Abstract
Despite the fact that most industrial processes for secondary metabolite production are performed with submerged cultures, a reliable developmental model for Streptomyces under these culture conditions is lacking. With the exception of a few species which sporulate under these conditions, it is assumed that no morphological differentiation processes take place. In this work, we describe new developmental features of Streptomyces coelicolor A3(2) grown in liquid cultures and integrate them into a developmental model analogous to the one previously described for surface cultures. Spores germinate as a compartmentalized mycelium (first mycelium). These young compartmentalized hyphae start to form pellets which grow in a radial pattern. Death processes take place in the center of the pellets, followed by growth arrest. A new multinucleated mycelium with sporadic septa (second mycelium) develops inside the pellets and along the periphery, giving rise to a second growth phase. Undecylprodigiosin and actinorhodin antibiotics are produced by this second mycelium but not by the first one. Cell density dictates how the culture will behave in terms of differentiation processes and antibiotic production. When diluted inocula are used, the growth arrest phase, emergence of a second mycelium, and antibiotic production are delayed. Moreover, pellets are less abundant and have larger diameters than in dense cultures. This work is the first to report on the relationship between differentiation processes and secondary metabolite production in submerged Streptomyces cultures.
Streptomyces is a soil bacterium that produces numerous clinically useful antibiotics (1, 54, 66), as well as many molecules that affect eukaryotic systems, such as inducers of eukaryotic cellular differentiation, inducers and inhibitors of apoptosis (59), and protein C kinase inhibitors with antitumor activity (such as staurosporine and others) (48, 57). Moreover, its remarkably complex developmental cycle makes this microorganism an interesting subject for study. The traditional developmental cycle of this bacterium describes two differentiated mycelial structures, a substrate (vegetative) mycelium and an aerial (reproductive) mycelium (10, 25, 33). In the substrate mycelium, septa are thought to be widely spaced and to define compartments containing several nucleoids (10, 63). After a short growth arrest phase characterized by reduced macromolecular synthesis (25), aerial hyphae develop from simple branching from substrate mycelium (29). Finally, the tips of the aerial mycelium differentiate into hydrophobic spore chains (8). Recently, we have been able to extend what is known about the developmental cycle in surface confluent cultures a great deal. Our main contribution has been to reveal the existence of a very young compartmentalized mycelium that dies following an orderly pattern, leaving alternating live and dead segments in the same hypha (37). Subsequently, the remaining live mycelium grows in successive waves that vary according to the density of the spore inocula. In the presence of dense inocula, the hyphae develop in regular circles approximately 0.5 cm in diameter (38). In contrast, with highly diluted spore inocula, hyphae develop initially in the form of smaller circles measuring 0.9 mm in diameter. Further mycelial development occurs between the second mycelium circles or islands until the plate surface is totally covered. The second mycelium is a syncytium (multinucleated structure) with sporadic septa, which presents the hydrophobic properties that characterize aerial hyphae (14, 41). This multinucleated mycelium suffers a second death round before to the sporulation phase (39-44), which corresponds to the one previously described by other authors that affects substrate hyphae (11, 25, 42, 62).
Although most industrial processes for secondary metabolite production are performed with liquid cultures, Streptomyces strains generally do not sporulate under these conditions (15, 45, 49); therefore, reliable morphological markers are lacking for liquid cultures and in-depth developmental studies have not been performed. Other published reports on Streptomyces in submerged cultures have focused on the relationships between culture media and/or microbial morphology and antibiotic production (5, 16, 19, 24, 32, 53, 68). Nutrient limitations and carbon or nitrogen sources can affect antibiotic production. Four morphological types of mycelial growth have been distinguished in submerged Streptomyces cultures: pellets (compact masses 950 μm in diameter), clumps (less compact masses 600 μm in diameter), branched hyphae, and nonbranched hyphae (16). Some of these structures develop in the form of a biofilm consisting of sticky extracellular polymers and insoluble substrates (35).
In this work, we have extended our previous investigations of Streptomyces development on solid cultures (37-39) to submerged conditions. Certain interesting features similar to those formerly described in surface cultures arise, such as the presence of different mycelial types, as well as a cell death process which precedes antibiotic production. Cell types functionally similar to the formerly termed second mycelium (the precursor of aerial mycelium in solid media) (37, 38, 41) are postulated to be the ones responsible for antibiotic synthesis.
MATERIALS AND METHODS
Strains and media.
Streptomyces coelicolor strain M145 was used in all of these studies (33). It was grown and maintained by following standard procedures (30). S. coelicolor strain M600 harboring the promoter of the redD gene fused with the enhanced green fluorescent protein (eGFP) gene was also used (54). Cultures were performed with R5A liquid medium lacking sucrose (21). Flasks of 100 ml with 20 ml of culture medium were inoculated directly with freshly prepared spores at different densities (1 × 107 or 1 × 105/ml) and incubated at 200 rpm and 30°C.
Viability staining.
Culture samples were obtained and processed for microscopy at different incubation time points, as previously described for Streptomyces cultures grown under submerged conditions (20). Cells were stained with a non-cell-permeating nucleic acid stain (propidium iodide, PI) in order to detect the dead cell population and with SYTO 9 green fluorescent nucleic acid stain (LIVE/DEAD Bac-Light bacterial viability kit, L-13152; Invitrogen) to detect viable cells. The SYTO 9 green fluorescent stain labels all of the cells, i.e., those with intact membranes, as well as those with damaged ones. In contrast, PI penetrates only bacteria with damaged membranes, decreasing SYTO 9 stain fluorescence when both dyes are present. Thus, in the presence of both stains, bacteria with intact cell membranes appear fluorescent green whereas bacteria with damaged membranes appear red (27). Samples were examined, after being left for at least 10 min in the dark, under a Leica TCS-SP2-AOBS confocal laser scanning microscope at excitation wavelengths of 488 and 568 nm and an emission wavelength of 530 nm (green) or 630 nm (red) (optical sections of about 0.2 μm). Images were mixed with the Leica Confocal Software. In some cases, samples were also examined in differential interference contrast mode, which is available with the same equipment. A significant number of images were analyzed in a minimum of three independent culture analyses.
Cell wall staining.
