to oyster larvae. The mortality rates of Pacific oyster larvae treated with supernatants from the mutant strain were significantly lower than those for larvae treated with the parent strain (Fig. 6C). Thus, vtpA appears to be responsible for the majority of protease production and toxicity in culture supernatants of V. tubiashii strain RE22.
FIG. 6.
Analyses of supernatants from a VtpA-deficient mutant of V. tubiashii strain RE22. Protease (A) and hemolysin (B) production, as well as toxicity to oyster larvae (C), was compared to that of the wild-type strain. Although the mutant strain, but not the wild-type strain, was grown in the presence of 50 μg/ml kanamycin, all cells were harvested at an OD600 of approximately 3.0. For the toxicity assay, filter-sterilized supernatants were added to a final concentration of 0.5%. The error bars indicate standard deviations (n = 3). Data for proteolytic and hemolytic activities were evaluated by Student's t test (*, P < 0.01 compared with the wild type). Data for larval mortality were evaluated by chi-square test (*, P < 0.05 compared with the wild type).
Metalloprotease and hemolysin production and toxicity by V. cholerae carrying the vtpA and vthA genes.
vtpA and vthA were introduced into a heterologous host to examine if individual expression of the V. tubiashii metalloprotease or hemolysin gene results in larval toxicity. For this purpose, we chose V. cholerae strain O395N1 and strain 638, a HapA-deficient mutant. These strains produce minimal amounts of protease or hemolysin (2, 15, 28). The vtpA and vthA genes from V. tubiashii strain RE22 were transformed into these V. cholerae strains. V. cholerae strains carrying these V. tubiashii genes successfully produced and secreted considerable amounts of protease and hemolysin, respectively, compared to the vector control (Fig. 7A and B). Interestingly, culture supernatants of both O395N1 and 638 strains carrying the vtpA gene showed much higher toxicity to Pacific oyster larvae than did those carrying either vector or the vthA plasmid (Fig. 7C). Thus, the metalloprotease alone, but not hemolysin, caused high levels of larval mortality.
FIG. 7.
Expression of vtpA and vthA in V. cholerae. Protease (A) and hemolysin (B) production, as well as toxicity to oyster larvae (C), was analyzed for V. cholerae strains O395N1 and 638 carrying the empty vector (pBAD-TOPO) or the V. tubiashii metalloprotease (pBAD-vtpA) or hemolysin (pBAD-vthA) gene. For the toxicity assay, filter-sterilized supernatants were added to a final concentration of 1%. The error bars indicate standard deviations (n = 3). Data for proteolytic and hemolytic activities were evaluated by Student's t test (*, P < 0.01 compared with the empty vector). Data for larval mortality were evaluated by the chi-square test (*, P < 0.05 compared with the empty vector).
DISCUSSION
Consistent with a previous report (48), we observed high toxicity of culture supernatants of V. tubiashii strains to C. gigas larvae. Supernatants of several bacterial strains isolated from Pacific Northwest hatcheries were tested for proteolytic and hemolytic activities. Two highly pathogenic V. tubiashii strains, but not a nonpathogenic bacterial isolate, produced high levels of extracellular protease. In contrast, all three strains produced similar levels of hemolysin, suggesting a possible correlation between protease, but not hemolysin, production and the toxicity of V. tubiashii supernatants to Pacific oyster larvae.
Although neither protease nor hemolysin is produced at a low cell density, hemolytic activity increased early during growth and decreased when the culture reached the stationary phase. In contrast, protease production increased steadily during growth and reached the highest level at stationary phase. These results indicate that expression of these proteins responds to the cell density of the culture. It has been documented that production of exoprotease and hemolysin is regulated by quorum sensing in V. vulnificus, as well as many other Vibrio species (19, 30, 43), in which the metalloprotease gene is activated at high cell population densities, whereas the hemolysin gene is negatively regulated under these conditions. Our data show a steep decrease in hemolysin production after the mid-stationary phase and a gradual increase in production of extracellular protease at the early stationary phase, suggesting that V. tubiashii may take advantage of a quorum-sensing system similar to that of V. vulnificus.
