Abstract
Clostridium perfringens food poisoning is caused mainly by enterotoxigenic type A isolates that typically possess high spore heat resistance. Previous studies have shown that α/β-type small, acid-soluble proteins (SASP) play a major role in the resistance of Bacillus subtilis and C. perfringens spores to moist heat, UV radiation, and some chemicals. Additional major factors in B. subtilis spore resistance are the spore's core water content and cortex peptidoglycan (PG) structure, with the latter properties modulated by the spm and dacB gene products and the sporulation temperature. In the current work, we have shown that the spm and dacB genes are expressed only during C. perfringens sporulation and have examined the effects of spm and dacB mutations and sporulation temperature on spore core water content and spore resistance to moist heat, UV radiation, and a number of chemicals. The results of these analyses indicate that for C. perfringens SM101 (i) core water content and, probably, cortex PG structure have little if any role in spore resistance to UV and formaldehyde, presumably because these spores’ DNA is saturated with α/β-type SASP; (ii) spore resistance to moist heat and nitrous acid is determined to a large extent by core water content and, probably, cortex structure; (iii) core water content and cortex PG cross-linking play little or no role in spore resistance to hydrogen peroxide; (iv) spore core water content decreases with higher sporulation temperatures, resulting in spores that are more resistant to moist heat; and (v) factors in addition to SpmAB, DacB, and sporulation temperature play roles in determining spore core water content and thus, spore resistance to moist heat.
Clostridium perfringens is a gram-positive, spore-forming, anaerobic bacterium that causes both gastrointenstinal and histotoxic diseases in humans and animals (19). C. perfringens food poisoning is caused mainly by enterotoxigenic type A isolates that produce spores that are highly resistant to heat and other environmental stress factors (32, 33, 36). These resistant spores can survive traditional cooking methods for meat and poultry products, as well as other processing treatments used in the food industry; the surviving spores will germinate and outgrow during improper cooling, and the resultant cells can then cause disease (2). Extensive work with Bacillus subtilis spores (23, 41, 42) has shown that factors involved in spore resistance include (i) the spore coats, (ii) the cortex peptidoglycan (PG) structure, (iii) the relatively impermeable spore inner membrane, (iv) the spore core's low water content, (v) the high levels of pyridine-2,6-dicarboxylic acid (DPA) in the spore core and the type and amount of cations chelated by DPA, and (vi) the saturation of spore DNA with α/β-type small, acid-soluble spore proteins (SASP). The most-important factors in the moist-heat resistance of B. subtilis spores are the cortex PG structure, the low core water content, the levels of DPA and associated cations in the core, and the α/β-type SASP (44). However, to date only the role of α/β-type SASP in C. perfringens spore resistance to moist heat, UV radiation, and some chemicals has been studied (26, 32, 33).
The spore cortex is composed of PG, with a structure similar to that of vegetative PG but with some cortex-specific modifications, as well as a lower degree of peptide cross-links (31, 50). The degree of PG cross-linking is determined by the low-molecular-weight penicillin-binding proteins (PBPs) (28) whose d,d-carboxypeptidase activity removes the terminal d-alanine of the PG's peptide side chain, preventing this side chain from serving as a donor in the formation of peptide cross-links. The B. subtilis genome encodes six low-molecular-weight PBPs, two of which, dacB and dacF, are expressed only during sporulation in the developing forespore under the control of the RNA polymerase sigma factors σE and σF, respectively (37, 45). DacB and, to a much-lesser extent, DacF determine the degree of cortex PG cross-linking (28), and the degree of cross-linking determines the amount of PG flexibility, with a loosely cross-linked PG exhibiting the greatest flexibility (25). Interestingly, B. subtilis spores lacking dacB have the same core water content but a much-lower moist-heat resistance than wild-type spores (28). In B. subtilis, dacB is the first gene in a three-gene operon with two genes (spmA and spmB) encoding proteins required for normal core dehydration during spore formation (Fig. 1A) (29). Mutations in either spmA or spmB result in B. subtilis spores with increased core water content and decreased resistance to moist heat (29, 30). The core water content of spores of Bacillus species is also affected by the sporulation temperature, with higher temperatures giving spores that have less core water and are more moist-heat resistant (21, 30). An increased core water content is also correlated with increased sensitivity of B. subtilis spores to some chemicals (21, 30). In this study, through the construction of mutations in spmA, spmB, and dacB and by preparation of spores at different temperatures, we have investigated the roles of cortex PG structure and core water content in the resistance of spores of a pathogenic C. perfringens isolate to moist heat, chemicals, and UV radiation.
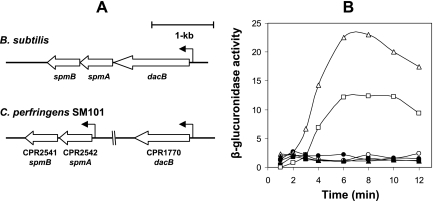
FIG. 1.
Organization of dacB and spmAB genes in B. subtilis and C. perfringens, and expression of these genes in C. perfringens. (A) Schematic representation of the genomic organization of dacB and spmAB in B. subtilis (15) and C. perfringens (22). (B) Expression of spmA-gusA (squares), spmB-gusA (circles), and dacB-gusA (triangles) fusions in C. perfringens wild-type SM101 grown in TGY vegetative (filled symbols) and DS sporulation (open symbols) media. GUS activity (in Miller units) was calculated as described in Materials and Methods. Data represent the averages of the results of three independent experiments, and time zero denotes the time of inoculation of cells into either TGY or DS medium.
MATERIALS AND METHODS
Bacterial strains and plasmids.