Streptomyces cells were collected by centrifugation at various time points. Harvested mycelium was processed as described elsewhere (52). Cells were fixed in 2.8% paraformaldehyde, 0.0045% glutaraldehyde in phosphate-buffered saline (PBS) (0.14 M NaCl, 2.6 mM KCl, 1.8 mM KH2PO4, 10 mM Na2HPO4) for 15 min at room temperature and washed twice with PBS. Wheat germ agglutinin conjugated with Texas Red (W-21405; Invitrogen), which binds selectively to N-acetylglucosamine and N-acetylneuraminic acid, was used to stain cell walls. Fixed cells were incubated for 1 min in 2 mg lysozyme ml−1 in glucose-Tris-EDTA (50 mM glucose, 20 mM Tris-HCl [pH 8], 10 mM EDTA). The samples were washed again with PBS and blocked in 2% bovine serum albumin in PBS for 5 min. Wheat germ agglutinin was added at a concentration of 100 mg ml−1 in 2% bovine serum albumin in PBS, and samples were incubated at room temperature for 3 h. Finally, the samples were washed eight times with PBS and observed under the confocal microscope at an excitation wavelength of 595 nm and an emission wavelength of 615 nm.
Vancomycin stains nascent peptidoglycan (23). BODIPY FL vancomycin (V34850; Invitrogen) was used at a concentration of 0.5 μg ml−1 in PBS to stain fixed cells (15 min). Samples were then washed twice with PBS and observed under the confocal microscope at an excitation wavelength of 505 nm and an emission wavelength of 513 nm.
Membrane staining.
Samples were processed and fixed as in the case of cell wall staining (see above). Lipophilic styryl dye, N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl)pyridinium dibromide (FM 4-64) (T-3166; Invitrogen), was added at a final concentration of 1 mg ml−1, and samples were incubated at room temperature for 3 h. Samples were washed twice and observed under the confocal laser-scanning microscope at an excitation wavelength of 550 nm and an emission wavelength of 700 nm.
Sampling of Streptomyces throughout the differentiation cycle.
Samples of S. coelicolor obtained from submerged cultures were centrifuged (77,406 × g, 30 min at 4°C). Supernatants were used to estimate extracellular proteins. To obtain cell extracts, the mycelium pellets were resuspended in a known volume of buffer A (Tris-HCl at 20 mM [pH 8], EDTA at 1 mM, β-mercaptoethanol at 7 mM, phenylmethylsulfonyl fluoride at 0.5 mM) and ruptured in an MSE Soniprep 150 by six cycles of 10 s on ice, after which the samples were centrifuged at 77,406 × g in an Eppendorf microcentrifuge for 30 min at 4°C. The supernatant fraction was used as the cellular fraction. For dry cell weight determinations, samples (5 ml in triplicate) of cellular suspensions were collected on preweighed 0.45-μm filters and dried at 100°C to a constant weight.
Activity gel analysis.
Nucleases were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (36) in a 12% gel containing 10 μg/ml denatured calf thymus DNA (Sigma). The sodium dodecyl sulfate used for these experiments was USB-US75819 from Amersham Biosciences. After electrophoresis, the proteins were renatured by repeatedly washing the gel with renaturation buffer (Tris-HCl at 25 mM [pH 8.8], EDTA at 1 mM, β-mercaptoethanol at 7 mM) for 2 h at 4°C. Nuclease activity was visualized by incubating the gels for 1 h at 37°C in 20 mM Tris-HCl (pH 8.0)-7 mM 2-mercaptoethanol-10 mM MgCl2-5 mM CaCl2-10% dimethyl sulfoxide buffer as reported elsewhere (44), followed by staining with ethidium bromide and analysis under UV light. Micrococcal nuclease (16.7 kDa) and bovine pancreatic DNase I (31 kDa) (Amersham Pharmacia Biotech) were included as positive controls. The reproducibility of the data shown has been corroborated by at least three independent cultures and nuclease analysis at various developmental time points.
Hexokinase activity and protein analysis.
The presence of hexokinase activity in the extracellular medium was tested in the supernatant fractions (see above) (39). Determination of protein concentrations was carried out by the Bradford assay (4) with a bovine serum albumin standard (Sigma).
Antibiotic quantification.
Undecylprodigiosin and actinorhodin were quantified spectrophotometrically according to Tsao et al. (57) and Bystrykh et al. (7). Cells were ruptured in the culture medium by adding 0.1 N KOH. After vortexing and centrifugation, actinorhodin was quantified in the supernatant (ɛ640 = 25,320). Undecylprodigiosin was measured after vacuum drying of the mycelium, followed by extraction with methanol, acidification with HCl (to 0.5 M), and a spectrophotometic assay (ɛ530 = 100,500). Reproducibility has been corroborated by at least three independent cultures and nuclease analysis at various developmental time points.
RESULTS
CLSM analysis of development-linked cell death processes of S. coelicolor A3(2) in submerged cultures.
S. coelicolor A3(2) was grown at two different inoculum densities, and the development processes were followed under confocal laser scanning fluorescence microscopy (CLSM) after SYTO 9 and PI staining (see Materials and Methods).
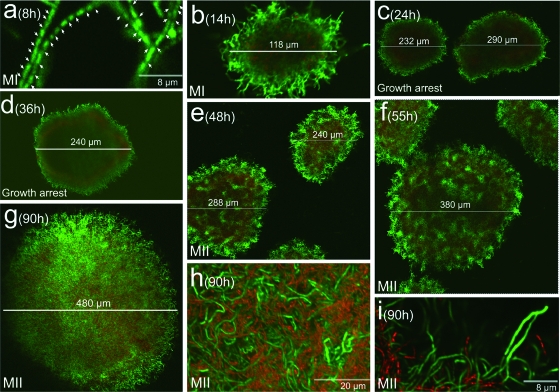
Figure 1 shows the developmental cycle of S. coelicolor A3(2) at an inoculum density of 107 spores/ml (see Materials and Methods). Hyphae with gaps separating nucleoids (compartmentalized first mycelium; see below) are perceptible at early time points (Fig. 1a; see also Fig. 3 and below). These hyphae form pellets which start to grow in a radial pattern and begin to die from the center outward at 14 h (Fig. 1b and c). Death processes inside mycelial masses coincide with a growth arrest, which is discernible by the fact that the pellet diameter ceases to increase (Fig. 1c and d; see also Fig. 4 and below). Later (approximately 50 h), development of the remaining viable mycelium inside the pellets resumes (Fig. 1e). This mycelium now develops as a syncytium with sporadic septa (Fig. 1i; see below and Fig. 3k, l, o, and p). Pellet diameters increase (Fig. 1f and g), thereby prompting culture growth to recommence.
FIG. 1.
CLSM analysis of the development-linked cell death processes of S. coelicolor A3(2) in submerged cultures (1 × 107 spores/ml). Images correspond to culture preparations stained with SYTO 9 and PI. Culture time points (hours), pellet diameters (micrometers), and the growth arrest phase are indicated. Arrows in panel a point to septa. MI, first mycelium; MII second mycelium. For details, see the text.
