Abstract
A simple, sensitive, and rapid cell-free assay system was developed for detection of N-acyl homoserine lactone (AHL) autoinducers involved in bacterial quorum sensing (QS). The present approach improves upon previous whole-cell biosensor-based approaches in its utilization of a cell-free assay approach to conduct bioassays. The cell-free assay was derived from the AHL biosensor bacterium Agrobacterium tumefaciens NTL4(pCF218)(pCF372), allowing the expression of β-galactosidase upon addition of exogenous AHLs. We have shown that β-galactosidase expression is possible in cell-free solution [lysate from Agrobacterium tumefaciens NTL4(pCF218)(pCF372) culture]. Assay detection limits with the use of chromogenic substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) ranged from approximately 100 nM to 300 nM depending on the specific AHL. Replacement (of X-Gal) with the luminescent substrate Beta-Glo increased sensitivity to AHLs by 10-fold. A major advantage of the cell-free assay system is elimination of time-consuming steps for biosensor cell culture conditioning, which are required prior to whole-cell bioassays. This significantly reduced assay times from greater than 24 h to less than 3 h, while maintaining high sensitivity. Assay lysate may be prepared in bulk and stored (−80°C) over 6 months for future use. Finally, the present protocol may be adapted for use with other biosensor strains and be used in high-throughput AHL screening of bacteria or metagenomic libraries.
Quorum sensing (QS) has been an emerging research focus in health and environmental sciences during the past decade (2, 5, 9, 37). QS is the population-dependent ability of bacteria to communicate and regulate gene expression through the production, release, and concentration-dependent sensing of signal molecules called autoinducers (9, 12, 38). A wide range of bacterial processes are now known to be influenced by QS and include bioluminescence, cell density control, toxin production, cell differentiation, exopolysaccharide production, motility, biofilm formation, and virulence factor production (20, 40).
Autoinducers are released by cells, diffuse through the extracellular environment, and are “detected” by neighboring cells, often resulting in concentration-dependent changes in gene expression. A major class of autoinducers is the N-acyl homoserine lactones (AHLs) (20). To date, many qualitative and quantitative approaches have been developed to detect AHLs. These include whole-cell-based bioassays using AHL-specific biosensors, thin-layer chromatography, gas chromatography-mass spectrometry (MS), high-performance liquid chromatography (HPLC), liquid chromatography-mass spectrometry (LC-MS), isotopic labeling, and absorbance-based assays (1, 6, 14, 19, 21, 26, 27, 34, 36, 39, 41, 42, 43).
A very useful and often applied approach for QS screening is the whole-cell bioassay, which utilizes specific bacterial biosensors (31). It is relatively sensitive and does not require extensive research instrumentation, such as HPLCs and LC-MS.
The β-galactosidase expression system has been used as a specific indicator of gene expression (17). The bacterial reporter strain Agrobacterium tumefaciens NTL4(pCF218)(pCF372) contains the β-galactosidase gene driven by a traI promoter, allowing the expression of β-galactosidase to be regulated by the presence of QS signals (AHLs). In the presence of the substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), β-galactosidase enzymatically cleaves X-Gal, which results in its conversion to a blue precipitate when active forms of AHLs are present. Accumulation of the blue precipitate is then detectable by spectral absorbance at 635 nm.
A rapidly developing area in the study of QS involves the detection of autoinducer activities from bacterial communities under natural conditions. Whole-cell bioassays are often used for screening gram-negative bacterial colonies that produce AHLs. However, whole-cell bioassays are constrained by several limitations: (i) the requirement of a time-consuming cell conditioning step prior to the start of the bioassay, (ii) the lengthy incubation times (i.e., at least 24 h for detection of AHLs), and (iii) the sensitive adjustments in cell densities needed (e.g., of each well of a 96-well plate or each test tube) to calculate relative activities of luminescence or absorbance. Therefore, a simple and rapid assay that is both sensitive and relatively robust (under environmental conditions) is needed for the detection of AHLs extracted from natural systems.