C. perfringens strains and plasmids used in this study are described in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic | Source/reference |
|---|---|---|
| C. perfringens strains | ||
| SM101 | Electroporatable derivative of type A food-poisoning isolate NCTC8798; carries a chromosomal cpe | 51 |
| NM101 | spmAB::catP | This study |
| NM102 | dacB::catP | This study |
| NM101(pDP24) | spmAB mutant expressing wild-type spmAB | This study |
| NM102(pDP52) | dacB mutant expressing wild-type dacB | This study |
| SM101(pDP73) | Wild-type strain carrying spmA-gusA fusion | This study |
| SM101(pDP74) | Wild-type strain carrying spmB-gusA fusion | This study |
| SM101(pDP75) | Wild-type strain carrying dacB-gusA fusion | This study |
| Plasmids | ||
| pJIR418 | C. perfringens/E. coli shuttle vector; Cmr Emr | 46 |
| pMRS127 | C. perfringens/E. coli shuttle vector which encodes erythromycin resistance (Emr), and has a promoterless gusA | 33 |
| pJIR751 | C. perfringens/E. coli shuttle vector; Emr | 1 |
| pMRS104 | Carries no origin of replication for C. perfringens; Emr | 12 |
| pMS1 | ∼3.1-kb PCR fragment containing spmAB operon in pCR-XL-TOPO | This study |
| pMS2 | ∼1.3-kb NaeI-SmaI catP fragment from pJIR418 cloned into the BglII site in the spmA ORF in pMS1 | This study |
| pMS3 | ∼4.4-kb KpnI-XhoI fragment from pMS2 in pMRS104 | This study |
| pMS4 | ∼3.4-kb PCR fragment containing dacB ORF in pCR-XL-TOPO | This study |
| pNM13 | ∼1.3-kb NaeI-SmaI catP fragment from pJIR418 cloned into the NdeI site in the dacB ORF in pMS4 | This study |
| pNM14 | ∼4.7-kb KpnI-XhoI fragment from pNM13 cloned into pMRS104 | This study |
| pDP23 | ∼1.7-kb PCR fragment containing spmAB operon and upstream region in pCR-XL-TOPO | This study |
| pDP24 | ∼1.7-kb KpnI-XhoI fragment containing spmAB operon and its upstream region between the KpnI-SalI sites in pJIR751 | This study |
| pDP52 | ∼3.2-kb KpnI-XhoI fragment containing dacB and its upstream region from pMS4 between the KpnI-SalI sites in pJIR751 | This study |
| pDP70 | 526-bp PCR fragment containing upstream region of spmA cloned into pCR-XL-TOPO | This study |
| pDP71 | 432-bp PCR fragment containing upstream region of spmB cloned into pCR-XL-TOPO | This study |
| pDP72 | 489-bp PCR fragment containing upstream region of dacB cloned into pCR-XL-TOPO | This study |
| pDP73 | 526-bp SalI-PstI fragment carrying spmA promoter region cloned into pMRS127 to create an spmA-gusA fusion construct | This study |
| pDP74 | 432-bp SalI-PstI fragment carrying spmB promoter region cloned into pMRS127 to create an spmB-gusA fusion construct | This study |
| pDP75 | 489-bp SalI-PstI fragment carrying dacB promoter region cloned into pMRS127 to create a dacB-gusA fusion construct | This study |
Construction of gusA fusion plasmids and GUS assay.
The expression of the C. perfringens spmA, spmB, and dacB genes was examined by fusing a large amount of DNA upstream of each gene to Escherichia coli gusA in pMRS127, an E. coli-C. perfringens shuttle vector (33). Briefly, 300- to 400-bp DNA fragments upstream of spmA, spmB, and dacB from C. perfringens SM101 were PCR amplified by using primers CPP376/CPP373, CPP374/CPP377, and CPP375/CPP378 (forward and reverse primers had SalI and PstI cleavage sites, respectively) and cloned into pCR-XL-TOPO (Invitrogen, Carlsbad, CA) to create pDP70, pDP71, and pDP72, respectively (Tables 1 and 2). The SalI-PstI fragments from pDP70, pDP71, and pDP72 were cloned between the SalI and PstI sites in pMRS127 to create spmA-gusA, spmB-gusA, and dacB-gusA fusion constructs in pDP73, pDP74, and pDP75, respectively. These plasmids were introduced into C. perfringens SM101 by electroporation (7), and erythromycin-resistant (Emr; 50 μg/ml) transformants were selected. The SM101 transformants carrying the plasmids with the spmA-gusA, spmB-gusA, and dacB-gusA fusions were grown in TGY (3% trypticase soy, 2% glucose, 1% yeast extract, 0.1% l-cysteine) vegetative (14) and Duncan-Strong (DS) sporulation (8) medium and assayed for β-glucuronidase (GUS) activity as described previously (11, 33).
TABLE 2.
Primers used in this study
| Primer name | Primer sequencea | Positionb | Gene | Use(s)c |
|---|---|---|---|---|
| CPP216 | ATTAGGTTTAACAGGAGTTTGG | −460 to −438 | spmAB | CP |
| CPP217 | CTGGAATATATTGTATTATTTCTGTA | +1190 to +1216 | spmAB | CP |
| CPP220 | CTGCAGTAATGGTGCTTGGA | −1196 to −1176 | spmAB | MP |
| CPP221 | AGCTCTTGGATGTGGTGAAAA | +1940 to +1961 | spmAB | MP |
| CPP222 | TTACTCCCTGCGAAGTAAGAAT | −1330 to −1308 | dacB | MP, CP |
| CPP223 | TTTCTAGGATCACTTTGCACCT | +2053 to +2075 | dacB | MP, CP |
| CPP376 | GCGTCGACATTAGGTTTAACAGGAGTTTGG | −460 to −438 | spmA | GUS |
| CPP373 | GCTGCAGGCTAAAAATCAATCCTAAGGC | +30 to +51 | spmA | GUS |
| CPP374 | GCGTCGACGGAGAGTTCTCATAGTGAC | −351 to −332 | spmB | GUS |
| CPP377 | GCTGCAGTCCCTTAAACATTCCATAGACT | +44 to +66 | spmB | GUS |
| CPP375 | GCGTCGACATAATGGTAAGGTTAGATGGAG | −386 to −364 | dacB | GUS |
| CPP378 | GCTGCAGTACAACTTTCCTTTCCAAGAAC | +66 to +88 | dacB | GUS |
Restriction sites are marked by underlining.