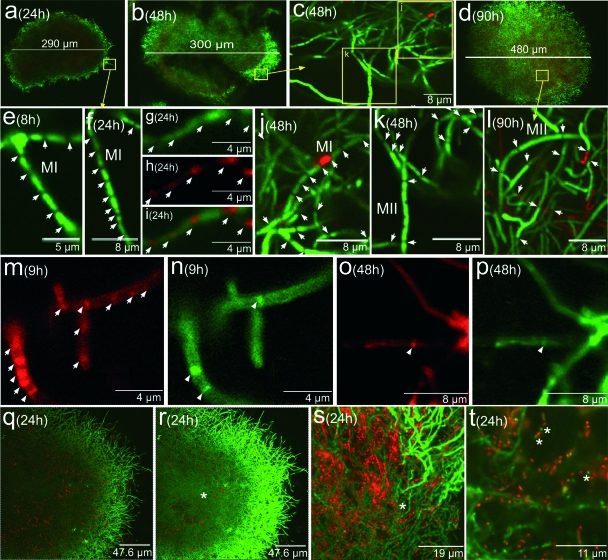
FIG. 3.
Analysis of hyphal compartmentalization with several fluorescent indicators in submerged S. coelicolor cultures (1 × 107 spores/ml). Letters, numbers, and arrows are as in Fig. 1 and 2. (a to f and j to l) SYTO 9-IP staining. (g) SYTO 9 staining. (h) The same field as in panel g, stained with FM 4-64. (i) Mixture of panels g and h. (m to p) Hyphae stained simultaneously with FM 4-64 (red) and vancomycin (green). Arrowheads indicate septa composed of membrane (FM 4-64 staining) and thick cell wall (vancomycin staining). (q) Twenty-four-hour point culture stained with SYTO 9-IP. (r) Same field as in panel q, taken with a higher laser intensity showing low-fluorescence hyphae in the pellet center (arrow). (s and t) Details of the former field. Asterisks point to the cellular segments which develop further as a syncytial mycelium. See the text for details.
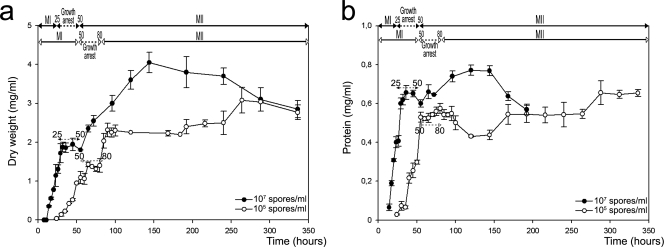
FIG. 4.
Growth curves of S. coelicolor A3(2) in submerged cultures at two different inocula, 1 × 107 spores/ml (solid circles) and 1 × 105 spores/ml (open circles). (a) Dry weight (milligrams per milliliter). (b) Total protein (milligrams per milliliter). Developmental phases observed with the concentrated inoculum (top) and diluted inoculum (below) are shown at the top. Error bars indicate standard deviations.
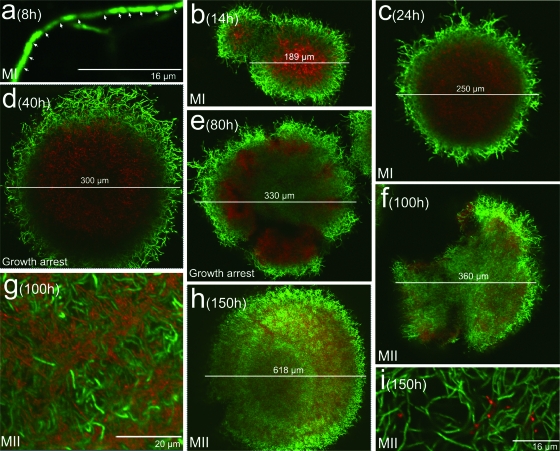
The developmental cycle of an S. coelicolor culture obtained with a diluted inoculum (105 spores/ml) is shown in Fig. 2. As observed in the higher-density inoculum culture, there is a compartmentalized mycelium (Fig. 2a) which begins to grow in the form of pellets. These masses continue to grow radially while undergoing conspicuous death processes in their centers (Fig. 2b and c) until about 50 h, at which point a growth arrest is observed (Fig. 2d and e). As described above, this is followed (80 h) by the development of a second, multinucleated, mycelium in the center and periphery of the pellets (Fig. 1f to i) until they reach their maximum diameter (Fig. 1h). Differences between dense- and diluted-inoculum cultures are therefore mainly noticed in the development onset times of the multinucleated mycelium following the growth delay, at 48 to 50 h for the hyphae in dense-inoculum cultures (Fig. 1e) and 80 to 100 h in diluted-inoculum cultures (Fig. 2e). Furthermore, pellets are more abundant in the dense-inoculum cultures (not shown) and have smaller diameters than those in diluted-inoculum cultures (compare Fig. 1g and 2h).
FIG. 2.
CLSM analysis of the development-linked cell death processes of S. coelicolor A3(2) in submerged cultures (1 × 105 spores/ml). Images correspond to culture preparations stained with SYTO 9 and PI. Letters, numbers, and arrows are as in Fig. 1.
Details of the first, compartmentalized, mycelium described above and the development of the second, multinucleated, mycelium are shown in Fig. 3. Data were obtained from a high-density inoculum culture; similar observations were made in the diluted-inoculum cultures, although at different time points (not shown). As previously stated, at 24 h (Fig. 3a) the pellets are composed of hyphae in which gaps separating the nucleoids are observed (Fig. 3f). Combination of a nucleic acid stain (SYTO 9) with a stain specific for membranes (FM 4-64) confirmed that this initial mycelium is compartmentalized by membranous septa (Fig. 3g to i) and that the gaps observed with the nucleic acid stain are always associated with at least one of these membranes (Fig. 3g to i). However, there are also membranous septa that are not associated with gaps (Fig. 3g to i). Transient growth arrest (25 and 48 h) is followed by the progressive growth of a second, multinucleated, mycelium (Fig. 3b; details in panels c, j, and k and below; and panels o and p). During this transition phase, growth-arrested, compartmentalized hyphae are visible (MI, Fig. 3j), together with cells resuming growth in the form of hyphae with more widely spaced septa (second, multinucleated, mycelium; MII, Fig. 3k). At later time points (90 h, Fig. 3d and l), all of the hyphae appear as a syncytium with sporadic septa (Fig. 3l). Further observations with stains specific for membrane (FM 4-64) or peptidoglycan (vancomycin) (see Materials and Methods) were carried out in order to better characterize the septa of the first- and second-mycelium hyphae. Figure 3m shows the early mycelium stained with membrane-specific fluorochrome FM 4-64. Structures transverse to the hyphae are clearly visible within the mycelium (arrows), which confirms the presence of a membrane separating the cellular compartments. As described previously in surface cultures (37), FM 4-64 stains some zones of the Streptomyces compartmentalized hyphae more intensely than others (Fig. 3m), a phenomenon also reported in Escherichia coli (22). The intensely red-fluorescent regions may be explained by the presence of membrane domains with greater affinity for the dye (22, 37). The same samples stained with vancomycin (Fig. 3n) displayed a majority of membrane septa that are not associated with a cell wall or at least cannot be detected by vancomycin staining. As expected, all of the cell wall septa are associated with a membrane (arrows in Fig. 3m and n). Multinucleated hyphae formed at later time points after the transient growth arrest display widely spaced septa composed of thick-walled membranes (Fig. 3o and p).