In the present approach, we have reduced these limitations by developing a cell-free assay system to detect AHL QS signals. Cell-free lysates were derived from the reporter bacterium A. tumefaciens NTL4(pCF218)(pCF372) and, without any addition, contained all the necessary cellular components for in vitro gene expression and translation (e.g., 70S ribosome; tRNAs; aminoacyl-tRNA synthetases; initiation, elongation, and termination factors; amino acids; ATP; GTP; and cofactors such as Mg2+ and K+) (28). The β-galactosidase expression system was shown to increase in stoichiometric proportion to the concentration of AHLs. Using this system, many samples could be rapidly screened for the presence of AHLs by simple addition of a cell-free lysate and reporter substrate. AHLs were then detected within 3 h using a microplate reader, spectrophotometer, or luminometer.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
The reporter strain A. tumefaciens NTL4(pCF218)(pCF372) lacks the Ti plasmid and contains two plasmids, pCF218 and pCF372, that encode the traR and traI-lacZ fusion genes, respectively (10, 11). This system provides extremely sensitive detection of AHLs (10). W. Clay Fuqua, Indiana University, generously provided the strain for our assay. Cultures of A. tumefaciens NTL4(pCF218)(pCF372) were grown in 250-ml vessels using AT minimal glucose medium (35) containing 0.5% (wt/vol) glucose, 0.079 M KH2PO4, 0.015 M (NH4)2SO4, 0.6 mM MgSO4·7H2O, 0.06 mM CaCl2·2H2O, 0.027 mM FeSO4·7H2O, and 0.0071 mM MnSO4·H2O in distilled H2O and adjusted with 1 N NaOH to pH 7.0. The antibiotics streptomycin (50 μg/ml) and tetracycline (5 μg/ml) were also added.
AHLs.
The AHLs N-butanoyl-homoserine lactone (C4-AHL), N-hexanoyl-homoserine lactone (C6-AHL), N-heptanoyl-homoserine lactone (C7-AHL), N-octanoyl-homoserine lactone (C8-AHL), N-decanoyl-homoserine lactone (C10-AHL), N-dodecanoyl-homoserine lactone (C12-AHL), N-tetradecanoyl-homoserine lactone (C14-AHL), N-3-oxo-hexanoyl-homoserine lactone (3-oxo-C6-AHL), and N-3-oxo-octanoyl-homoserine lactone (3-oxo-C8-AHL) were purchased from Sigma-Aldrich (St. Louis, MO) and used for development of the cell-free assay system. C8-AHL was used for final optimization of the cell-free assay system.
Preparation of cell-free lysate.
One liter of the reporter strain was grown in AT minimum glucose medium (30°C with shaking for 18 h) to early exponential phase and then harvested by centrifugation (12,000 × g; 10 min). Cell pellets were collected and resuspended in 5 ml of KH2PO4 buffer (100 mM; pH 7.4) and then sonicated for 30 s three times and centrifuged (12,000 × g) at 4°C for 30 min to remove particulate cell fragments. The supernatant was collected as the “cell-free lysate” reagent and was stored (−80°C) until use.
Confirmation of β-galactosidase expression in vitro in cell-free solution.
To confirm β-galactosidase expression in cell-free solution, the following simple absorbance assay for AHLs was used. The protocol involved (i) addition of 50 μl of sample solution containing AHLs into a 96-well plate; (ii) addition of 50 μl of cell extract and 100 μl of 20 mM KH2PO4 (pH 7.0) into each well, followed by mixing and incubation at 30°C for 2 h; (iii) addition of 1 μl of X-Gal (20 mg/ml) into each well and then mixing and incubation at 30°C for 1 h; and (iv) measurement of absorbance in each well at 635 nm by spectrophotometry (Shimadzu UV-2401PC UV-VIS; Shimadzu Corp.). To determine the detection limits, 1:1 serial dilutions of each AHL in 20 mM KH2PO4 (pH 7.0) buffer were performed in triplicate. Induction of β-galactosidase activity was calculated by dividing the absorbance of samples by those of controls. An induction ratio greater than 3 was considered active (27).
Components required for reactions 1 through 3.
In order for the assay to detect AHLs in a concentration-dependent manner, a series of reactions that are normally restricted to the confines of the cell must also be able to occur in the cell-free assay. These reactions include binding of AHLs with the receptor protein TraR (reaction 1), binding of the TraR/AHL complex to the traR promoter driving the expression of the β-galactosidase gene (reaction 2), and translation of mRNA for the synthesis of β-galactosidase (reaction 3). The following experiments were conducted to confirm that the above reactions were occurring in the cell-free assay.