The nucleotide position numbering begins from the first codon and refers to the relevant position within the respective gene sequence.
GUS, construction of plasmid for GUS assays; CP, construction of complementing plasmid; MP, construction of mutator plasmid.
Construction of a C. perfringens spmAB mutant.
To isolate a derivative of C. perfringens SM101 with an insertion of catP giving chloramphenicol resistance (Cmr; 20 μg/ml) in the spmAB operon, an spmAB mutator plasmid was constructed as follows. A 3,157-bp fragment carrying the spmAB operon plus 1,196 bp upstream of spmA and 851 bp downstream of spmB was PCR amplified by using primers CPP220/CPP221 (Table 2). The ∼3.2-kb PCR fragment was cloned into pCR-XL-TOPO (Invitrogen) in E. coli, giving plasmid pMS1. This plasmid was digested with BglII, which cuts only once within the spmA open reading frame (ORF); the ends were filled; and the ∼1.3-kb SmaI-NaeI catP gene from plasmid pJIR418 (46) was inserted, giving plasmid pMS2. This plasmid was digested with KpnI and XhoI, and the ∼4.5-kb KpnI-XhoI fragment was cloned into pMRS104 digested with KpnI and SalI to create pMS3. This latter plasmid contains an inactivated spm operon, and since it contains no C. perfringens origin of replication, cannot replicate in this host. Plasmid pMS3 was introduced into C. perfringens strain SM101 by electroporation (7), and an spmAB mutant, strain NM101, was selected by allelic exchange as described previously (35). The replacement of wild-type spmA with the mutant allele in strain NM101 and the loss of the plasmid from this strain were confirmed by PCR and Southern blot analyses (data not shown).
Construction of a C. perfringens dacB mutant.
To isolate a derivative of C. perfringens SM101 with an insertion of catP in the dacB ORF, a dacB mutator plasmid was constructed as follows. A 3,405-bp fragment carrying the dacB ORF plus 1,330 bp upstream and 1,199 bp downstream was PCR amplified with primers CPP222/CPP223 (Table 2). The ∼3.4-kb PCR fragment was cloned into pCR-XL-TOPO (Invitrogen) in E. coli, giving plasmid pMS4. This plasmid was digested with NdeI, which cuts only once within the dacB ORF; the ends were filled; and the ∼1.3-kb SmaI-NaeI catP gene from plasmid pJIR418 (46) was inserted, giving plasmid pNM13. This plasmid was digested with KpnI and XhoI, and the ∼4.7-kb KpnI-XhoI fragment was cloned into pMRS104 digested with KpnI and SalI to give plasmid pNM14. This latter plasmid contains an inactivated dacB gene, and since it contains no C. perfringens origin of replication, cannot replicate in this host. Plasmid pNM14 was introduced into C. perfringens strain SM101 by electroporation (7), and a dacB mutant, strain NM102, was selected by allelic exchange as described previously (35). The expected genomic structure in the dacB region of strain NM102 was confirmed by PCR and Southern blot analyses (data not shown).
Construction of complemented strains.
An ∼1.7-kb fragment containing the spmA and spmB ORFs plus 460 bp upstream of spmA was PCR amplified by using primers CPP216/CPP217 (Table 2) and cloned into pCR-XL-TOPO (Invitrogen), giving pDP23. The ∼1.7-kb KpnI-XhoI fragment of pDP23 was cloned between the KpnI and SalI sites of plasmid pJIR751 (1) to create the spmAB-complementing plasmid pDP24. A 3,405-bp fragment carrying the dacB ORF plus 1,330 bp upstream and 1,199 bp downstream was PCR amplified by using primers CPP222/CPP223 (Table 2) and cloned into pCR-XL-TOPO (Invitrogen), giving pMS4. The ∼3.4-kb KpnI-XhoI fragment of pMS4 was then cloned between the KpnI and SalI sites of plasmid pJIR751 (1) to create the dacB-complementing plasmid pDP52. Plasmids pDP24 and pDP52 were introduced into C. perfringens strains NM101 and pNM102, respectively, by electroporation (7), and Emr transformants were selected. The presence of plasmids pDP24 and pDP52 in NM101(pDP24) and NM102(pDP52), respectively, was confirmed by PCR (data not shown).
Spore preparation and purification.
Starter cultures (10 ml) of C. perfringens strains were prepared by overnight growth at 37°C in fluid thioglycolate broth (FTG) (Difco) as described previously (14). Sporulating cultures of C. perfringens were prepared by inoculating 0.2 ml of an FTG starter culture into 10 ml of DS sporulation medium (8), and this culture was incubated for 24 h at 37°C to form spores as confirmed by phase-contrast microscopy. Spore preparations were prepared by scaling up the latter procedure. Spore preparations were cleaned by repeated centrifugation and washing with sterile distilled water until the spores were >99% free of sporulating cells, cell debris, and germinated spores; suspended in distilled water at a final optical density at 600 nm (OD600) of ∼6; and stored at −20°C (27).
Measurement of spore properties.
Spore core DPA content was determined by incubating 1 ml of spores at an OD600 of 6 in a water bath at 100°C for 60 min, cooling the sample on ice, centrifuging for 5 min, and measuring the DPA in the supernatant fluid as described previously (34). For the determination of spore core wet densities by equilibrium density gradient centrifugation (17), spore coats were first removed by the extraction of 3 to 5 mg dry weight of spores with 1 ml of 50 mM Tris-HCl (pH 8.0)-8 M urea-1% (wt/vol) sodium dodecyl sulfate-50 mM dithiothreitol for 90 min at 37°C and then spores were washed three times with 150 mM NaCl and twice with water (17, 30). Decoated spores were suspended in 100 μl of 30% Histidenz (Nycodenz) (Sigma, St. Louis, MO), incubated for 60 min on ice, and loaded into the top of a 2-ml linear gradient of 51 to 70% Histidenz in ultraclear tubes, and the tubes centrifuged for 45 min at 11,200 × g and 20°C in a swinging bucket rotor in a Beckman TL-100 ultracentrifuge. Spore core water content was calculated according to the formula y = −0.00254x + 1.460 (17), where y is the spore core wet density and x is the core water content in g per 100 g of wet protoplast (core).