The development of the second, multinucleated, mycelium is described in Fig. 3q to t. Figure 3q shows a mycelial pellet stained as usual; dead processes are located in their center (red fluorescence). Other areas of this section are not stained and appear in black. However, when the same image was taken at a higher laser intensity (Fig. 3r and t), a noticeable proportion of the viable compartmentalized hyphae located there showed a less prominent green fluorescence (asterisks). These cellular green compartments (asterisks in Fig. 3s and t) are the precursors of the second, multinucleated, mycelium which start to grow at later time points (see above).
Differentiation of submerged S. coelicolor A3(2) cultures determines biphasic growth curves.
Growth curves (total protein and dry weight) were obtained from concentrated or diluted inocula (Fig. 4). There are two distinct stages of exponential growth separated by a temporary growth arrest phase (better shown in the dry-weight curve in Fig. 4a). The two waves of exponential growth correspond to the development of the first, compartmentalized, mycelium and the growth of the second, multinucleated, mycelium (Fig. 3 and above). As observed by confocal microscopy (see above), the main difference between the dense- and diluted-inoculum cultures is delayed development in the latter case. Thus, a close correlation exists between the morphological phases described above and the growth curves. Streptomyces development under the above-described submerged conditions could be summarized as the emergence of two types of mycelia separated by a transient growth arrest phase.
Cell lysis and nuclease activities during S. coelicolor death processes in submerged cultures.
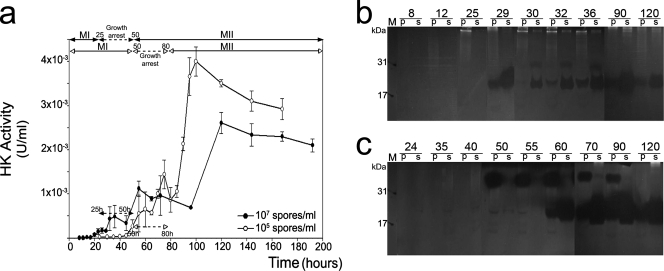
The presence of cytosolic enzymes in the culture medium is an indicator of cell lysis (39). Hexokinase, an enzyme which plays an important role in glycolysis, has been analyzed in the extracellular medium of Streptomyces surface cultures in order to monitor cell lysis subsequent to culture death processes (39). Variations in extracellular hexokinase activity with respect to culture incubation times were also studied in submerged cultures obtained from concentrated or diluted inocula (Fig. 5a). The results confirm the presence of this enzyme in the extracellular medium, thereby corroborating the existence of a lytic process. Lysis was preceded by the alteration of membrane permeability, as demonstrated by PI staining (Fig. 1, 2, and 3). The release of hexokinase gradually increases during the transient growth arrest phase, followed by a subsequent massive increase, coinciding with the widespread disintegration of dead hyphae (Fig. 5a).
FIG. 5.
(a) Appearance of hexokinase (HK) activity in the extracellular medium as an indicator of cell lysis in submerged cultures of S. coelicolor. Solid circles, dense-inoculum culture (1 × 107 spores/ml); open circles, diluted-inoculum culture (1 × 105 spores/ml). Error bars indicate standard deviations. Developmental phases are shown as in Fig. 4. (b and c) Activity gel analysis of nucleases from dense-inoculum culture (b) and diluted-inoculum culture (c) measured in the cell pellet (p) and culture medium supernatant (s). M, DNase I (31 kDa) and micrococcal nuclease (17 kDa), which were used as controls.
Programmed cell death (PCD) in Streptomyces has been described as a genetically determined process around which the differentiation events are structured (37-40). Nucleases are fairly stable enzymes that participate in the death processes and remain active until late culture time points. Activity gel analysis of these enzymes is a simple and reliable way to detect lytic processes associated with PCD (39). Figure 5b and c show the deoxyribonucleolytic activities detected during S. coelicolor development in submerged cultures. The pattern of nuclease appearance correlates with the aforementioned death indicators, and the activity in cultures with a dense inoculum (Fig. 5b) is detected earlier than in cultures with a diluted inoculum (Fig. 5c). As clearly shown in the diluted-inoculum culture (Fig. 5c), a nuclease band of high molecular weight is detected first in the cellular extracts (pellet; p in Fig. 5c), and then 20- to 22-kDa nucleolytic forms are detected in both cellular extracts and the extracellular medium, where they were released by lysis. The high-molecular-weight protein corresponds to the previously described nuclease precursor present only in cells at early death times (39, 40). The smaller nucleases are likely formed by proteolytic processing of the precursor within the cytoplasm of the dying cells (Fig. 5c) (39).
Together, the appearance of hexokinase and the nuclease kinetics during death processes correlate well with the CLSM observations, revealing that the phenomena observed are similar to what has been reported in solid cultures (38, 39).
Antibiotics are produced by the second, multinucleated, mycelium which develops after cell death.