Hemolysin production by V. tubiashii ATCC 19105 has been described previously (20), and the authors identified the first 17 amino acid residues of the purified protein. They revealed that 12 of the 17 residues are identical to those of the V. vulnificus hemolysin, VvhA. Here we report the nucleotide sequences of the hemolysin genes (vthA) from two V. tubiashii strains. Our study revealed that the N-terminal region of the ATCC 19105 hemolysin was identical to that of VthA produced by V. tubiashii strains RE22 and RE98. Our amino acid sequence analysis of the entire gene product further clarified that VthA indeed shares significant homology with VvhA. In V. vulnificus, vvhA and the smaller upstream gene vvhB are clustered as an operon, yet the function of vvhB is unknown to date (6). We found a putative homolog of vvhB (vthB) upstream of vthA, indicating that V. tubiashii produces a similar hemolysin to that of V. vulnificus. The functional role of the vthB gene in V. tubiashii is currently under investigation.
A previous study with the extracellular protease of V. tubiashii ATCC 19105 demonstrated that 20 residues of the N-terminal sequence of secreted and processed metalloprotease are similar to those in other varieties of Vibrio species (9). In this study, we isolated the gene (vtpA) encoding a zinc metalloprotease from two pathogenic V. tubiashii strains. Our data showed that the 20 residues of the metalloprotease purified from strain ATCC 19105 are identical to those of VtpA. Sequence alignment of VtpA with several proteases produced by different Vibrio species showed high sequence similarity to these zinc metalloproteases. In addition, Delston et al. (9) reported that the proteolytic activity of the purified protease from ATCC 19105 was impaired by the zinc metalloprotease-specific inhibitor Zincov. Our results showed that metalloprotease inhibitors such as EDTA, TEP, and PTL dramatically reduced proteolytic activity in culture supernatants of V. tubiashii. Taken together, the results show that it is very likely that VtpA functions as a zinc metalloprotease.
Here we examined the roles of the V. tubiashii extracellular hemolysin and protease in toxicity of culture supernatants to Pacific oyster larvae. We concluded that the metalloprotease VtpA, but not the hemolysin VthA, acts as one of the critical factors for the toxicity of V. tubiashii supernatants on Pacific oyster larvae, based on the following evidence: (i) treatment of V. tubiashii culture supernatants with metalloprotease inhibitors severely diminished the toxicity to Pacific oyster larvae, whereas other classes of protease inhibitors or a specific inhibitor of hemolysin did not affect the lethality; (ii) strains of V. cholerae expressing the vtpA gene, but not vthA or the vector plasmid, caused high larval mortality; and (iii) a VtpA-negative mutant strain of V. tubiashii showed a significant loss of toxicity to the oyster larvae.
Extracellular metalloproteases produced by marine Vibrio spp. have been well documented as pathogenicity factors in several cases. For example, the zinc metalloprotease is involved in the invasive mechanism of the fish pathogen V. anguillarum (33). In V. splendidus, the metalloprotease is essential for toxicity when the extracellular products are injected into oysters (25). Moreover, it has been reported that V. aestuarianus zinc metalloprotease, which shares homology with that of V. cholerae, is responsible for virulence against the oyster Crassostrea gigas (50). A subsequent study by Labreuche et al. (21) supported the observation that total protease levels in V. aestuarianus-injected oysters are well correlated with a decrease in phagocytic activities by the host, suggesting that a variety of marine Vibrio species take advantage of similar proteases to cause disease in oysters and humans.
With VtpA described as a critical factor in V. tubiashii toxicity, the functional role of VtpA as a pathogenicity factor has yet to be characterized. Previous studies reported that toxicity of culture supernatants of V. proteolyticus to bivalve larvae of the native oyster (Ostrea edulis) was due to an extracellular protease, since the enzyme rapidly broke down gill tissues of blue mussels (Mytilus edulis) (35, 36). A subsequent study by the same group revealed that production of the protease was maximal during the late exponential phase of growth, and the exoprotease was inhibited by EDTA but not by pepstatin A or PMSF (34). Although it is still uncertain how VtpA in V. tubiashii is involved in the bacillary necrosis disease mechanism, these previous findings and our present data suggest that VtpA contributes to pathogenicity by degrading tissues. Interestingly, Takahashi et al. (48) found that treatment of V. tubiashii strain ATCC 19106 with ovoglobulin, a protein derived from hen egg whites which also acts as a strong metalloprotease inhibitor, significantly lowered the mortality rate of Pacific oyster larvae, suggesting that the extracellular metalloprotease is critical for the toxicity of the strain. They further demonstrated that the addition of ovoglobulins suppressed the growth of ATCC 19106 in gelatin-seawater broth, which indicates that the extracellular protease is required for bacterial growth under these conditions. Therefore, VtpA may contribute to the destruction of the host tissues, which may in turn provide the bacteria with nutrients under poor growth conditions.
There are three other classes of proteases based on catalytic mechanisms (serine, cysteine, and aspartic proteases), some of which have been reported as important pathogenicity factors in marine Vibrio species. For instance, for Vibrio harveyi, a cysteine protease acts as a major exotoxin in the tiger prawn (26), while for Vibrio alginolyticus, a serine protease is reported to be the dominant protease secreted as well as a major pathogenicity factor in the tiger prawn (5). In this study, however, only metalloprotease inhibitors severely impaired the toxicity of culture supernatants to Pacific oyster larvae. Interestingly, culture supernatants of a VtpA-deficient mutant of V. tubiashii strain RE22 still produced approximately 30% proteolytic activity compared to that of the wild type, indicating the presence of several other proteases produced by V. tubiashii. The facts that multiple proteolytic bands were observed in our zymography assay and that a putative second extracellular metalloprotease gene is present in the Pacific oyster pathogen V. splendidus strain LPG32 (GenBank accession no. ZP_00989149.1) suggest that there might be a redundant vtpA homolog in addition to other types of protease genes in V. tubiashii. Similarly, the metalloprotease-deficient mutant of LPG32 also produced approximately 20% of the proteolytic activity of the wild-type parent (25). An intriguing future study might attempt to define the roles of these other putative proteases in the toxicity of V. tubiashii culture supernatants to oyster larvae.
Although we have shown that the metalloprotease, not hemolysin, is the major pathogenicity factor in supernatants of V. tubiashii, we should acknowledge that hemolysin as well as some of the other secreted proteins may contribute to the overall pathogenicity of V. tubiashii. Nottage and Birbeck (37) described a heat-stable ciliostatic toxin, a lethal exotoxin produced by V. tubiashii and V. alginolyticus, which degrades gill segments of blue mussels. It has also been described that a heat-stable toxin produced by Vibrio pectenicida is toxic to king scallop hemocytes (22). Moreover, the presence of a type III secretion system has been reported for V. tubiashii (38), suggesting that the bacterium produces host-interacting effector proteins. Therefore, further studies are essential to fully understand the disease mechanisms of shellfish larval vibriosis.
In summary, we have focused on identifying the critical pathogenicity factors produced in supernatants of V. tubiashii. Our data revealed that pathogenic strains of V. tubiashii produced both extracellular protease and hemolysin in vitro, while a nonpathogenic isolate did not produce any detectable levels of extracellular protease. Sequence analyses of genes encoding these proteins revealed that the protease belongs to a family of zinc metalloproteases which is widespread among Vibrio species, whereas the hemolysin shared significant homology only with the hemolysin/cytolysin of V. vulnificus. Moreover, we have concluded that the metalloprotease (VtpA) acts as one of the critical virulence factors for the pathogenicity of V. tubiashii to Pacific oyster larvae.
Acknowledgments
We thank Ralph Elston for providing the V. tubiashii strains as well as many helpful insights. We thank Anisia Silva for generously providing V. cholerae strain 638. We are also grateful to Coast Seafoods Company for generously providing Pacific oyster larvae.
This work was supported in part by a USDA-SBIR grant and NIH grant AI-063121-02.
Footnotes
Published ahead of print on 2 May 2008.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. S. Truhl. 1991. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 2.Benitez, J. A., A. J. Silva, and R. A. Finkelstein. 2001. Environmental signals controlling production of hemagglutinin/protease in Vibrio cholerae. Infect. Immun. 69:6549-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, C. 1981. A study of two shellfish-pathogenic Vibrio strains isolated from a Long Island hatchery during a recent outbreak of disease. J. Shellfish Res. 1:83-87. [Google Scholar]
- 4.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, F. R., P. C. Liu, and K. K. Lee. 2000. Lethal attribute of serine protease secreted by Vibrio alginolyticus strains in kuruma prawn Penaeus japonicus. Z. Naturforsch. Sect. C 55:94-99. [DOI] [PubMed] [Google Scholar]
- 6.Choi, H. K., N. Y. Park, D. Kim, H. J. Chung, S. Ryu, and S. H. Choi. 2002. Promoter analysis and regulatory characteristics of vvhBA encoding cytolytic hemolysin of Vibrio vulnificus. J. Biol. Chem. 277:47292-47299. [DOI] [PubMed] [Google Scholar]
- 7.Croxatto, A., J. Lauritz, C. Chen, and D. L. Milton. 2007. Vibrio anguillarum colonization of rainbow trout integument requires a DNA locus involved in exopolysaccharide transport and biosynthesis. Environ. Microbiol. 9:370-382. [DOI] [PubMed] [Google Scholar]
- 8.Dehio, C., and M. Meyer. 1997. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179:538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delston, R. B., M. H. Kothary, K. A. Shangraw, and B. D. Tall. 2003. Isolation and characterization of zinc-containing metalloprotease expressed by Vibrio tubiashii. Can. J. Microbiol. 49:525-529. [DOI] [PubMed] [Google Scholar]
- 10.Elston, R. A. 1999. Development, histology and health management of seed oysters. World Aquaculture Society, Baton Rouge, LA.
- 11.Elston, R. A. 1984. Prevention and management of infectious diseases in intensive mollusc husbandry. J. World Maricult. Soc. 15:284-300. [Google Scholar]
- 12.Elston, R. A., and L. Leibovitz. 1980. Pathogenesis of experimental vibriosis in larval American oysters, Crassostrea gigas. Can. J. Fish. Aquat. Sci. 37:964-978. [Google Scholar]
- 13.Elston, R. A., L. Leibovitz, D. Relyea, and J. Zatila. 1981. Diagnosis of vibriosis in a commercial oyster hatchery epizootic, a case history. Aquaculture 24:53-62. [Google Scholar]
- 14.Estes, R. M., C. S. Friedman, R. A. Elston, and R. P. Herwig. 2004. Pathogenicity testing of shellfish hatchery bacterial isolates on Pacific oyster Crassostrea gigas larvae. Dis. Aquat. Org. 58:223-230. [DOI] [PubMed] [Google Scholar]
- 15.Figueroa-Arredondo, P., J. E. Heuser, N. S. Akopyants, J. H. Morisaki, S. Giono-Cerezo, F. Enriquez-Rincon, and D. E. Berg. 2001. Cell vacuolation caused by Vibrio cholerae hemolysin. Infect. Immun. 69:1613-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garland, C. D., G. V. Nash, C. E. Summer, and T. A. McMeekin. 1983. Bacterial pathogens of oyster larvae (Crassostrea gigas) in a Tasmanian hatchery. Aust. J. Mar. Freshw. Res. 34:483-487. [Google Scholar]
- 17.Hada, H. S., P. A. West, J. V. Lee, J. Stemmler, and R. R. Colwell. 1984. Vibrio tubiashii sp. nov., a pathogen of bivalve mollusks. Int. J. Syst. Bacteriol. 34:1-4. [Google Scholar]
- 18.Halpern, M., H. Gancz, and Y. Kashi. 2003. Vibrio cholerae hemagglutinin/protease degrades chironomid egg masses. Appl. Environ. Microbiol. 69:4200-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, S. Y., S. E. Lee, Y. R. Kim, C. M. Kim, P. Y. Ryu, H. E. Choy, S. S. Chung, and J. H. Rhee. 2003. Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol. Microbiol. 48:1647-1664. [DOI] [PubMed] [Google Scholar]
- 20.Kothary, M. H., R. B. Delston, S. K. Curtis, B. A. McCardell, and B. D. Tall. 2001. Purification and characterization of vulnificolysin-like cytolysin produced by Vibrio tubiashii. Appl. Environ. Microbiol. 67:3707-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labreuche, Y., C. Lambert, P. Soudant, V. Boulo, A. Huvet, and J. L. Nicolas. 2006. Cellular and molecular hemocyte responses of the Pacific oyster, Crassostrea gigas, following bacterial infection with Vibrio aestuarianus strain 01/32. Microbes Infect. 8:2715-2724. [DOI] [PubMed] [Google Scholar]
- 22.Lambert, C., J. L. Nicolas, and V. Bultel. 2001. Toxicity to bivalve hemocytes of pathogenic Vibrio cytoplasmic extract. J. Invertebr. Pathol. 77:165-172. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J. H., G. T. Kim, J. Y. Lee, H. K. Jun, J. H. Yu, and I. S. Kong. 1998. Isolation and sequence analysis of metalloprotease gene from Vibrio mimicus. Biochim. Biophys. Acta 1384:1-6. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. E., P. Y. Ryu, S. Y. Kim, Y. R. Kim, J. T. Koh, O. J. Kim, S. S. Chung, H. E. Choy, and J. H. Rhee. 2004. Production of Vibrio vulnificus hemolysin in vivo and its pathogenic significance. Biochem. Biophys. Res. Commun. 324:86-91. [DOI] [PubMed] [Google Scholar]
- 25.Le Roux, F., J. Binesse, D. Saulnier, and D. Mazel. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 73:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, P. C., and K. K. Lee. 1999. Cysteine protease is a major exotoxin of pathogenic luminous Vibrio harveyi in the tiger prawn, Penaeus monodon. Lett. Appl. Microbiol. 28:428-430. [DOI] [PubMed] [Google Scholar]
- 27.Mekalanos, J. J., D. J. Swartz, G. D. N. Pearson, N. Harford, F. Groyne, and M. de Wilde. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551-557. [DOI] [PubMed] [Google Scholar]
- 28.Mel, S. F., K. J. Fullner, S. Wimer-Mackin, W. I. Lencer, and J. J. Mekalanos. 2000. Association of protease activity in Vibrio cholerae vaccine strains with decreases in transcellular epithelial resistance of polarized T84 intestinal epithelial cells. Infect. Immun. 68:6487-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 30.Milton, D. L. 2006. Quorum sensing in vibrios: complexity for diversification. Int. J. Med. Microbiol. 296:61-71. [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi, S., and S. Shinoda. 2000. Microbial metalloproteases and pathogenesis. Microbes Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 32.Miyoshi, S., H. Wakae, K. Tomochika, and S. Shinoda. 1997. Functional domains of a zinc metalloprotease from Vibrio vulnificus. J. Bacteriol. 179:7606-7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norqvist, A., B. Norrman, and H. Wolf-Watz. 1990. Identification and characterization of a zinc metalloprotease associated with invasion by the fish pathogen Vibrio anguillarum. Infect. Immun. 58:3731-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nottage, A. S., and T. H. Birkbeck. 1987. Purification of a proteinase produced by the bivalve pathogen Vibrio alginolyticus NCMB 1339. J. Fish Dis. 10:211-220. [Google Scholar]
- 35.Nottage, A. S., and T. H. Birkbeck. 1987. The role of toxins in Vibrio infections of bivalve mollusca. Aquaculture 67:244-246. [Google Scholar]
- 36.Nottage, A. S., and T. H. Birkbeck. 1986. Toxicity of marine bivalves of culture supernatant fluids of the bivalve-pathogenic Vibrio strain NCMB 1338 and other marine vibrios. J. Fish Dis. 9:249-256. [Google Scholar]
- 37.Nottage, A. S., P. D. Sinclair, and T. H. Birkbeck. 1989. Role of low-molecular weight ciliostatic toxins in vibriosis of bivalve mollusks. J. Aquat. Anim. Health 1:180-186. [Google Scholar]
- 38.Park, K. S., T. Ono, M. Rokuda, M. H. Jang, K. Okada, T. Iida, and T. Honda. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72:6659-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perkins, F. O. 1993. Infectious diseases of mollusks, p. 255-287. In J. A. Couch and J. W. Fournie (ed.), Pathobiology of marine and estuarine organisms. CRC Press, Boca Raton, FL.
- 40.Prado, S., J. L. Romalde, J. Montes, and J. L. Barja. 2005. Pathogenic bacteria isolated from disease outbreaks in shellfish hatcheries. First description of Vibrio neptunius as an oyster pathogen. Dis. Aquat. Organ. 67:209-215. [DOI] [PubMed] [Google Scholar]
- 41.Robert, A., A. Silva, J. A. Benitez, B. L. Rodriguez, R. Fando, J. Campos, D. K. Sengupta, M. Boesman-Finkelstein, and R. A. Finkelstein. 1996. Tagging a Vibrio cholerae El Tor candidate vaccine strain by disruption of its hemagglutinin/protease gene using a novel reporter enzyme: Clostridium thermocellum endoglucanase A. Vaccine 14:1517-1522. [DOI] [PubMed] [Google Scholar]
- 42.Saltikov, C. W., and D. K. Newman. 2003. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. USA 100:10983-10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao, C. P., and L. I. Hor. 2001. Regulation of metalloprotease gene expression in Vibrio vulnificus by a V. harveyi LuxR homologue. J. Bacteriol. 183:1369-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinoda, S. 1999. Protein toxins produced by pathogenic vibrios. J. Nat. Toxins 8:259-269. [PubMed] [Google Scholar]
- 45.Sindermann, C. J. 1988. Vibriosis of larval oysters, p. 271-274. In C. J. Sindermann and D. V. Lightner (ed.), Disease diagnosis and control in North American aquaculture. Elsevier, Amsterdam, The Netherlands.
- 46.Su, J. H., M. C. Chang, Y. S. Lee, I. C. Tseng, and Y. C. Chuang. 2004. Cloning and characterization of the lipase and lipase activator protein from Vibrio vulnificus CKM-1. Biochim. Biophys. Acta 1678:7-13. [DOI] [PubMed] [Google Scholar]
- 47.Sugumar, G., T. Nakai, Y. Hirata, D. Matsubara, and K. Muroga. 1998. Pathogenicity of Vibrio splendidus biovar II, the causative bacterium of bacillary necrosis of Japanese oyster larvae. Fish Pathol. 33:79-84. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi, K. G., A. Nakamura, and K. Mori. 2000. Inhibitory effects of ovoglobulins on bacillary necrosis in larvae of the pacific oyster, Crassostrea gigas. J. Invertebr. Pathol. 75:212-217. [DOI] [PubMed] [Google Scholar]
- 49.Tang, B., S. Nirasawa, M. Kitaoka, and K. Hayashi. 2002. The role of the N-terminal propeptide of the pro-aminopeptidase processing protease: refolding, processing, and enzyme inhibition. Biochem. Biophys. Res. Commun. 296:78-84. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, F. L., K. E. Klose, and the AVIB Group. 2005. Vibrio2005: the first international conference on the biology of vibrios. J. Bacteriol. 188:4592-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tubiash, H. S., P. E. Chanley, and E. Leifson. 1965. Bacillary necrosis, a disease of larval and juvenile bivalve mollusks. I. Etiology and epizootiology. J. Bacteriol. 90:1036-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tubiash, H. S., R. R. Colwell, and R. Sakazaki. 1970. Marine vibrios associated with bacillary necrosis, a disease of larval and juvenile bivalve mollusks. J. Bacteriol. 103:2721-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyckoff, E. E., A. R. Mey, and S. M. Payne. 2007. Iron acquisition in Vibrio cholerae. Biometals 20:405-416. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, X. H., and B. Austin. 2005. Haemolysins in Vibrio species. J. Appl. Microbiol. 98:1011-1019. [DOI] [PubMed] [Google Scholar]