Measurement of spore resistance.
The resistance of C. perfringens spores to moist heat was determined as previously described (33, 36). Briefly, 10 ml of 24-hr-grown DS medium cultures of C. perfringens strains were heat treated at 75°C for 20 min to kill vegetative cells. An aliquot of each heat-treated DS medium culture was serially diluted in phosphate-buffered saline (140 mM NaCl-25 mM Na2HPO4 [pH 7.0]), plated on brain heart infusion (BHI) agar, and incubated anaerobically for 24 h at 37°C to determine initial CFU/ml; these values were routinely 106 to 107. 75°C-treated DS medium cultures were then immediately heated at 100°C for various times (20, 40, and 60 min), and aliquots of appropriate dilutions were plated and incubated anaerobically for 24 h at 37°C as described above. Plots of CFU/ml versus time at 100°C were used to determine decimal reduction times (D100° values), which are the time cultures need to be kept at 100°C to get a 90% reduction in CFU/ml. All experiments measuring spore moist-heat resistance were repeated at least three times.
The resistance of C. perfringens spores to chemicals was determined as previously described (26). Briefly, spore suspensions at an OD600 of ∼1 (∼4 × 107 spores/ml) were (i) treated with 2 M hydrogen peroxide (Mallinckrodt Baker, Inc., Phillipsburg, NJ) at room temperature (20°C), and aliquots were neutralized with catalase (Sigma) as described previously (40); (ii) treated with 300 mM HCl at room temperature, and aliquots were diluted 100-fold in 25 mM in Na2HPO4 (pH 7.0); (iii) treated with 400 mM NaNO2-400 mM Na acetate (pH 4.5) at room temperature, and aliquots were diluted 10-fold in 500 mM Na2HPO4 (pH 8.5); or (iv) treated with 25 g liter−1 formaldehyde (Sigma) at 30°C, and aliquots were diluted 10-fold in 400 mM glycine (pH 7.0) and incubated for 20 min at room temperature prior to analysis. For the analysis of spore killing, untreated and treated spores were serially diluted in phosphate-buffered saline and plated on BHI agar and the plates were incubated anaerobically for 24 h at 37°C.
The UV resistance of C. perfringens spores was determined as described previously (30, 32). Briefly, purified C. perfringens spores at an OD600 of 2 (∼8 × 107 spores/ml) were diluted 100-fold in 25 mM Na2HPO4 buffer (pH 6.8) and UV irradiated at 254 nm with a UVGL-25 Mineralight lamp (UVP, Inc., Upland, CA) for various times (0.5, 1.0, 2.5, and 5.0 min). Appropriate dilutions were spread onto BHI plates and incubated as described above prior to the assessment of colony formation.
Statistical analyses.
Student's t test was used for statistical analyses.
RESULTS
Identification of putative C. perfringens dacB and spmAB genes.
In B. subtilis, a tricistronic operon (Fig. 1A) encodes proteins involved in determining both the degree of cortex PG cross-linking (DacB) and the spore's core water content (SpmA and SpmB); all three genes are expressed only during sporulation (29, 30). When we scanned the C. perfringens SM101 genome (22) by BLASTP analysis, three ORFs (CPR1770, CPR2541, and CPR2542) with high similarity to B. subtilis dacB, spmA, and spmB were found (Fig. 1A). However, these genes are organized differently in C. perfringens (Fig. 1A), with CPR1770 being monocistronic and located ∼800 kb upstream of a bicistronic operon containing CPR2541 and CPR2542 (Fig. 1A). This genetic organization is not unique to C. perfringens, as dacB is also monocistronic and located hundreds of kb from spmAB in the Clostridium acetobutylicum, Clostridium difficile, Clostridium novyi, and Clostridium tetani genomes (3, 4, 24, 38).
Analysis of the amino acid sequence of C. perfringens CPR1770 revealed a high similarity (62%) with B. subtilis DacB (47). Despite the absence of 114 residues from the carboxy terminus of CPR1770 present in B. subtilis DacB, four conserved regions typical of PBPs are present in the C. perfringens protein (data not shown). Near the amino terminus, a highly hydrophobic region probably facilitates anchoring the protein to the outer membrane during sporulation (9). The other well-conserved regions are a catalytic serine domain (S-X-X-K), an S-X-N domain typical of type A β-lactamases (48), and a K-T-G sequence that is essential for the tertiary structure of the active site (13).
The first gene in the bicistronic operon, CPR2542 (Fig. 1A), encodes a 192-amino-acid-residue protein that is highly similar to B. subtilis SpmA (62%), with an estimated molecular mass of 20.4 kDa and four putative transmembrane alpha-helical domains (TMH). The second gene, CPR2541 (Fig. 1A), encodes a protein that is highly similar to B. subtilis SpmB (62%), with 172 residues, a molecular mass of 18.8 kDa, and five TMH. The high number of TMH in both putative C. perfringens Spm proteins suggests that they may be anchored in either of the forespore membranes as suggested for B. subtilis DacB (28).
Evaluation of the expression of C. perfringens dacB, spmA, and spmB.
To assess whether the C. perfringens spm and dacB genes are expressed during sporulation, 300 to 400 bp upstream of each gene's coding sequence, containing the putative promoter region of each gene, was fused to E. coli gusA and GUS activity was measured after the fusions were introduced into C. perfringens SM101. When vegetative cells of strain SM101 carrying pDP73 (spmA-gusA), pDP74 (spmB-gusA), and pDP75 (dacB-gusA) were assayed for GUS activity, no significant expression of spmA, spmB, or dacB was observed (Fig. 1B). However, sporulating cultures carrying spmA-gusA and dacB-gusA exhibited significant GUS activity, although no GUS activity was detected in sporulating cultures carrying spmB-gusA (Fig. 1B), consistent with spmB being the second gene in an operon with spmA. The expression of spmA-gusA began ∼2 h after the initiation of sporulation and reached a maximum specific activity ∼4 to 6 h later (Fig. 1B). The expression of dacB-gusA also began ∼2 h after the induction of sporulation and reached a maximum specific activity 4 to 8 h later (Fig. 1B). Collectively, these results suggest that the C. perfringens spm and dacB genes are expressed only during sporulation and led us to hypothesize that their products might be involved in resistance of C. perfringens spores, as they are in spores of B. subtilis (28).