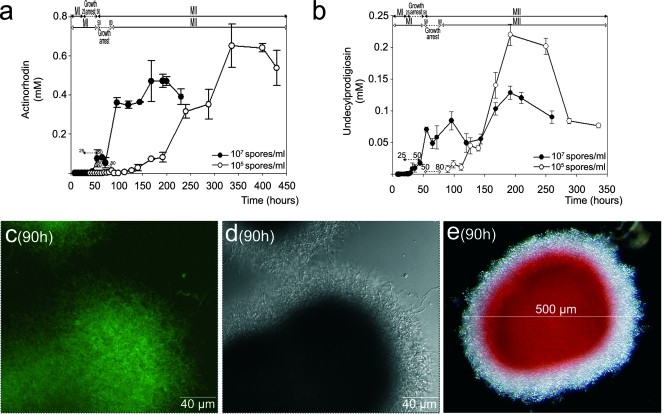
S. coelicolor A3(2) produces two well-characterized antibiotics, undecylprodigiosin and actinorhodin (30). The kinetics involved in the production of these two antibiotics were analyzed within the context of the developmental phases described above (Fig. 6a and b). Antibiotic (undecylprodigiosin and actinorhodin) production commences following the aforementioned transient growth arrest (Fig. 1, 2, and 4), and antibiotic appearance correlates strongly with the development of the second, multinucleated, mycelium. In fact, in the diluted-inoculum culture, antibiotic production is delayed until the second mycelium is formed (compare Fig. 6a and b with Fig. 2).
FIG. 6.
Antibiotic production in submerged S. coelicolor A3(2) cultures. (a) Actinorhodin. (b) Undecylprodigiosin. Solid circles, dense-inoculum culture (1 × 107 spores/ml); open circles, diluted-inoculum culture (1 × 105 spores/ml). Letters, numbers, and error bars are as in Fig. 4 and 5. Error bars indicate standard deviations. Developmental phases are shown as in Fig. 4. (c) CLSM analysis of redD-eGFP expression in a dense-inoculum culture of S. coelicolor. (d) The same microscope field as in panel c in differential interference contrast mode. (e) A pellet from the same culture time observed by phase-contrast microscopy; red corresponds to antibiotic. The values in parentheses are growth times. See the text for details.
Additional data that support the idea that the second, multinucleated, mycelium is the antibiotic-producing structure were obtained by the analysis of an S. coelicolor strain harboring the redD gene promoter (which encodes a pathway-specific regulatory protein for the production of undecylprodigiosin) fused with eGFP (54; see Materials and Methods). Under the culture conditions used in this work, S. coelicolor exhibits a high degree of autofluorescence, which hinders the analysis of eGFP expression. Despite this, it was possible to detect an increase in the green fluorescence in the strain harboring the eGFP gene at those time points (around 90 h in the dense-inoculum culture) at which antibiotic production peaked (Fig. 6c; data from diluted-inoculum culture not shown). Antibiotics are produced by the mycelium located along the periphery of the pellets, as well as in their center (Fig. 6c, d, and e). As shown above, this is the developmental pattern of the second, multinucleated, mycelium emergence (compare, for example, Fig. 1g and 6c).
DISCUSSION
Most authors assume that no differentiation processes occur in submerged cultures and that antibiotics are produced by the substrate mycelium when it reaches the stationary phase (13, 25, 43, 46, 61, 67). On the other hand, it is known that mycelial morphology correlates with the production of secondary metabolites, and most authors state that cellular aggregation, and therefore pellet and clump formation, is fundamental to obtaining good production. This is the case for, among others, retamycin in Streptomyces olindensis (17), nikkomycin in Streptomyces tendae (60), or hybrid antibiotics in Streptomyces lividans (51). The pellet characteristics (i.e., the pellet diameter and/or the existence of an active mycelium inside) have been otherwise related to the rate of antibiotic production, as in the case of nystatin in Streptomces noursei (32), tylosin in Streptomyces fradiae (47), or actinorhodin in S. coelicolor A3(2) (18, 59). Erythromycin production by Saccharopolyspora erythraea has been reported to take place in pellets 80 to 90 μm in diameter or larger, supporting the idea that the antibiotic is produced at a fixed distance from the hyphal end. Consequently, mycelia that are too small and have not developed to this length would be incapable of producing antibiotics (64). In some cases, as in the production of virginiamycin by Streptomyces virginiae (66), no clear relationship between morphology and secondary metabolite production has been encountered. All of these observations together make it difficult to establish a general relationship between morphology and antibiotic production (12). Complexity increases when physicochemical and genetic factors that also influence aggregation are considered (6). The lack of a coherent developmental model in streptomycetes has made it impossible to precisely describe reliable phenotypes in order to analyze and optimize fermentation processes.
We have recently described new aspects of the Streptomyces developmental cycle in surface cultures which substantially extend the classical developmental cycle (37-39). The existence of a previously unidentified young compartmentalized mycelium (first mycelium), which precedes a syncytial mycelium with sporadic septa (second mycelium) that develops from the compartmentalized mycelium after PCD, is the most relevant aspect of the new Streptomyces developmental life cycle we have proposed (37). The main objective of this work was to expand what we have learned about the developmental model of Streptomyces on solid cultures to submerged culture conditions, paying special attention to the relationship between differentiation and antibiotic production. We have studied the development and differentiation of S. coelicolor A3(2), a strain whose genome has been fully sequenced (2), in submerged cultures. The most important differences between surface and submerged development reside in the absence of a second death round and sporulation in the case of liquid cultures (see above). However, as in surface cultures, there is a first, compartmentalized, mycelium (first mycelium) which begins to group into pellets at early time points and which starts to die from the center outward. There is also a transient growth arrest which precedes the emergence of a multinucleated mycelium (second mycelium) that grows from the remaining viable segments of the first, compartmentalized, mycelium. Additionally, a PCD also occurs that displays the same biochemical characteristics described in solid cultures (see above).
There is a general consensus that a transient arrest of growth characterizes Streptomyces grown in submerged cultures (13, 25, 43, 46, 52, 67). This growth arrest precedes antibiotic production (28) and can activate antibiotic biosynthetic genes (31, 43), antibiotic resistance genes (50), and stress response stimulons (48), as well as arrest of ribosomal protein synthesis (3). In this work, we demonstrate that this growth arrest corresponds to the transition phase from the first vegetative compartmentalized mycelium to the second multinucleated differentiated mycelium, which is the one which produces antibiotics. We have also shown that the transient growth arrest is the consequence of the death of the first mycelium in the center of the pellets and that the resumption of growth reflects the emergence of a second, multinucleated, mycelium (see above). Death is a gradual process which starts at earlier time points than the growth arrest (14 h, Fig. 1 and 2) and peaks during that phase (see above; Fig. 5). The growth arrest prior to the appearance of the second mycelium has also been detected in solid cultures (25, 38, 55), conditions under which nutritional stresses have been postulated to activate the formation of aerial mycelium, secondary metabolism, and even lysis of the mycelium (9, 11, 63). Also comparable to surface growth conditions is the attenuation of the green fluorescence intensity of some of the hyphae from the first, compartmentalized, mycelium, which remains alive when death processes inside the pellets take place (Fig. 3q to t) (37). As suggested in previous work (37), low-fluorescence viable hyphae are conceivably in a quiescence-like state until they resume growth as a multinucleated mycelium (second mycelium). Entrance into this quiescence-like phase could be triggered by local oxygen and/or nutrient depletion in the pellet center. This would be in accordance with recent results describing how S. coelicolor hyphae, which remain viable during several weeks in anaerobiosis, resume growth after oxygen replacement (58). Further experiments are necessary to completely clarify all of these aspects.