In order to determine if constitutive production (reactions 1 through 3) of β-galactosidase was occurring within the cell extract (i.e., in the absence of added AHLs), assays were carried out with/without added N-octanoyl-homoserine lactone (C8-AHL), X-Gal, and C8-AHL-plus-X-Gal combinations. The treatments (in triplicate) consisted of added C8-AHL, X-Gal, C8-AHL plus X-Gal, cell lysate plus C8-AHL, cell lysate plus X-Gal, and cell lysate plus C8-AHL plus X-Gal. The concentration of C8-AHL in 20 mM KH2PO4 (pH 7.0) was 1 μM. Absorbance was measured at 635 nm, and values were compared for each treatment.
Inhibition of reaction 1.
In order to show that blocking TraR binding of AHL inhibits β-galactosidase expression, high temperature (i.e., 60°C) was used to degrade most proteins including the TraR protein, which was required for transcription of the β-galactosidase gene. Cell-free lysates were heated for 30 min at 30°C, 40°C, 50°C, 55°C, and 60°C prior to the assay. Once the cell-free lysates had cooled to 30°C, C8-AHL standards were added at a concentration of 1 μM with replicates and incubated. Then, expression of β-galactosidase was measured using the absorbance assay protocol.
Inhibition of reactions 2 and 3.
To artificially inhibit binding of the AHL/TraR complex to the β-galactosidase promoter, we utilized the aminoglycoside antibiotic streptomycin, which precipitates DNA, therefore inhibiting the binding of the AHL/TraR complex to the β-galactosidase promoter (28). First, 50-μl aliquots of 1 μM C8-AHL solution were added to each tube, followed by addition of cell extract. Further, a series of increasing concentrations of streptomycin (i.e., 0, 50, 125, 250, and 300 μg) were added to the cell extract and incubated for 30 min prior to the assay (4, 22, 29). Then, β-galactosidase expression was measured by absorbance.
Optimization of cell-free assay.
In order to optimize cell-free assay conditions, the following experiments were conducted. All optimization experiments used the C8-AHL (1 μM).
Crude protein concentration in a cell-free assay solution.
Initially, protein concentrations of undiluted cell extract were determined by the Bradford protein assay (3). Then, cell extract (30 μg/ml) was diluted with 20 mM KH2PO4 (pH 7.0) buffer to 12 μg/ml and 6 μg/ml. Absorbance-based measurements were conducted using the cell-free assay system to determine the effect of protein concentration in cell extracts on β-galactosidase expression.
Optimum pH.
In order to assess the effect of pH on the cell-free assay, triplicate assay solutions were adjusted to pH 4.5, 5.0, 5.4, 6.5, 7.0, 8.0, and 8.8. Then, cell-free assay absorbances were measured.
Incubation time.
To determine the effect of incubation time of assay measurements, triplicate cell extract solutions were spiked with C8-AHL (1 μM) and then incubated for different time periods (1, 2, 3, and 4 h). Then, X-Gal was added to each solution, the solution was incubated at 37°C for 1 h, and absorbance (635 nm) was measured.
Comparison of absorbance and luminescence assays.
To improve the sensitivity of the cell-free approach, a luminescence assay was developed for AHLs. The protocol involved (i) addition of 50 μl of sample solution containing AHLs into a 96-well plate; (ii) addition of 50 μl of cell extract, diluted in 20 mM KH2PO4 buffer (pH 7.0), resulting in a protein concentration of 80 μg/ml for each well, and then mixing and incubation at 30°C for 2 h; (iii) addition of 100 μl of Beta-Glo (Promega) into each well, followed by mixing and incubation (30°C for 1 h); and (iv) measurement of luminescence using a Veritus microplate luminometer (Turner BioSystems). For control wells, 20 mM KH2PO4 (pH 7.0) buffer was used. To determine the detection limit, a 1:1 serial dilution of each AHL was performed in triplicate using 20 mM KH2PO4 (pH 7.0) buffer. Then, absorbance assays were conducted. Induction of β-galactosidase activity was calculated by dividing the sample absorbance or luminescence by that of controls. An induction ratio greater than 3 was considered active (27).
Application of cell-free assay for high-throughput screening of genomic clones for the identification of putative AHL genes.