Effect of spmAB mutation on spore properties.
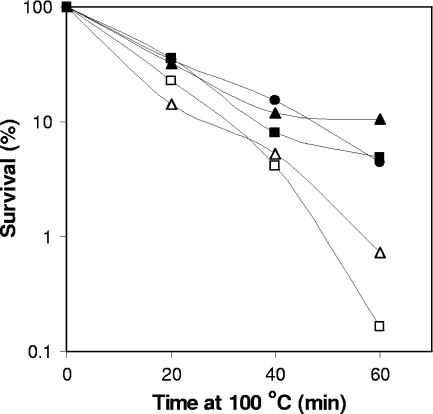
Studies with B. subtilis indicate that the spore core water content is determined during sporulation at least in part by the SpmA and SpmB proteins (29, 30). To assess whether SpmA and -B have any role in C. perfringens spore core water content and thus, perhaps, spore moist-heat resistance, we constructed an insertion mutation in spmA, giving strain NM101. Since no spmB-gusA activity was detected in sporulating SM101 cells (Fig. 1B), it is most likely that disruption of spmA has a polar effect on the downstream spmB and thus, strain NM101 in effect carries both spmA and spmB mutations. As expected, the moist-heat resistance of NM101 spores was significantly lower (P < 0.01) than that of the wild-type spores, as the D100 value for NM101 spores was twofold lower than for SM101 spores (Fig. 2; Table 3). Although the levels of DPA in NM101 and SM101 spores were similar, the core wet density of NM101 spores was significantly lower than that of SM101 spores and thus, NM101 spores have a higher core water content (Table 3). The moist-heat resistance and core water content of NM101 spores were restored to wild-type levels when strain NM101 was complemented with wild-type spmAB (Fig. 2; Table 3). These results suggest that SpmAB proteins are involved in the determination of C. perfringens spore core water content and further indicate that C. perfringens spores with higher water content have lower moist-heat resistance, in agreement with the results of studies with B. subtilis spores (30).
FIG. 2.
Thermal-death curves of spores of various C. perfringens strains. Spores of strains SM101 (wild type; filled circles), NM101 (spmAB; open squares), NM101(pDP24) (spmAB strain complemented with wild-type spmAB; filled squares), NM102 (dacB; open triangles), and NM102(pDP53) (dacB strain complemented with wild-type dacB; filled triangles) were heated at 100°C for various times, and spore survival was determined as described in Materials and Methods. The data are the results from a representative experiment.
TABLE 3.
Effects of spmAB and dacB mutations on C. perfringens spore properties
| Strain (genotype) | Heat resistance (D100° [min])a | Amt of DPA (μg/ml/ OD600)b | Core wet density (g/ml)c | Water content (g/100 g of wet protoplast)d |
|---|---|---|---|---|
| SM101 (wild type) | 49.1 ± 6 | 16.7 | 1.378 | 32.4 |
| NM101 (spmAB::catP) | 19.2 ± 3 | 16.4 | 1.370 | 35.4 |
| NM101(pDP24) | 45.2 ± 2 | NDe | 1.382 | 30.7 |
| NM102 (dacB::catP) | 28.8 ± 1 | 17.2 | 1.382 | 30.7 |
| NM102(pDP52) | 52.2 ± 7 | ND | 1.383 | 30.3 |
Values are averages ± standard deviations of determinations for three spore preparations.
Values are averages of determinations for three different spore preparations.
Values are averages of determinations for two to four different spore preparations, and the standard deviation was ± 0.001 g/ml.
Spore core water contents were calculated as described in Materials and Methods.
ND, not determined.
Effect of dacB mutation on spore properties.
Another factor that has been suggested to be important in the moist-heat resistance of B. subtilis spores is the cortex PG structure, specifically, the degree of cortex PG cross-linking that is determined primarily by the d,d-carboxypeptidase DacB (29). To test the role of DacB in C. perfringens spore resistance, we constructed an insertion mutation in dacB, giving strain NM102. As expected, the moist-heat resistance of NM102 spores was significantly lower than that of SM101 spores, although the levels of DPA in both wild-type and dacB spores were almost identical (Fig. 2; Table 3). The core wet density of dacB spores was also similar to that of SM101 spores (Table 3). Again, the defects in strain NM102 were complemented with wild-type dacB [strain NM102(pDP52)] (Fig. 2; Table 3).
Effect of sporulation temperature on C. perfringens spore heat resistance and core water content.
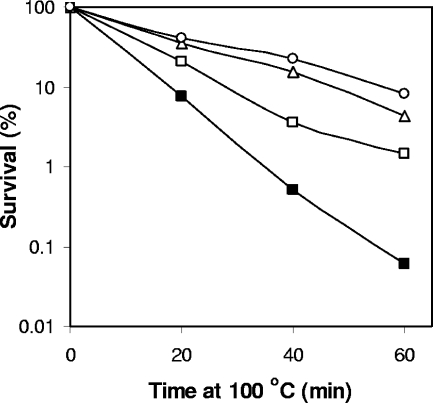
An additional factor that can significantly influence the core water content and moist-heat resistance of B. subtilis spores is the sporulation temperature, with higher sporulation temperatures giving spores with lower core water content and higher moist-heat resistance (10, 21), although how the sporulation temperature affects the core water content and moist-heat resistance of these spores is not known. C. perfringens spores of strains SM101 (wild type), NM101 (spmAB), and NM102 (dacB) were prepared at different sporulation temperatures, and the spore core water content and moist-heat resistance were determined. In agreement with results with B. subtilis spores (10, 21), C. perfringens wild-type spores prepared at higher temperatures exhibited higher resistance to moist heat and had a lower core water content (Fig. 3; Table 4). This trend was also observed for spmAB and dacB spores (Table 4). However, there was no obvious quantitative relationship between the precise core water content and the moist-heat resistance of the spores of these different strains (Table 4), suggesting that additional factors besides core water content influence the moist-heat resistance of C. perfringens spores and that these unknown factors are in turn influenced in some way by DacB and SpmAB.