Second, multinucleated, mycelium emergence after the death processes under surface growth conditions has been postulated to respond to diffusible activator signals (38). Interestingly, several authors have reported that pellets of S. coelicolor (56), S. fradiae (47), or S. noursei (32) suffer lytic processes at their centers in liquid cultures, and it has been suggested that these lytic processes could release secondary metabolism activators (6). These data are consistent with some experimental observations with cell-free conditioned culture medium obtained from dense-inoculum cultures which was capable of accelerating the emergence of the second mycelium, and antibiotic production (from 100 h to 40 h), in diluted-inoculum cultures (A. Manteca and J. Sanchez, unpublished results).
Analysis of antibiotic (undecylprodigiosin and actinorhodin) production, as well as genetic expression of the undecylprodigiosin redD-eGFP gene, has revealed that antibiotics are produced by the second, multinucleated, mycelium. This mycelium, as previously seen in the presporulating aerial mycelium in solid cultures, emerges after a death process which affects a compartmentalized mycelium (41). These results clarify data from other authors who have claimed that redD expression is correlated with secondary metabolism, but they were unable to associate it with any specific mycelial structure (67). Consequently, from a phenotypic point of view, S. coelicolor hyphae can be considered to experience a differentiation process under submerged conditions until the presporulation phase; this corresponds to the antibiotic-producing second, multinucleated, mycelium. In order to characterize these types of mycelium, proteomic and genomic analyses of the second, multinucleated, mycelium under liquid and solid conditions, as well as of the aerial mycelium (i.e., mycelium harboring chaplin and rodlin proteins) (14, 41) in solid cultures, are necessary.
The new perspective on the development of S. coelicolor A3(2) in submerged cultures provides a framework in which hyphal differentiation and PCD processes appear to have a profound influence on antibiotic production. This will have interesting biotechnological implications for the fermentation industries that use this microorganism; for example, the results open the possibility of designing a monitoring system (26, 65) based on the analysis of hyphal differentiation. Besides this, the developmental features described will contribute to a better understanding and/or interpretation of previously reported data on submerged cultures. For instance, proteomic experiments on the transient growth arrest phase of S. coelicolor have detected several stress-induced proteins whose biological role could not be explained (46); according to our work, these proteins would be a consequence of mycelium death. Other authors have reported that the culture medium is enriched with proteins when the growth phase is complete (34), suggesting that protein secretion is mainly a stationary-phase phenomenon. Our data open a possible different interpretation, i.e., that during the stationary phase a significant fraction of the proteins present in the culture medium is released by lysis and not by active excretion (Fig. 5).
Acknowledgments
This research was funded by a grant from the DGI, Subdireccion General de Proyectos de Investigacion, MEC, Spain (BIO2007-66313).
We thank M. Bibb, Department of Molecular Microbiology, John Innes Centre, Norwich, United Kingdom, for providing the S. coelicolor M600 redD-eGFP strain and Priscilla A. Chase for revising the text.
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Baltz, R. H. 1998. Genetic manipulation of antibiotic-producing Streptomyces. Trends Microbiol. 6:76-83. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Blanco, G., M. R. Rodicio, A. M. Puglia, C. Mendez, C. J. Thompson, and J. A. Salas. 1994. Synthesis of ribosomal proteins during growth of Streptomyces coelicolor. Mol. Microbiol. 12:375-385. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Brana, A. F., S. Wolfe, and A. L. Demain. 1986. Relationship between nitrogen assimilation and cephalosporin synthesis in Streptomyces clavuligerus. Arch. Microbiol. 146:46-51. [DOI] [PubMed] [Google Scholar]
- 6.Braun, S., and S. E. Vecht-Lifshitz. 1991. Mycelial morphology and metabolite production. Trends Biotechnol. 9:63-68. [Google Scholar]
- 7.Bystrykh, L. V., M. A. Fernández-Moreno, J. K. Herrema, F. Malpartida, D. A. Hopwood, and L. Dijkhuizen. 1996. Production of actinorhodin-related “blue pigments” by Streptomyces coelicolor A3(2). J. Bacteriol. 178:2238-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chater, K. F. 1984. Morphological and physiological differentiation in Streptomyces in microbial development. Cold Spring Harbor Monogr. Ser. 16:89-115. [Google Scholar]
- 9.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685-713. [DOI] [PubMed] [Google Scholar]
- 10.Chater, K. F., and R. Losick. 1997. The mycelial life-style of Streptomyces coelicolor A3(2) and its relatives in bacteria as multicellular organisms, p. 149-182. In J. A. Shapiro and M. Dworkin (ed.), Bacteria as multicellular organisms. Oxford University Press, New York, NY.
- 11.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 12.Chater, K. F., and G. Chandra. 2006. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol. Rev. 30:651-672. [DOI] [PubMed] [Google Scholar]
- 13.Chouayekh, H., H. Nothaft, S. Delaunay, M. Linder, B. Payrastre, N. Seghezzi, F. Titgemeyer, and M. J. Virolle. 2007. Phosphoinositides are involved in control of the glucose-dependent growth resumption that follows the transition phase in Streptomyces lividans. J. Bacteriol. 189:741-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claessen, D., L. Stokroos, H. J. Deelstra, N. A. Penninga, C. Bormann, J. A. Salas, L. Dijkhuizen, and H. A. Wösten. 2004. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol. Microbiol. 53:433-443. [DOI] [PubMed] [Google Scholar]
- 15.Daza, A., J. F. Martin, A. Dominguez, and J. A. Gil. 1989. Sporulation of several species of Streptomyces in submerged cultures after nutritional downshift. J. Gen. Microbiol. 135:2483-2491. [DOI] [PubMed] [Google Scholar]
- 16.Denser, C. R., L. M. Guimaraes, and M. Candida. 2002. Applications of image analysis in the characterization of Streptomyces olindensis in submerged culture. Braz. J. Microbiol. 33:17-21. [Google Scholar]
- 17.Denser, C. R., D. Pamboukian, and M. Candida. 2004. Production of the antitumoral retamycin during continuous fermentations of Streptomyces olindensis. Process Biochem. 39:2249-2255. [DOI] [PubMed] [Google Scholar]
- 18.Doull, J. L., and L. C. Vining. 1989. Culture conditions promoting dispersed growth and biphasic production of actinorhodin in shaken cultures of Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 53:265-268. [DOI] [PubMed] [Google Scholar]
- 19.Elibol, M. 2004. Optimization of medium composition for actinorhodin production by Streptomyces coelicolor A3(2) with response surface methodology. Process Biochem. 39:1057-1062. [Google Scholar]
- 20.Fernandez, M., and J. Sanchez. 2001. Viability staining and terminal deoxyribonucleotide transferase-mediated dUTP nick end labelling of the mycelium in submerged cultures of Streptomyces antibioticus ETH7451. J. Microbiol. Methods 47:293-298. [DOI] [PubMed] [Google Scholar]
- 21.Fernández, E., U. Weissbach, C. Sanchez-Reillo, A. F. Braña, C. Méndez, J. Rohr, and J. A. Salas. 1998. Identification of two genes from Streptomyces argillaceus encoding two glycosyltransferases involved in the transfer of a disaccharide during the biosynthesis of the antitumor drug mithramycin. J. Bacteriol. 180:4929-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fishov, I., and C. L. Woldringh. 1999. Visualization of membrane domains in Escherichia coli. Mol. Microbiol. 32:1166-1172. [DOI] [PubMed] [Google Scholar]
- 23.Flärdh, K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 49:1523-1536. [DOI] [PubMed] [Google Scholar]
- 24.Gesheva, V., V. Ivanova, and R. Gesheva. 2005. Effects of nutrients on the production of AK-111-81 macrolide antibiotic by Streptomyces hygroscopicus. Microbiol. Res. 160:243-248. [DOI] [PubMed] [Google Scholar]
- 25.Granozzi, C., R. Billeta, R. Passantino, M. Sollazzo, and A. M. Puglia. 1990. A breakdown in macromolecular synthesis preceding differentiation in Streptomyces coelicolor A3(2). J. Gen. Microbiol. 136:713-716. [DOI] [PubMed] [Google Scholar]
- 26.Hantelmann, K., M. Kollecker, D. Hüll, B. Hitzmann, and T. Scheper. 2006. Two-dimensional fluorescence spectroscopy: a novel approach for controlling fed-batch cultivations. J. Biotechnol. 121:410-417. [DOI] [PubMed] [Google Scholar]
- 27.Haugland, R. P. 2002. Nucleic acid detection and genomics technology, p. 265-352. In J. Gregory (ed.), Handbook of fluorescent probes and research chemicals, 9th ed. Molecular Probes, Inc., Eugene, OR.
- 28.Holt, T. G., C. Chang, C. Laurent-Winter, T. Murakami, J. I. Garrels, J. E. Davies, and C. J. Thompson. 1992. Global changes in gene expression related to antibiotic synthesis in Streptomyces hygroscopicus. Mol. Microbiol. 6:969-980. [DOI] [PubMed] [Google Scholar]
- 29.Hopwood, D. A., and A. M. Glauert. 1961. Electron microscope observations on the surface structures of Streptomyces violaceoruber. J. Gen. Microbiol. 26:325-330. [DOI] [PubMed] [Google Scholar]
- 30.Hopwood, D. A., K. F. Chater, and M. J. Bibb. 1995. Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Bio/Technology 28:65-102. [DOI] [PubMed] [Google Scholar]
- 31.Huang, J., C. J. Lih, K. H. Pan, and S. N. Cohen. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15:3183-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jonsbu, E., M. McIntyre, and J. Nielsen. 2002. The influence of carbon sources and morphology on nystatin production by Streptomyces noursei. J. Biotechnol. 95:133-144. [DOI] [PubMed] [Google Scholar]
- 33.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics, p.43-61. John Innes Foundation, Norwich, United Kingdom.
- 34.Kim, D.-W., K. Chater, K.-J. Lee, and A. Hesketh. 2005. Changes in the extracellular proteome caused by the absence of the bldA gene product, a developmentally significant tRNA, reveal a new target for the pleiotropic regulator AdpA in Streptomyces coelicolor. J. Bacteriol. 187:2957-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, Y. M., and J. H. Kim. 2004. Formation and dispersion of mycelial pellets of Streptomyces coelicolor A3(2). J. Microbiol. 42:64-67. [PubMed] [Google Scholar]
- 36.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 37.Manteca, A., M. Fernandez, and J. Sanchez. 2005. A death round affecting a young compartmentalized mycelium precedes aerial mycelium dismantling in confluent surface cultures of Streptomyces antibioticus. Microbiology 151:3689-3697. [DOI] [PubMed] [Google Scholar]
- 38.Manteca, A., M. Fernandez, and J. Sanchez. 2005. Mycelium development in Streptomyces antibioticus ATCC11891 occurs in an orderly pattern which determines multiphase growth curves. BMC Microbiol. 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manteca, A., M. Fernandez, and J. Sanchez. 2006. Cytological and biochemical analysis of two lytic programmed cellular dismantling rounds accompanying the development of Streptomyces antibioticus in surface cultures. Res. Microbiol. 157:143-152. [DOI] [PubMed] [Google Scholar]
- 40.Manteca, A., U. Mäder, B. A. Connolly, and J. Sanchez. 2006. A proteomic analysis of Streptomyces coelicolor programmed cell death. Proteomics 6:6008-6022. [DOI] [PubMed] [Google Scholar]
- 41.Manteca, A., D. Claessen, C. Lopez-Iglesias, and J. Sanchez. 2007. Aerial hyphae in surface cultures of Streptomyces lividans and Streptomyces coelicolor originate from viable segments surviving an early programmed cell death event. FEMS Microbiol. Lett. 274:118-125. [DOI] [PubMed] [Google Scholar]
- 42.Méndez, C., A. F. Braña, M. B. Manzanal, and C. Hardisson. 1985. Role of substrate mycelium in colony development in Streptomyces. Can. J. Microbiol. 31:446-450. [DOI] [PubMed] [Google Scholar]
- 43.Neumann, T., W. Piepersberg, and J. Distler. 1996. Decision phase regulation of streptomycin production in Streptomyces griseus. Microbiology 142:1953-1963. [Google Scholar]
- 44.Nicieza, R. G., J. Huergo, B. A. Connolly, and J. Sanchez. 1999. Purification, characterization and role of nucleases and serine proteases in Streptomyces differentiation: analogies with the biochemical processes described in late steps of eukaryotic apoptosis. J. Biol. Chem. 274:20366-20375. [DOI] [PubMed] [Google Scholar]
- 45.Novella, I. S., C. Barbes, and J. Sanchez. 1992. Sporulation of Streptomyces antibioticus ETHZ 7451 in submerged culture. Can. J. Microbiol. 38:769-773. [DOI] [PubMed] [Google Scholar]
- 46.Novotna, J., J. Vohradsky, P. Berndt, H. Gramajo, H. Langen, X. M. Li, W. Minas, L. Orsaria, D. Roeder, and C. J. Thompson. 2003. Proteomic studies of diauxic lag in the differentiating prokaryote Streptomyces coelicolor reveal a regulatory network of stress-induced proteins and central metabolic enzymes. Mol. Microbiol. 48:1289-1303. [DOI] [PubMed] [Google Scholar]
- 47.Park, Y., S. Tamura, Y. Koike, M. Toriyama, and M. Okabe. 1997. Mycelial pellet intrastructure visualization and viability prediction in a culture of Streptomyces fradiae using confocal scanning laser microscopy. J. Ferment. Bioeng. 84:483-486. [Google Scholar]
- 48.Puglia, A. M., J. Vohradsky, and C. J. Thompson. 1995. Developmental control of the heat-shock stress regulon in Streptomyces coelicolor. Mol. Microbiol. 17:737-746. [DOI] [PubMed] [Google Scholar]
- 49.Rueda, B., E. M. Miguelez, C. Hardisson, and M. B. Manzanal. 2001. Mycelial differentiation and spore formation by Streptomyces brasiliensis in submerged culture. Can. J. Microbiol. 47:1042-1047. [PubMed] [Google Scholar]
- 50.Salah-Bey, K., V. Blanc, and C. J. Thompson. 1995. Stress-activated expression of a Streptomyces pristinaespiralis multidrug resistance gene (ptr) in various Streptomyces spp. and Escherichia coli. Mol. Microbiol. 17:1001-1012. [DOI] [PubMed] [Google Scholar]
- 51.Sarra, M., C. Casas, and F. Godia. 1997. Continuous production of a hybrid antibiotic by Streptomyces lividans TK21 pellets in a three-phase fluidized-bed bioreactor. Biotechnol. Bioeng. 53:601-610. [DOI] [PubMed] [Google Scholar]
- 52.Schwedock, J., J. R. McCormick, E. R. Angert, J. R. Nodwell, and R. Losick. 1997. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 25:847-858. [DOI] [PubMed] [Google Scholar]
- 53.Stocks, S. M., and C. R. Thomas. 2001. Viability, strength, and fragmentation of Saccharopolyspora erythraea in submerged fermentation. Biotechnol. Bioeng. 75:702-709. [DOI] [PubMed] [Google Scholar]
- 54.Sun, J., G. H. Kelemen, J. M. Fernandez-Abalos, and M. J. Bibb. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145:2221-2227. [DOI] [PubMed] [Google Scholar]
- 55.Süsstrunk, U., J. Pidoux, S. Taubert, A. Ullmann, and C. J. Thompson. 1998. Pleiotropic effects of cAMP on germination, antibiotic biosynthesis and morphological development in Streptomyces coelicolor. Mol. Microbiol. 30:33-46. [DOI] [PubMed] [Google Scholar]
- 56.Tough, A. J., and J. I. Prosser. 1996. Experimental verification of a mathematical model for pelleted growth of Streptomyces coelicolor A3(2) in submerged batch culture. Microbiology 142:639-648. [DOI] [PubMed] [Google Scholar]
- 57.Tsao, S. W., B. A. Rudd, X. G. He, C. J. Chang, and H. G. Floss. 1985. Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J. Antibiot. (Tokyo) 38:128-131. [DOI] [PubMed] [Google Scholar]
- 58.van Keulen, G., J. Alderson, J. White, and R. G. Sawers. 2007. The obligate aerobic actinomycete Streptomyces coelicolor A3(2) survives extended periods of anaerobic stress. Environ. Microbiol. 9:3143-3149. [DOI] [PubMed] [Google Scholar]
- 59.van Wezel, G. P., P. Krabben, B. A. Traag, B. J. Keijser, R. Kerste, E. Vijgenboom, J. J. Heijnen, and B. Kraal. 2006. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl. Environ. Microbiol. 72:5283-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vecht-Lifshitz, S. E., Y. Sasson, and S. Braun. 1992. Nikkomycin production in pellets of Streptomyces tendae. J. Appl. Bacteriol. 72:195-200. [DOI] [PubMed] [Google Scholar]
- 61.Vohradský, J., X. M. Li, and C. J. Thompson. 1997. Identification of procaryotic developmental stages by statistical analyses of two-dimensional gel patterns. Electrophoresis 18:1418-1428. [DOI] [PubMed] [Google Scholar]
- 62.Waksman, S. A., and H. B. Woodruff. 1940. The soil as a source of microorganisms antagonistic to disease-producing bacteria. J. Bacteriol. 40:581-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waksman, S. A. 1967. The actinomycetes: a summary of current knowledge, p. 101-112. Ronald Press, New York, NY.
- 64.Wardell, J. N., S. M. Stocks, C. R. Thomas, and M. E. Bushell. 2002. Decreasing the hyphal branching rate of Saccharopolyspora erythraea NRRL 2338 leads to increased resistance to breakage and increased antibiotic production. Biotechnol. Bioeng. 78:141-146. [DOI] [PubMed] [Google Scholar]
- 65.Wei, N., J. You, K. Friehs, E. Flaschel, and T. W. Nattkemper. 2007. An in situ probe for on-line monitoring of cell density and viability on the basis of dark field microscopy in conjunction with image processing and supervised machine learning. Biotechnol. Bioeng. 97:1489-1500. [DOI] [PubMed] [Google Scholar]
- 66.Yang, Y. K., M. Morikawa, H. Shimizu, S. Shioya, K. I. Suga, T. Nihira, and Y. Yamada. 1996. Image analysis of mycelial morphology in virginiamycin production by batch culture of Streptomyces virginiae. J. Ferment. Bioeng. 81:7-12. [Google Scholar]
- 67.Zhou, L.-H., Y.-Q. Li, Y.-Q. Li, and D. Wu. 2005. Spatio-temporal expression of the pathway-specific regulatory gene redD in S. coelicolor. J. Zhejiang Univ. Sci. 6:464-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhuang, Y. P., B. C. J. Chu, and S. Zhang. 2006. Medium optimization for meilingmycin production by Streptomyces nanchangensis using response surface methodology. Process Biochem. 41:405-409. [Google Scholar]