To test the applicability of the cell-free assay for detecting AHL genes, we used the assay to screen a genomic library derived from a sulfate-reducing bacterium (SRB) isolated from marine stromatolites at Highborne Cay, Bahamas. The SRB strain was identified as Desulfovibrio sp. strain H2.3jLac (GenBank accession no. DQ822786). To identify the genes involved in Desulfovibrio sp. strain H2.3jLac QS, genomic DNA was extracted and a fosmid-based genomic library was constructed. The fosmid library was subsequently screened using the cell-free assay, described above, to identify regions within the Desulfovibrio genome that may play a role in QS. Five hundred fosmid clones were grown overnight in 96-well plates followed by the addition of 20 μl of cell extract. Plates were incubated for 2 h followed by the addition of 1 μl (20 mg/ml) of X-Gal and then further incubated overnight at 37°C, and clones producing a blue color were scored as positive for putative QS gene activity. Production of AHLs by positive clones was confirmed by LC-MS. Briefly, positive clones were grown in marine broth at 37°C. Culture supernatants were extracted with acidified ethyl acetate, dried, and reconstituted with 50% acetonitrile. AHLs in samples were separated by HPLC using a 2.1-mm × 150-mm Aquasep C18 column (ES Industries). The separation was performed using a binary gradient of two solvents (solvent A, H2O with 0.1% [wt/vol] formic acid; solvent B, acetonitrile with 0.1% formic acid). Initially, gradient conditions were 20% solvent A for 2 min and then linear ramping (28 min) to 100% solvent B. AHLs were detected using a Waters Premier XE triple quadrupole mass spectrometer with positive-ion electrospray ionization. The triple quad mass spectrometer was operated in multiple reaction monitoring mode utilizing two characteristic fragment transitions per analyte.
RESULTS
Confirmation of β-galactosidase expression upon addition of AHLs in vitro to cell-free solution.
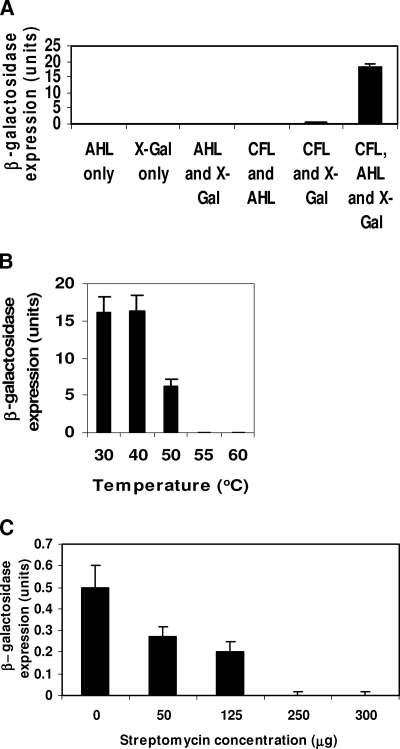
Results of negative and positive controls, consisting of cell-free assays conducted with and without X-Gal and C8-AHL (Fig. 1A), respectively, showed that, as expected, unless both C8-AHL and X-Gal were added into cell-free assay solution, β-galactosidase activity remained minimal. Controls, which had addition of X-Gal but no AHL, exhibited minimal background β-galactosidase activity in the cell extract.
FIG. 1.
Confirmation of β-galactosidase expression in in vitro cell-free solution. (A) Components required for reactions 1 through 3. These reactions include binding of AHLs with the reporter protein TraR (reaction 1), binding of the TraR/AHL complex to the traR promoter driving the expression of β-galactosidase (reaction 2), and translation of mRNA for the synthesis of β-galactosidase (reaction 3). CFL, cell-free lysate. (B) Inhibition of reaction 1 with high temperature. (C) Inhibition of reactions 2 and 3 by streptomycin. Bars indicates means ± standard deviations (n = 3).
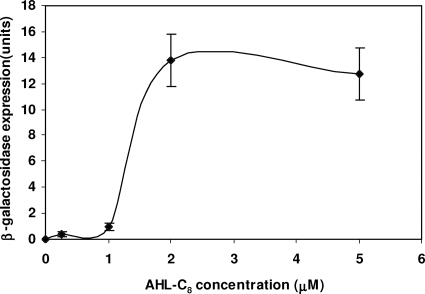
Results of β-galactosidase activities showed that detection of AHLs using the cell-free assay with the cell extract was deactivated above 55°C (Fig. 1B). Therefore, β-galactosidase expression did not occur when pretreatment of cell extracts with high temperature (e.g., 55°C and 60°C) was followed by addition (at 30°C) of C8-AHL. Further, streptomycin additions above 125 μg/ml reduced assay activities (Fig. 1C). Results of C8-AHL additions showed that absorbance changes (i.e., blue color at 635 nm), resulting from the enzymatic cleavage of the chromogenic substrate X-Gal, were proportional to concentration, up to 2 μM C8-AHLs (Fig. 2).
FIG. 2.
Dose-response curve of β-galactosidase expression in a cell-free lysate with addition of C8-AHL. Points represent mean values of three samples. Error bars represent standard deviations.
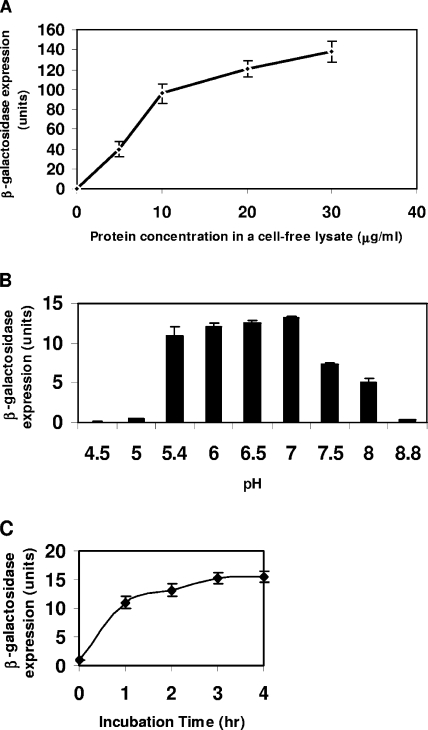
Optimization of cell-free assay. (i) Crude protein concentration in cell extract solution.
Crude protein concentrations, ranging from 6 μg/ml to 30 μg/ml, resulted in a nonlinear increase of β-galactosidase expression with time (Fig. 3A).
FIG. 3.
Optimization of cell-free assay conditions. (A) Crude protein concentrations in a cell-free extract solution. (B) Optimum pH for a cell-free assay. (C) Incubation times for cell-free assays. Values (points or bars) indicate means ± standard deviations (n = 3).
(ii) Determination of optimum pH.
Results of incubations examining the effect of pH on β-galactosidase expression in the cell-free assay (Fig. 3B) showed that the highest β-galactosidase expression occurred between pH 6 and 7, while expression declined dramatically at pH 7.5 or higher. Therefore, pH 6.5 was chosen for the measurement conditions for the cell-free assay. Since the assay was very sensitive to pH, reducing the volume of sample to maintain the pH of the whole assay solution was important.
(iii) Incubation time.
Incubation time affected β-galactosidase expression (Fig. 3C). Increasing assay incubation times from 1 to 4 hours resulted in an asymptotic increase in expression of β-galactosidase activity for a given concentration of added AHL. In order to make the cell-free assay system rapid and practical, a 2-h incubation was chosen for our assays.
Comparison of absorbance and luminescence assays.
The cell-free assay was not able to detect C4- and C14-AHLs. Using the luminescence substrate Beta-Glo, detection of various AHLs was approximately 10-fold more sensitive than that with absorbance assays (Table 1).
TABLE 1.
Minimum concentration of each AHL resulting in an induction ratio greater than 3 in cell-free absorbance and luminescence assays
| AHL | Minimum concn (nM) (mean ± SD [n = 3])
|
|
|---|---|---|
| Luminescence assay | Absorbance assay | |
| C4-AHL | NDa | ND |
| C6-AHL | 30 ± 3 | 335 ± 10 |
| C7-AHL | 25 ± 5 | 245 ± 8 |
| C8-AHL | 20 ± 2 | 225 ± 11 |
| C10-AHL | 100 ± 5 | 1,300 ± 20 |
| C12-AHL | 200 ± 6 | 2,400 ± 30 |
| C14-AHL | ND | ND |
| oxo-C6-AHL | 17 ± 2 | 180 ± 25 |
| oxo-C8-AHL | 10 ± 3 | 120 ± 10 |
ND, not detectable.
Application of cell-free assay for screening genomic clones for the identification of putative luxI homologs.
To demonstrate one possible application of the newly developed cell-free assay, a genomic library was constructed from the SRB isolate Desulfovibrio strain H2.3jLac and screened for putative QS activity by the cell-free assay. Results of screening (∼5× genome coverage) revealed that 24/500 fosmid clones demonstrated positive reactions ranging from a high to a low level of activity. Then, production of several AHLs (C6-AHL, oxo-C6-AHL, C8-AHL, C10-AHL, and C12-AHL) by positive clones was confirmed by MS (results not shown). Further studies are under way to identify the genes involved in the production of AHLs by Desulfovibrio strain H2.3jLac.
DISCUSSION
Cell-free assay for AHLs.
In practice, only a few cell-free-based expression systems have been developed and have been utilized largely for in vitro protein synthesis. The most frequently used cell-free translation systems consist of extracts from rabbit reticulocytes, wheat germ cells, insect cells, and Escherichia coli (7, 8, 10, 13, 15, 23, 30, 32). In the present study, we present evidence for the occurrence of in vitro gene expression and protein translation in a cell-free lysate system derived from the bacterium Agrobacterium tumefaciens NTL4(pCF218)(pCF372) and have used the system for the purpose of detecting AHL autoinducers involved in QS. We demonstrated that the sensitivity of the cell-free assay is comparable to that of whole-cell-based approaches (27) but additionally the assay is more simplified and shortened. Typically, an assay with a whole-cell bioreporter such as A. tumefaciens NTL4(pZLR4) requires 16 to 18 h to culture the bioreporter strain and an additional 16 to 18 h for incubation of the assay mixture (27). In contrast, our cell-free assay system takes 2 to 3 h in total. Therefore, the cell-free assay system for detecting QS signals eliminates the time-consuming cell conditioning step in biosensor cell cultures that is required before each whole-cell bioassay.
In contrast to eukaryotic systems, where transcription and translation occur sequentially, bacterial systems have transcription and translation occurring simultaneously within the cell. Our in vitro reactions likely occurred in the same way and allowed the assay system to proceed.
We succeeded in showing that active β-galactosidase was produced in the cell-free system. We also showed that, while there was an inherent basal level of β-galactosidase expression, an upregulation of enzyme expression occurred in response to exogenously added AHL.
An important step in maintaining the accuracy and precision of the assay is the ability of the cell-free lysate to achieve consistent in vitro transcription of the β-galactosidase gene and protein expression. Without this, the lower concentrations of AHLs that will be detectable using the assay will be limited by basal (i.e., constitutive) concentrations of β-galactosidase.
In our cell-free assays, relatively short incubation times (e.g., 2 h) still resulted in highly sensitive detection of AHLs. It is interesting that in some A. tumefaciens reporter strains which contain the wild-type Ti plasmid there was an 8-h delay in gene activation when AHLs were added (24). This was found to be due to the presence of a negative regulator protein, TraM, whose expression is located on the Ti plasmid (18, 25). TraM tightly binds to TraR and inhibits activation of target genes of TraR (16, 33). The A. tumefaciens NTL4(pCF218)(pCF372) strain used in our study lacked the Ti plasmid, and this likely contributed to the more rapid (<2-h) gene activation and autoinduction that were observed in our assays.
Previously, the regulator protein TraR was thought to be loosely associated with the inner leaflet of the cytoplasmic membrane when it was not complexed to an AHL (25). Complexation of TraR with a cognate AHL converts this transcription factor to an active form (18). Our results suggested either that membrane-bound TraR was present in our cell-free assay, which could be activated by complexation with an AHL, or alternatively that TraR was not associated with large membrane fragments removed by fractionation. It is likely that our centrifugation protocol (e.g., 12,000 × g for 30 min) allowed cell membrane fragments (and their attached TraR) to remain in the supernatant.
Application of cell-free assay.
The cell-free assay protocol is very suitable for the rapid screening of bacterial isolates and extracts for the detection of QS compounds and QS inhibitors. It is additionally valuable for the rapid screening of putative QS activity in genomic clones, as demonstrated in our study. Also, the cell-free assay may be potentially linked with other types of reporters. In our study, replacement (of X-Gal) with the luminescent substrate Beta-Glo increased assay sensitivity to AHLs by 10-fold, compared with the absorbance-based assay. Also, the method is simple and cost-effective and can be easily applied to test for the presence of AHLs in environmental samples under field conditions. Since the present assay using A. tumefaciens NTL4(pCF218)(pCF372) was not sensitive to C4-AHL and C14-AHL, development of a cell-free assay that utilizes another strain, Agrobacterium tumefaciens KYC55(pJZ372)(pJZ384)(pJZ410), is in progress. This strain is much more sensitive to a wider range of AHLs (43), and the overall sensitivity of the assay may be improved.
Acknowledgments
This work was supported by grants from the U.S. National Science Foundation's Collaborative Research in Chemistry Program (CHE-0526821), the NSF Biocomplexity Program (BE/CBC-0221796), and the NSF Environmental Genomics Program (EF-0723707).
We kindly thank Clay Fuqua for providing the reporter strain, Agrobacterium tumefaciens NTL4(pCF218)(pCF372), that was used in the study. We also thank Pieter Visscher for providing SRB isolate Desulfovibrio H2.3jLac and appreciate the comments from anonymous reviewers.
The manuscript represents contribution 42 to the Research Initiative on Bahamian Stromatolites (RIBS) project.
Footnotes
Published ahead of print on 18 April 2008.
REFERENCES
- 1.Andersen, J. B., A. Heydorn, M. Hentzer, L. Eberl, O. Geinsenberger, B. B. Christensen, S. Molin, and M. Givskov. 2001. gfp-based N-acyl homoserine lactone sensor system for detection of bacterial communication. Appl. Environ. Microbiol. 67:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L. 2002. Small talk: cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantity of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brock, T. D. 1964. Action of streptomycin and related antibiotics. Fed. Proc. 23:965-975. [PubMed] [Google Scholar]
- 5.Camara, M., P. Williams, and A. Hardman. 2002. Controlling infection by tuning in and tuning down the volume of bacterial small-talk. Lancet Infect. Dis. 2:667-676. [DOI] [PubMed] [Google Scholar]
- 6.Cataldi, T. R. I., G. Bianco, L. Palazzo, and V. Quaranta. 2007. Occurrence of N-acyl-l-homoserine lactones in extracts of some gram-negative bacteria evaluated by gas chromatography-mass spectrometry. Anal. Biochem. 361:226-235. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H. Z., and G. Zubay. 1983. Prokaryotic coupled transcription-translation. Methods Enzymol. 101:674-690. [DOI] [PubMed] [Google Scholar]
- 8.Ezure, T., T. Suzuki, S. Higashide, E. Shintami, K. Endo, S. Kobayashi, M. Shikata, M. Ito, K. Tanimizu, and O. Nishimura. 2006. Cell-free protein synthesis system prepared from insect cells by freeze-thawing. Biotechnol. Prog. 22:1570-1577. [DOI] [PubMed] [Google Scholar]
- 9.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 10.Fuqua, C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González, J. E., and N. D. Keshavan. 2006. Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 70:859-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson, R. J., and T. Hunt. 1983. Preparation and use of nuclease-treated rabbit reticulocyte lysates for translation of eukaryotic messenger RNA. Methods Enzymol. 96:50-74. [DOI] [PubMed] [Google Scholar]
- 14.Jafra, S., and J. M. van der Wolf. 2004. Fast screening method for detection of acyl-HSL-degrading soil isolates. J. Microbiol. Methods 57:415-420. [DOI] [PubMed] [Google Scholar]
- 15.Kim, D. M., T. Kigawa, C. Y. Choi, and S. Yokaoyama. 1996. A highly efficient cell-free protein synthesis system from Escherichia coli. Eur. J. Biochem. 239:881-886. [DOI] [PubMed] [Google Scholar]
- 16.Luo, Z. Q., Y. Quin, and S. K. Farrand. 2000. The antiactivator traM interferes with the autoinducer-dependent binding of traR to DNA by interacting with the C-terminal region of the quorum sensing activator. J. Biol. Chem. 275:7713-7722. [DOI] [PubMed] [Google Scholar]
- 17.Luo, Z. Q., T. E. Clemente, and S. K. Farrand. 2001. Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol. Plant-Microbe Interact. 14:98-103. [DOI] [PubMed] [Google Scholar]
- 18.Luo, Z. Q., S. Su, and S. K. Farrand. 2003. In situ activation of the quorum-sensing transcription factor TraR by cognate and noncognate acyl-homoserine lactone ligands: kinetics and consequences. J. Bacteriol. 185:5665-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. A. B. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacin production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 20.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 21.Morin, D., B. Grasland, K. Vallée-Réhel, C. Dufau, and D. Haras. 2003. On-line high-performance liquid chromatography-mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum sensing signal molecules in the presence of biological matrices. J. Chromatogr. A 1002:79-92. [DOI] [PubMed] [Google Scholar]
- 22.Moskowitz, M. 1963. Differences in the precipitability of nucleic acids with streptomycin and dihydrostreptomycin. Nature 200:335-337. [DOI] [PubMed] [Google Scholar]
- 23.Olliver, L., and C. D. Boyd. 1998. In vitro translation of messenger RNA in a rabbit reticulocyte lysate cell-free system. Methods Mol. Biol. 86:221-227. [DOI] [PubMed] [Google Scholar]
- 24.Piper, K. R., and S. K. Farrand. 2000. Quorum sensing but not autoinduction of Ti plasmid conjugal transfer requires control by opine regulon and the antiactivator TraM. J. Bacteriol. 182:1080-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin, Y., S. Su, and S. K. Farrand. 2007. Molecular basis of transcriptional antiactivation. J. Biol. Chem. 282:19979-19991. [DOI] [PubMed] [Google Scholar]
- 26.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh, M. P., and M. Greenstein. 2006. A simple, rapid, sensitive method detecting homoserine lactone (HSL)-related compounds in microbial extracts. J. Microbiol. Methods 65:32-37. [DOI] [PubMed] [Google Scholar]
- 28.Snyder, L., and W. Champness. 2002. Molecular genetics of bacteria, 2nd ed. ASM Press, Washington, DC.
- 29.Spelsberg, T. C. 1983. A rapid method for analysis of ligand binding to deoxyribonucleic acid and soluble nucleoproteins using streptomycin: application to steroid receptor ligands. Biochemistry 22:12-21. [DOI] [PubMed] [Google Scholar]
- 30.Spirin, A. S., V. I. Baranov, L. A. Ryabova, S. Y. Ovodov, and Y. B. Alakhov. 1988. A continuous cell-free translation system capable of producing polypeptides in high yield. Science 242:1162-1164. [DOI] [PubMed] [Google Scholar]
- 31.Steindler, L., and V. Venturi. 2007. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 266:1-9. [DOI] [PubMed] [Google Scholar]
- 32.Swerdel, M. R., and A. M. Fallon. 1989. Cell-free translation in lysates from Spodoptera frugiperda (Lepidoptera: Noctuidae) cells. Comp. Biochem. Physiol. 93B:803-806. [DOI] [PubMed] [Google Scholar]
- 33.Swiderska, A., A. K. Berndtson, M. R. Cha, L. Li, G. M. Beaudoin, J. Zhu, and C. Fuqua. 2001. Inhibition of the Agrobacterium tumefaciencs traR quorum sensing regulator—interactions with the traM anti-activator. J. Biol. Chem. 276:49449-49458. [DOI] [PubMed] [Google Scholar]
- 34.Tait, K., I. Joint, M. Daykin, D. L. Milton, P. Williams, and M. Cámara. 2005. Disruption of quorum sensing in seawater abolishes attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ. Microbiol. 7:229-240. [DOI] [PubMed] [Google Scholar]
- 35.Tempe, J., A. Petit, M. Holsters, M. Montagu, and J. Schell. 1977. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc. Natl. Acad. Sci. USA 74:2848-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, Y. J., and J. R. Leadbetter. 2005. Rapid acyl-homoserine lactone quorum signal decomposition by diverse soils. Appl. Environ. Microbiol. 71:1291-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 38.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 39.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jørgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. A. B. Stewart. 1998. Construction of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone mediated quorum sensing. FEMS Microbiol. Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]
- 40.Wu, H., Z. Song, N. Hoiby, and M. Givskov. 2004. Quorum sensing in Gram-negative bacteria. Prog. Nat. Sci. 14:377-387. [Google Scholar]
- 41.Yan, L., M. S. Allen, M. L. Simpson, G. S. Sayler, and C. D. Cox. 2007. Direct quantification of N-(3-oxo-hexanoyl)-l-homoserine lactone in culture supernatant using a whole-cell bioreporter. J. Microbiol. Methods 68:40-45. [DOI] [PubMed] [Google Scholar]
- 42.Yang, Y.-H., T. H. Lee, J. H. Kim, E. J. Kim, H. S. Joo, C. S. Lee, and B. G. Kim. 2006. High-throughput detection method of quorum-sensing molecules by colorimetry and its applications. Anal. Biochem. 356:297-299. [DOI] [PubMed] [Google Scholar]
- 43.Zhu, J., Y. Chai, Z. Zhong, S. Li, and S. C. Winans. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 69:6949-6953. [DOI] [PMC free article] [PubMed] [Google Scholar]