FIG. 3.
Effect of sporulation temperature on thermal-death curves of C. perfringens spores. SM101 (wild type) spores were prepared at 26°C (filled squares), 32°C (open squares), 37°C (open triangles), and 42°C (open circles) and heated at 100°C for various times, and spore survival was determined as described in Materials and Methods. The data are the results from a representative experiment.
TABLE 4.
Effect of sporulation temperature on C. perfringens spore properties
| Strain (genotype) |
D100° values of spores prepared ata:
|
Water content of spores prepared atb:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 26°C | 32°C | 37°Cc | 42°C | 26°C | 32°C | 37°Cc | 42°C | |
| SM101 (wild type) | 15 ± 3 | 26 ± 4 | 49 ± 6 | 54 ± 4 | 39.0 | 38.6 | 32.4 | 30.7 |
| NM101 (spmAB) | 12 ± 2 | 12 ± 0 | 19 ± 3 | 34 ± 2 | 36.2 | 36.2 | 35.4 | 31.3 |
| NM102 (dacB) | 8 ± 2 | 12 ± 2 | 29 ± 1 | 71 ± 6 | 39.0 | 36.6 | 30.7 | 33.3 |
Spores of various strains were prepared at different sporulation temperatures, and heat resistance was determined and expressed as D100° values as described in Materials and Methods. Values are averages ± standard deviations of the results for three spore preparations.
Spore core wet densities were determined for spores of various strains prepared at different temperatures, and core water contents were calculated as described in Materials and Methods. Values presented are averages of determinations for two different spore preparations.
The 37°C data are from Table 1.
Effects of spmAB and dacB mutations on chemical resistance of C. perfringens spores.
To gain insight into the roles of SpmAB and DacB in C. perfringens spore resistance to treatments other than heat, we also measured the resistance of spmAB and dacB spores to a variety of chemicals. Previous work indicated that B. subtilis spores with an altered cortex and higher core water content have lower hydrogen peroxide resistance than wild-type spores (20, 30). In contrast, C. perfringens spmAB, dacB, and wild-type spores had similar hydrogen peroxide resistance (Fig. 4A), suggesting that low core water content and a wild-type cortex PG structure have no significant role in C. perfringens spore resistance to hydrogen peroxide. This suggestion is consistent with the results of previous work indicating that α/β-type SASP are a major factor in hydrogen peroxide resistance of B. subtilis and C. perfringens spores (26, 44). The spmAB, dacB, and wild-type spores also exhibited similar resistance to HCl (Fig. 4B), a chemical that kills B. subtilis spores by somehow disrupting the inner-membrane permeability barrier (39).
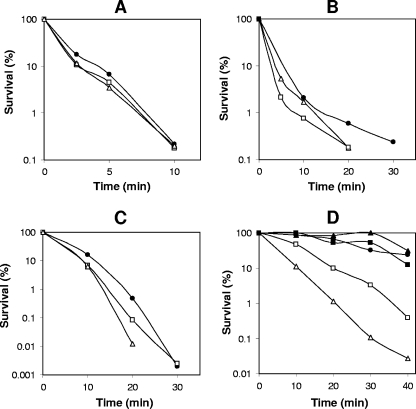
FIG. 4.
Resistance of spores of various C. perfringens strains to hydrogen peroxide (A), hydrochloride acid (B), formaldehyde (C), and nitrous acid (D). Spores of strains SM101 (wild type; filled circles), NM101 (spmAB; open squares), NM101(pDP2) (spmAB strain complemented with wild-type spmAB; filled squares), NM102 (dacB mutant; open triangles), and NM102(pDP53) (dacB strain complemented with wild-type dacB; filled triangles) were purified, and their survival after various treatments was determined as described in Materials and Methods. The variability in survival values in these experiments was ±15%.
The other two chemicals tested, formaldehyde and nitrous acid, kill B. subtilis spores at least in large part by DNA damage, and α/β-type SASP are very important in spore resistance to these agents (18, 49). Wild-type and spmAB C. perfringens spores exhibited no difference in formaldehyde resistance, although dacB spores were slightly more sensitive (Fig. 4C). Surprisingly, spmAB and dacB spores were much more sensitive (P < 0.01) than wild-type spores to nitrous acid (Fig. 4D). However, wild-type levels of nitrous acid resistance were restored to spores of strains NM101 (spmAB) and NM102 (dacB) when the latter strains were complemented with wild-type spmAB and dacB genes, respectively (Fig. 4D). These results clearly indicate that in addition to α/β-type SASP (26), core water content and cortex PG structure are also likely to be major determinants in the nitrous acid resistance of C. perfringens spores.
Effect of spmAB and dacB mutations on UV resistance of C. perfringens spores.
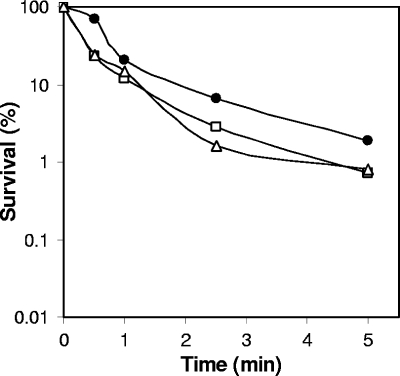
Previous work (42) has shown that with B. subtilis spores, binding of α/β-type SASP to spore DNA is the predominant factor in spore resistance to UV radiation, a treatment commonly used to sterilize surfaces in the food industry. C. perfringens spores with decreased levels of α/β-type SASP are also more sensitive to UV treatment than the parental wild-type spores (32). Consistent with the predominant role for α/β-type SASP in spore UV resistance, spores of wild-type, spmAB, and dacB C. perfringens strains showed essentially identical UV resistance (Fig. 5), as was found previously for B. subtilis spores lacking SpmAB and DacB (30).
FIG. 5.
UV resistance of spores of various C. perfringens strains. Spores of strains SM101 (wild type; filled circles), NM101 (spmAB; open squares), and NM102 (dacB; open triangles) were purified, and their survival after UV treatment was determined as described in Materials and Methods.
DISCUSSION
Dormant spores of C. perfringens type A food poisoning isolates are the causative agent of type A C. perfringens-caused food-borne illness. These spores have high heat resistance that favors spore survival during inadequate cooking or holding temperatures (19, 36). Consequently, an understanding of the molecular basis of C. perfringens spore resistance is crucial to the development of better strategies for spore inactivation. In this communication, we report a number of observations leading to new conclusions about the mechanisms of C. perfringens spore resistance to a variety of agents, an area that has generally received very little study. While many of these conclusions reinforce previous conclusions about the mechanisms of resistance of spores of Bacillus species, primarily B. subtilis, there are some significant differences in the details of the resistance of spores of C. perfringens and B. subtilis.
One straightforward conclusion is that dacB and spmAB are sporulation genes in both B. subtilis and C. perfringens, and the products of these genes have very similar effects on spore properties in these two species. In B. subtilis, the mutation of spmA and -B or spmB alone results in a significant increase in spore core wet density and an ∼eightfold decrease in the D90° value for moist-heat killing (29). The effects of the spmAB mutation on C. perfringens spores are similar, albeit of lower magnitude (∼twofold decrease in the D100°). In B. subtilis, an in-frame dacB mutation has no effect on core water content but does cause an ∼fivefold decrease in the D90° value (28, 29). Again, the effects of a dacB mutation on C. perfringens spores are similar, although the decrease in the D100° value in dacB C. perfringens spores is smaller than the effect of a dacB mutation on D90° values for B. subtilis spores (28-30). A dacB mutation also has a large effect on B. subtilis spore cortex PG structure, as cortex PG cross-linking increases two- to fourfold, since DacB is by far the major d,d-carboxypeptidase affecting the degree of cross-linking (28). We have not examined the cross-linking of cortex PG in C. perfringens wild-type and dacB spores but would suggest that by analogy with results in B. subtilis, C. perfringens dacB spores will also exhibit a significantly higher degree of cortex PG cross-linking than wild-type spores.
A second significant conclusion is that neither the level of core water nor, most likely, the degree of cortex cross-linking plays any role in C. perfringens spore UV resistance, as is also the case with B. subtilis spores (28, 30). The UV resistance of B. subtilis spores is due almost exclusively to the saturation of spore DNA with α/β-type SASP, proteins that are also present in C. perfringens spores (42, 44). Presumably, small changes in core water content do not affect α/β-type SASP-DNA binding appreciably and thus do not affect spore UV resistance. Spore core water content and, probably, the degree of cortex PG cross-linking also play no role in C. perfringens spore resistance to formaldehyde, as is also the case at least for changes in core water content in B. subtilis spores (21). Again, the saturation of spore DNA with α/β-type SASP is a major factor in spore resistance to formaldehyde (26, 44).
A third conclusion is that the lack of appreciable effects of core water content and, probably, the degree of cortex cross-linking on C. perfringens spore hydrogen peroxide resistance are consistent with α/β-type SASP binding as a major factor protecting C. perfringens spore DNA from this reagent (26). An in-frame dacB mutation giving increased cortex PG cross-linking also has only a minimal effect on B. subtilis spore resistance to hydrogen peroxide (30). However, spmAB mutations decreased B. subtilis spore resistance to hydrogen peroxide significantly (30). We do not understand the reason for this difference between the spores of these two species. However, perhaps the α/β-type SASP in C. perfringens are more effective in preventing DNA damage due to hydrogen peroxide than in B. subtilis spores, such that the effects of changes in core water content on the hydrogen peroxide resistance of C. perfringens spores are minimal. Another explanation is that perhaps the structure of the complex between α/β-type SASP and DNA is somewhat different in B. subtilis and C. perfringens spores and this alters DNA protection against hydrogen peroxide in these two types of spores. While the amino acid sequences of α/β-type SASP from B. subtilis and C. perfringens are quite similar, they do have a significant difference in the spacing between the two most-highly conserved structural elements (41). Since the structure of DNA bound to a B. subtilis α/β-type SASP has now been determined (16), it may be possible to use this structure to model the effects of a C. perfringens α/β-type SASP on DNA structure and properties.
A fourth conclusion is that it appears that core water content and, likely, the degree of cortex PG cross-linking play major roles in determining C. perfringens spore resistance to nitrous acid. While this may initially seem surprising, since α/β-type SASP-DNA binding is a major factor in B. subtilis and C. perfringens spore resistance to this agent (26, 41), B. subtilis spores made at higher temperatures that have the lowest core water content are significantly more nitrous acid resistant than spores made at low temperatures that have the highest core water content (21). However, the low permeability of the spore's inner membrane is also an important factor in B. subtilis spore resistance to nitrous acid, and this membrane's permeability decreases significantly in spores made at higher temperatures (6). It is also possible that there are significant differences in the structure of DNA when saturated with α/β-type SASP from B. subtilis and C. perfringens, as noted above, and perhaps these differences result in different protection of DNA against nitrous acid in the complexes formed with the proteins of the two species. Clearly, more work is needed with C. perfringens spores to identify all factors involved in spore resistance to nitrous acid.
The final and perhaps the most-significant conclusion is that core water content is a major factor in C. perfringens spore resistance to moist heat, as is also the case with B. subtilis spores (10, 44). Presumably a higher core water content results in more-rapid inactivation of one or more key proteins in the spore core whose loss results in spore death, as recent work has provided strong evidence that it is through core protein inactivation that moist heat kills B. subtilis spores (5). The effects of core water content on C. perfringens spore moist-heat resistance appear to be relatively independent of the cause of alterations in core water content, whether via spmAB mutations or sporulation at different temperatures. However, factors in addition to core water content can be affected by sporulation temperature and the spmAB mutations, as the response of C. perfringens spmAB and dacB spore resistance to sporulation temperature is quantitatively different than that of wild-type spores. Thus, while it is clear that core water content is a major factor in determining C. perfringens spore resistance to moist heat, this is clearly not the only factor. As noted above, α/β-type SASP-DNA binding is one additional factor in C. perfringens spore moist-heat resistance (32, 33), and it appears likely that the degree of cortex PG cross-linking is an additional factor, as dacB spores had lower moist-heat resistance than wild-type spores, although both had similar core water contents. It has been suggested for spores of B. subtilis (28) that more-loosely cross-linked PG in the spore cortex may be less able to maintain the spore core's low water content upon heating at elevated temperatures, and we suggest that the same is true with C. perfringens spores. However, data on the degree of cross-linking of cortex PG in spores of these various strains is needed before this conclusion can be made definitive.
Acknowledgments
This research was supported by a fellowship from MIDEPLAN (CHILE) to D. Paredes-Sabja and grants from the U.S. Department of Agriculture (2002-35201-12643) and Army Research Office to M. Sarker and from the National Institutes of Health (GM19698) and Army Research Office to P. Setlow.
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Bannam, T. L., and J. I. Rood. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29:233-235. [DOI] [PubMed] [Google Scholar]
- 2.Bean, N. H., P. M. Griffin, J. S. Goulding, and C. B. Ivey. 1990. Foodborne disease outbreaks, 5-year summary, 1983-1987. MMWR Surveill. Summ. 39:15-57. [PubMed] [Google Scholar]
- 3.Bettegowda, C., X. Huang, J. Lin, I. Cheong, M. Kohli, S. A. Szabo, X. Zhang, L. A. Diaz, Jr., V. E. Velculescu, G. Parmigiani, K. W. Kinzler, B. Vogelstein, and S. Zhou. 2006. The genome and transcriptomes of the anti-tumor agent Clostridium novyi-NT. Nat. Biotechnol. 24:1573-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruggemann, H., S. Baumer, W. F. Fricke, A. Wiezer, H. Liesegang, I. Decker, C. Herzberg, R. Martinez-Arias, R. Merkl, A. Henne, and G. Gottschalk. 2003. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl. Acad. Sci. USA 100:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman, W. H., D. Chen, Y. Q. Li, A. E. Cowan, and P. Setlow. 2007. How moist heat kills spores of Bacillus subtilis. J. Bacteriol. 189:8458-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortezzo, D. E., and P. Setlow. 2005. Analysis of factors that influence the sensitivity of spores of Bacillus subtilis to DNA damaging chemicals. J. Appl. Microbiol. 98:606-617. [DOI] [PubMed] [Google Scholar]
- 7.Czeczulin, J. R., R. E. Collie, and B. A. McClane. 1996. Regulated expression of Clostridium perfringens enterotoxin in naturally cpe-negative type A, B, and C isolates of C. perfringens. Infect. Immun. 64:3301-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan, C. L., and D. H. Strong. 1968. Improved medium for sporulation of Clostridium perfringens. Appl. Microbiol. 16:82-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frere, J. M., and B. Joris. 1984. Penicillin-sensitive enzymes in peptidoglycan biosynthesis. Crit. Rev. Microbiol. 11:299-396. [DOI] [PubMed] [Google Scholar]
- 10.Gerhardt, P., and R. E. Marquis. 1989. Spore thermoresistance mechanisms, p. 43-63. In R. S. I. Smith and P. Setlow (ed.), Regulation of prokaryotic development, 1st ed. American Society for Microbiology, Washington, DC.
- 11.Griffith, K. L., and R. E. Wolf, Jr. 2002. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assay in 96-well arrays. Biochem. Biophys. Res. Commun. 290:397-402. [DOI] [PubMed] [Google Scholar]
- 12.Huang, I. H., M. Waters, R. R. Grau, and M. R. Sarker. 2004. Disruption of the gene (spo0A) encoding sporulation transcription factor blocks endospore formation and enterotoxin production in enterotoxigenic Clostridium perfringens type A. FEMS Microbiol. Lett. 233:233-240. [DOI] [PubMed] [Google Scholar]
- 13.Joris, B., J. M. Ghuysen, G. Dive, A. Renard, O. Dideberg, P. Charlier, J. M. Frere, J. A. Kelly, J. C. Boyington, P. C. Moews, et al. 1988. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem. J. 250:313-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kokai-Kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 16.Lee, K. S., D. Bumbaca, J. Kosman, P. Setlow, and M. J. Jedrzejas. 2008. Structure of a protein-DNA complex essential for DNA protection in spores of Bacillus species. Proc. Natl. Acad. Sci. USA 105:2806-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsay, J. A., T. C. Beaman, and P. Gerhardt. 1985. Protoplast water content of bacterial spores determined by buoyant density sedimentation. J. Bacteriol. 163:735-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loshon, C. A., P. C. Genest, B. Setlow, and P. Setlow. 1999. Formaldehyde kills spores of Bacillus subtilis by DNA damage and small, acid-soluble spore proteins of the alpha/beta-type protect spores against this DNA damage. J. Appl. Microbiol. 87:8-14. [DOI] [PubMed] [Google Scholar]
- 19.McClane, B. A. 2007. Clostridium perfringens, p. 423-444. In M. P. Doyle and L. R. Beuchat (ed.), Food microbiology: fundamentals and frontiers, 3rd ed. ASM Press, Washington, DC.
- 20.Melly, E., A. E. Cowan, and P. Setlow. 2002. Studies on the mechanism of killing of Bacillus subtilis spores by hydrogen peroxide. J. Appl. Microbiol. 93:316-325. [DOI] [PubMed] [Google Scholar]
- 21.Melly, E., P. C. Genest, M. E. Gilmore, S. Little, D. L. Popham, A. Driks, and P. Setlow. 2002. Analysis of the properties of spores of Bacillus subtilis prepared at different temperatures. J. