Abstract
In recent years, the absence of acquired antimicrobial resistance has become an important criterion to evaluate the biosafety of lactobacilli used as industrial starter or probiotic cultures. At present, however, standards for susceptibility testing of Lactobacillus strains or approved guidelines for interpreting the test results are not available. Hence, this study was carried out to contribute to the establishment of a standardized procedure for antimicrobial susceptibility testing of lactobacilli. The results obtained by testing 104 strains of the Lactobacillus acidophilus group were compared based on broth microdilution, disk diffusion, and Etest. Except for some specific agent-related effects, agreement between MICs resulting from the broth microdilution method and the Etest was good. In addition, inhibition zone diameters determined with disk diffusion correlated well with MICs from Etest and broth microdilution.
Within the lactic acid bacteria (LAB), the Lactobacillus acidophilus group, consisting of the six closely related species L. acidophilus, L. amylovorus, L. crispatus, L. gallinarum, L. gasseri, and L. johnsonii, represents one of the three major groups of lactobacilli hosted in the intestinal tracts of healthy humans (10). Strains of this group have not only been widely used as dairy starter cultures but are also applied as probiotics (10).
Especially in the case of probiotic products, where the concentration of viable bacteria is a key criterion used to define the microbiological product's quality (14), lactobacilli and other LAB are usually consumed in high numbers. The close contact with other bacteria in the human intestine is an excellent precondition for horizontal gene transfer with the aid of conjugative transposons and plasmids (26). Therefore, it is of particular importance to reassess the safety of bacterial cultures intended for use as food additives (28), even though most strains of the L. acidophilus group are classified as “generally recognized as safe” bacteria due to their long history of proven health benefits and safe use (11, 19). There is a growing interest in the possible role of LAB as vectors for antibiotic resistance genes (26). Hence, the safety of these microorganisms should be verified with respect to their ability to acquire and disseminate resistance determinants (14).
A prerequisite for the identification of resistant genotypes is the assessment of resistant phenotypes by standardized methods (7). Although there are well-established standard procedures and breakpoints available for susceptibility testing of bacteria with clinical significance, they are poorly validated or unsuitable for susceptibility testing of nonpathogenic bacteria such as nonenterococcal LAB and bifidobacteria (20). Only recently were guidelines published providing recommendations on how to test Lactobacillus isolates that cause endocarditis (13). To our knowledge, no standards for susceptibility testing of Lactobacillus isolates or approved guidelines for interpreting the test results exist (6, 21, 23). Consequently, a broad variety of techniques have been reported for the in vitro susceptibility testing of lactobacilli (1, 2, 4, 12, 15, 22, 24, 25, 29). Owing to the different testing conditions applied (e.g., methodologies, basal media, temperature, time, and atmosphere), both the comparability and the interpretation of results for the detection of resistance determinants across different studies are limited.
Among the recent efforts undertaken to establish a standardized method for antimicrobial susceptibility testing of LAB, the development and evaluation of LAB susceptibility test medium (LSM) proved a first major step forward (16). Largely based on the use of the LSM formulation, standard operating procedures for the antimicrobial susceptibility testing of LAB by Etest or the broth microdilution method have been proposed within the 6th framework EU project ACE-ART (G. Huys, unpublished data).
In the scope of these developments, the present study aimed to establish a standardized procedure for the antimicrobial susceptibility testing of members of the L. acidophilus group. For this purpose, the antimicrobial susceptibility of 104 isolates encompassing the six species of this group was tested by the Etest and the broth microdilution method according to the recently proposed standard operating procedures. Additionally, the disk diffusion method was performed under Etest conditions. Finally, the suitability and precision of the three methods were evaluated.
MATERIALS AND METHODS
Bacterial strains.
A total of 104 isolates of the L. acidophilus group were included in this study, i.e., 10 strains of L. acidophilus, 31 strains of L. amylovorus, 7 strains of L. crispatus, 7 strains of L. gallinarum, 26 strains of L. gasseri, and 23 strains of L. johnsonii. The origin and identification of these strains by species-specific PCR or by amplified ribosomal DNA restriction analysis were described by Danielsen et al. (5). Enterococcus faecalis ATCC 29212 was included as a quality control strain.
Antimicrobial susceptibility testing.
For all three susceptibility testing methods, inocula of the isolates tested were prepared by suspending colonies from LSM agar plates (16), incubated for 24 h at 37°C in an anaerobic cabinet (Scholzen Technik, Kriens, Switzerland), in 5 ml 0.85% NaCl solution to a turbidity of McFarland standard 1.
(i) Broth microdilution.
VetMIC 96-well microtiter plates (National Veterinary Institute, Uppsala, Sweden) were used for determining the MICs of the antimicrobial agents ampicillin (0.12 to 8 μg ml−1), clindamycin (0.12 to 8 μg ml−1), erythromycin (0.12 to 16 μg ml−1), gentamicin (0.5 to 32 μg ml−1), streptomycin (2 to 256 μg ml−1), and tetracycline (0.5 to 128 μg ml−1). Additionally, vancomycin resistance was tested in separate microdilution assays in a concentration range of 0.12 to 128 μg ml−1. Therefore, a vancomycin stock solution of 5.12 mg ml−1 was prepared in sterile water and diluted in LSM broth (16) to obtain solutions with preliminary vancomycin concentrations in a range of 0.25 to 256 μg ml−1. A 50-μl volume of each solution was dispensed into each well of the microtiter plates. The inoculated saline suspension, prepared as described above, was diluted 1:1,000 in LSM broth for inoculation of the VetMIC plates and 1:500 for inoculation of the vancomycin plates. Subsequently, 100 and 50 μl of the diluted inoculum was added to each well of the VetMIC and vancomycin plates, respectively. Plates were incubated under anaerobic conditions at 37°C for 48 h. Subsequently, MICs were read as the lowest concentration of an antimicrobial agent at which visible growth was inhibited.
(ii) Etest.
Bacterial suspensions with a turbidity equivalent to McFarland standard 1 were swabbed evenly onto LSM agar plates with a sterile cotton swab. After drying the surfaces of the plates, the Etest strips (AB Biodisk, Sweden) of all of the antimicrobial agents tested (ampicillin, clindamycin, erythromycin, gentamicin, streptomycin, tetracycline, and vancomycin; 0.016 to 256 μg ml−1) were applied. The plates were incubated under the same condition as for the broth microdilution method. MICs were read directly from the test strip according to the instructions of the manufacturer.
(iii) Disk diffusion.
LSM agar plates were inoculated with the bacterial suspension as described above for the Etest. Antibiotic disks containing 10 μg ampicillin, 2 μg clindamycin, 15 μg erythromycin, 10 μg gentamicin, 10 μg streptomycin, 30 μg tetracycline, or 30 μg vancomycin (Oxoid) were placed on LSM agar plates. Plates were incubated under anaerobic conditions for 48 h at 37°C, followed by measurement of the inhibition zone diameters (IZDs), including the diameter of the disk (in millimeters).
Statistical analysis. (i) Comparison of broth microdilution and Etest.
MIC agreement between the broth microdilution and Etest was defined as the same MIC ±1 log2 dilution based on the MICs of the broth microdilution method. Etest results that fell between the standard twofold dilution MICs of the broth microdilution method were rounded up to the next higher twofold concentration. Isolates with off-scale MICs, i.e., MICs less than or equal to the lowest concentration or greater than the highest concentration tested, were excluded for this agreement calculation.
(ii) Correlation between disk diffusion and broth microdilution or Etest.
To define the linear functions between IZDs (in millimeters) and MICs (in micrograms per milliliter), regression analysis was applied after logarithmic conversion (log2) of the MICs. Correlation between the two variables was established by the correlation coefficient (r). P values of <0.01 were considered statistically significant.
(iii) Precision of the methods.
As an important validation parameter to characterize the three methods, their respective precision was expressed in terms of repeatability. The repeatability of each method was evaluated by testing a set of 10 strains in three independent assays for each antimicrobial agent. The resulting MIC and IZD variations were expressed as standard deviations of repeatability. Generally, the acceptable repeatability of broth microdilution and Etest is within a range of ±1 log2 MICs or lies within a ±3- to 4-mm IZD variation for disk diffusion (8).
RESULTS
(i) Comparison of broth microdilution and Etest.
Table 1 displays the agreement of the MICs of seven antimicrobial agents for 104 isolates of the L. acidophilus group generated by broth microdilution and Etest. The levels of agreement between the two methods were high for the antimicrobial agents ampicillin, gentamicin, streptomycin, and vancomycin (>90%). Lower levels of agreement were obtained for clindamycin (71%), erythromycin (80%), and especially tetracycline (34%).
TABLE 1.
Comparison of broth microdilution and Etest MICs of seven antimicrobial agents for 104 isolates of the L. acidophilus groupa
| Antimicrobial agent | No. of isolates | No. of isolates with the same (0) or deviating Etest results in comparison with the broth microdilution method
|
% Agreementb | ||||||
|---|---|---|---|---|---|---|---|---|---|
| −4 | −3 | −2 | −1 | 0 | 1 | 2 | |||
| Ampicillin | 102 | 1 | 7 | 48 | 46 | 92 | |||
| Clindamycin | 80 | 2 | 6 | 15 | 21 | 28 | 8 | 71 | |
| Erythromycin | 45 | 1 | 8 | 9 | 20 | 7 | 80 | ||
| Gentamicin | 101 | 1 | 6 | 51 | 40 | 3 | 96 | ||
| Streptomycin | 93 | 3 | 38 | 35 | 15 | 2 | 95 | ||
| Tetracycline | 76 | 16 | 34 | 11 | 8 | 7 | 34 | ||
| Vancomycin | 104 | 12 | 87 | 5 | 95 | ||||
Only isolates with on-scale MICs are included.
Agreement (bold values) between the two methods is defined as the difference between MICs falling within a 1-dilution MIC range (0, 1, and −1).
In general, lower MICs of ampicillin, clindamycin, erythromycin, and streptomycin were obtained by Etest. Conversely, Etest tended to yield higher MICs of gentamicin and vancomycin. MICs of tetracycline obtained by Etest were lower at the susceptible end of the MIC range and higher at the resistant end of the MIC range compared to those obtained by broth microdilution.
(ii) Correlation between disk diffusion and broth microdilution or Etest.
The linear functions between IZDs (in millimeters) of the disk diffusion method and MICs (in micrograms per milliliter) of the broth microdilution method or Etest were characterized by a relatively strong correlation coefficient (r = −0.84 to −0.98) for the antimicrobial agents clindamycin, erythromycin, streptomycin, and tetracycline. Concerning ampicillin, gentamicin, and vancomycin, the correlation coefficient was only moderately strong (r = −0.63 to −0.83). The P values of all correlation analyses were less than 10−4, which expresses a highly significant relationship between the methods compared.
(iii) Precision of the methods.
MIC variations, expressed as standard deviations of repeatability and obtained for the broth microdilution method and Etest by testing a set of 10 strains in three independent assays, were within the expected range of ±1 log2 dilution variation of MICs. Correspondingly, IZD variations of the agar disk diffusion method lay within an IZD variation of ±3 to 4 mm. Interestingly, lower variations (±0.32 to 0.45 log2 μg ml−1 and ±1 to 1.5 mm) could be obtained for the antibiotics gentamicin, streptomycin, and vancomycin by Etest and the agar disk diffusion method, respectively, whereas the variation was higher (±0.48 to 0.63 log2 μg ml−1 and ±2.2 to 2.9 mm) for ampicillin, clindamycin, erythromycin, and tetracycline for these two agar-based methods.
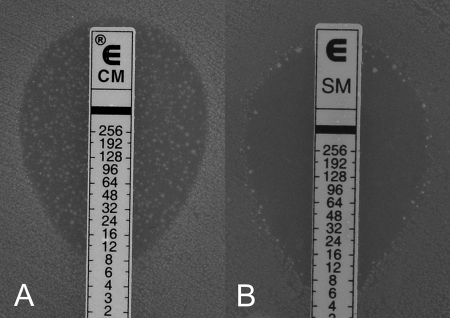
By testing a clindamycin-resistant L. gasseri strain several times, a less stable type of resistance appeared as substantial ingrowth in Etest elliptical inhibition zones (Fig. 1A). In general, this type of resistance could be observed for some strains of the species L. gasseri and L. johnsonii and the antimicrobial agents clindamycin and erythromycin.
FIG. 1.
(A) Ingrowth of isolated resistant colonies in elliptical inhibition zones when testing the susceptibility of an L. gasseri strain to clindamycin by Etest. According to the manufacturer's instructions, the MIC has to be read as the concentration at total inhibition, including discrete colonies (MIC, >256 μg ml−1), despite the apparent inhibition ellipse at a MIC of 8 μg ml−1. (B) Occurrence of isolated resistant colonies at the margin of the inhibition zone, when testing the susceptibility of an L. johnsonii strain to streptomycin by Etest.
Another finding, which was not repeatable and occurred arbitrarily, was inconsistent growth with the broth microdilution method and the aminoglycoside antibiotics streptomycin and gentamicin. Irrespective of the occurrence of this phenomenon, isolated resistant colonies could be detected at the inhibition zone margins for these two antimicrobial agents in Etest and the agar disk diffusion method (Fig. 1B).
DISCUSSION
The recent description of the LSM formulation (16) is considered a first step toward the development of a standard protocol for the susceptibility testing of industrially important LAB. Still, very few data are currently available on the suitability of this medium when using different testing methods. Thus, the Etest, agar disk diffusion, and broth microdilution results obtained by LSM-based antimicrobial susceptibility testing of LAB strains of the L. acidophilus group were compared within this study.
In general, the broth microdilution and Etest results were in good agreement. The overall agreement between these two susceptibility testing procedures, which should be higher than 90% (27), was sufficiently achieved for the bactericidal drugs ampicillin, gentamicin, streptomycin, and vancomycin. The minor satisfying agreement between the results obtained with the bacteriostatic agents clindamycin, erythromycin, and tetracycline may be due to the diffuse edges of their inhibition zones observed with agar-based methods. Additionally, reading at 80% inhibition of growth, as indicated by the manufacturer, further complicates the interpretation of data. In contrast, accurate reading of endpoints was possible for all of the bactericidal agents tested except ampicillin because inhibition zones were much better delineated. Moreover, results were read at the point of total inhibition for bactericidal agents, thus simplifying the readings. Concerning the bacteriostatic agents, the greatest discrepancy between Etest and broth microdilution was observed with tetracycline. This may be due to the fact that only for this antimicrobial agent were the Etest values lower than those of the broth microdilution method at the susceptible end of the MIC range and higher at the resistant end.
Repeated measurements were included to determine precision as a key parameter to characterize each method. Broth microdilution and Etest results were highly reproducible, as for all of the antimicrobial agents the MIC dilution variations fell within the expected range of ±1 log2 dilution (8). As mentioned above, the higher variations obtained by Etest for the bacteriostatic agents tested and ampicillin could be traced back to the difficult reading of results. As the Etest and the disk diffusion method are based on the same principle, also a higher variation between the results was found for the same antimicrobial agents concerning the disk diffusion method. Nevertheless, all IZD variations fell within a range of ±3 to 4 mm, which corresponds to the intrinsic variation of the testing system (8).
With respect to the comparison of the disk diffusion method with broth microdilution or Etest, it was found that the IZDs correlated well with the MICs determined by the two other methods. An increase in MICs was accompanied by a decrease in IZD sizes and vice versa. As a result of the simultaneous presence of low- or high-level resistant and susceptible strains (5), the correlation coefficients (r) for clindamycin, erythromycin, streptomycin, and tetracycline indicated a relatively strong relationship between the variables. On the contrary, very few strains displayed phenotypic resistance to ampicillin, gentamicin, or vancomycin (5). As a result, the MIC distribution mainly consisted of low values which resulted in an unreliable regression line and lower correlation coefficients.
Because quantitative MIC information is usually preferred above classification into resistant, intermediate, and susceptible phenotypes for evaluation of the biosafety of industrial LAB cultures, dilution methods and the Etest are favored over the disk diffusion test (17). However, if criteria for interpreting the results of this technique were established, a useful method for rapid tentative differentiation among susceptible and resistant isolates could be developed.
In addition to the evaluation of the suitability and precision of all three methods, some other important observations, like substantial ingrowth in elliptical inhibition zones regarding Etest and the antimicrobial agents clindamycin and erythromycin could be noted. Isolated resistant colonies within the inhibition zone of the Etest strip were also observed by Danielsen and Wind in the testing of Lactobacillus sp. susceptibility to imipenem and nitrofurantoin (6). They concluded that this phenomenon may be explained by the high spontaneous frequency of mutation to antibiotic resistance (6), which is not uncommon in lactobacilli (3). Additionally, the loss of antibacterial activity of unstable antimicrobial agents during incubation may result in subinhibitory concentrations that could promote the emergence of resistant strains during prolonged exposure (9). This assumption is supported by the detection of erythromycin- and clindamycin-resistant colonies after 48 h but never after 24 h of incubation within the present study. To avoid major discrepancies between the Etest and the broth microdilution method due to substantial ingrowth in elliptical inhibition zones, the Etest MIC should be read at the point of intersection instead of at the point of total growth inhibition. In any case, however, the appearance of isolated resistant colonies should be recorded.
Possibly, inconsistent growth of some isolates in broth microdilution and the appearance of resistant colonies at the margin of the inhibition zones of Etest or agar disk diffusion during gentamicin and streptomycin susceptibility testing could also be explained by a high rate of spontaneous mutation to antibiotic resistance in lactobacilli (3). However, it is also known that some antibiotics possess the capability to increase mutability (18). The appearance of isolated resistant colonies at the margin of the inhibition zone does not substantially influence Etest or agar disk diffusion results. For broth microdilution, the lowest MIC was always read, as this effect, which was only observed for aminoglycosides, seems to be more drug specific.
Isolated resistant colonies are an important observation that might not be detectable by applying the broth microdilution method (6). This highlights an advantage of using agar-based methods (especially the Etest) over broth-based methods.
In conclusion, this study has clearly indicated that each susceptibility test has inherent advantages and limitations. Agar-based methods like Etest and agar disk diffusion represent valid methods compared to the broth microdilution method, using the new protocol and test medium for antimicrobial susceptibility testing of lactobacilli. Nevertheless, further performance evaluations are required to develop a standardized method for antimicrobial susceptibility testing of lactobacilli used as starter cultures or probiotics to evaluate acquired antibiotic resistances more easily.
Acknowledgments
This work was supported by a grant from the European Commission (CT-2003-506214, ACE-ART, 6th Framework).
G. Huys is a postdoctoral fellow of the Fund for Scientific Research—Flanders, Belgium (F.W.O.—Vlaanderen).
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Charteris, W. P., P. M. Kelly, L. Morelli, and J. K. Collins. 1998. Antibiotic susceptibility of potentially probiotic Lactobacillus species. J. Food Prot. 61:1636-1643. [DOI] [PubMed] [Google Scholar]
- 2.Charteris, W. P., P. M. Kelly, L. Morelli, and J. K. Collins. 2001. Gradient diffusion antibiotic susceptibility testing of potentially probiotic lactobacilli. J. Food Prot. 64:2007-2014. [DOI] [PubMed] [Google Scholar]
- 3.Curragh, H. J., and M. A. Collins. 1992. High levels of spontaneous drug resistance in Lactobacillus. J. Appl. Bacteriol. 73:31-36. [Google Scholar]
- 4.D'Aimmo, M. R., M. Modesto, and B. Biavati. 2007. Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products. Int. J. Food Microbiol. 115:35-42. [DOI] [PubMed] [Google Scholar]
- 5.Danielsen, M., S. Mayrhofer, K. J. Domig, E. Amtmann, H. K. Mayer, A. B. Florez, B. Mayo, J. Korhonen, and L. Tosi. 2008. Assessment of the antimicrobial wild type minimum inhibitory concentration of species of the Lactobacillus delbrueckii group. Dairy Sci. Technol. 88:183-191. [Google Scholar]
- 6.Danielsen, M., and A. Wind. 2003. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 82:1-11. [DOI] [PubMed] [Google Scholar]
- 7.FEEDAP Panel. 2005. Opinion of the Scientific Panel on Additives and Products or Substances Used in Animal Feed on the updating of the criteria used in assessment of bacteria for resistance to antibiotics of human and veterinary importance. EFSA J. 223:1-12. [Google Scholar]
- 8.Fuchs, P. C., A. L. Barry, and S. D. Brown. 2002. Selection of zone size interpretive criteria for disk diffusion susceptibility test of three antibiotics against Streptococcus pneumoniae, using new guidelines of the National Committee for Clinical Laboratory Standards. Antimicrob. Agents Chemother. 46:398-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herra, C. M., M. T. Cafferkey, and C. T. Keane. 1995. The in-vitro susceptibilities of vaginal lactobacilli to four broad-spectrum antibiotics, as determined by the agar dilution and Etest methods. J. Antimicrob. Chemother. 35:775-783. [DOI] [PubMed] [Google Scholar]
- 10.Holzapfel, W. H. 2006. Introduction to prebiotics and probiotics, p. 1-33. In I. Goktepe, V. K. Juneja, and M. Ahmedna (ed.), Probiotics in food safety and human health. CRC Press, Inc., Boca Raton, FL.
- 11.Holzapfel, W. H., P. Haberer, R. Geisen, J. Bjorkroth, and U. Schillinger. 2001. Taxonomy and important features of probiotic microorganisms in food and nutrition. Am. J. Clin. Nutr. 73(Suppl.):S365-S373. [DOI] [PubMed] [Google Scholar]
- 12.Huys, G., K. D'Haene, and J. Swings. 2002. Influence of the culture medium on antibiotic susceptibility testing of food-associated lactic acid bacteria with the agar overlay disc diffusion method. Lett. Appl. Microbiol. 34:402-406. [DOI] [PubMed] [Google Scholar]
- 13.Jorgensen, J. H., and J. F. Hindler. 2007. New consensus guidelines from the Clinical and Laboratory Standards Institute for antimicrobial susceptibility testing of infrequently isolated or fastidious bacteria. Clin. Infect. Dis. 44:280-286. [DOI] [PubMed] [Google Scholar]
- 14.Kastner, S., V. Perreten, H. Bleuler, G. Hugenschmidt, C. Lacroix, and L. Meile. 2006. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst. Appl. Microbiol. 29:145-155. [DOI] [PubMed] [Google Scholar]
- 15.Katla, A. K., H. Kruse, G. Johnsen, and H. Herikstad. 2001. Antimicrobial susceptibility of starter culture bacteria used in Norwegian dairy products. Int. J. Food Microbiol. 67:147-152. [DOI] [PubMed] [Google Scholar]
- 16.Klare, I., C. Konstabel, S. Muller-Bertling, R. Reissbrodt, G. Huys, M. Vancanneyt, J. Swings, H. Goossens, and W. Witte. 2005. Evaluation of new broth media for microdilution antibiotic susceptibility testing of lactobacilli, pediococci, lactococci, and bifidobacteria. Appl. Environ. Microbiol. 71:8982-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klare, I., C. Konstabel, G. Werner, G. Huys, V. Vankerckhoven, G. Kahlmeter, B. Hildebrandt, S. Muller-Bertling, W. Witte, and H. Goossens. 2007. Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J. Antimicrob. Chemother. 59:900-912. [DOI] [PubMed] [Google Scholar]
- 18.Martinez, J. L., and F. Baquero. 2000. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44:1771-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathur, S., and R. Singh. 2005. Antibiotic resistance in food lactic acid bacteria—a review. Int. J. Food Microbiol. 105:281-295. [DOI] [PubMed] [Google Scholar]
- 20.Mättö, J., M.-L. Suihko, and M. Saarela. 2006. Comparison of three test media for antimicrobial susceptibility testing of bifidobacteria using the Etest method. Int. J. Antimicrob. Agents 28:42-48. [DOI] [PubMed] [Google Scholar]
- 21.Ocana, V., C. Silva, and M. E. Nader-Macias. 2006. Antibiotic susceptibility of potentially probiotic vaginal lactobacilli. Infect. Dis. Obstet. Gynecol. 2006:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salminen, M. K., H. Rautelin, S. Tynkkynen, T. Poussa, M. Saxelin, V. Valtonen, and A. Jarvinen. 2004. Lactobacillus bacteremia, clinical significance, and patient outcome, with special focus on probiotic L. rhamnosus GG. Clin. Infect. Dis. 38:62-69. [DOI] [PubMed] [Google Scholar]
- 23.Salminen, M. K., H. Rautelin, S. Tynkkynen, T. Poussa, M. Saxelin, V. Valtonen, and A. Jarvinen. 2006. Lactobacillus bacteremia, species identification, and antimicrobial susceptibility of 85 blood isolates. Clin. Infect. Dis. 42:e35-e44. [DOI] [PubMed] [Google Scholar]
- 24.Sozzi, T., and M. B. Smiley. 1980. Antibiotic resistances of yogurt starter cultures Streptococcus thermophilus and Lactobacillus bulgaricus. Appl. Environ. Microbiol. 40:862-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Temmerman, R., B. Pot, G. Huys, and J. Swings. 2003. Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int. J. Food Microbiol. 81:1-10. [DOI] [PubMed] [Google Scholar]
- 26.Teuber, M., L. Meile, and F. Schwarz. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie van Leeuwenhoek 76:115-137. [PubMed] [Google Scholar]
- 27.Thornsberry, C. 1985. Automated procedures for antimicrobial susceptibility tests, p. 1015-1018. In E. H. Lennette, A. Balows, W. J. Hausler, and H. J. Shadomy (ed.), Manual of clinical microbiology, 4th ed. American Society for Microbiology, Washington, DC.
- 28.Vizoso Pinto, M. G., C. M. Franz, U. Schillinger, and W. H. Holzapfel. 2006. Lactobacillus spp. with in vitro probiotic properties from human faeces and traditional fermented products. Int. J. Food Microbiol. 109:205-214. [DOI] [PubMed] [Google Scholar]
- 29.Zhou, J. S., C. J. Pillidge, P. K. Gopal, and H. S. Gill. 2005. Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int. J. Food Microbiol. 98:211-217. [DOI] [PubMed] [Google Scholar]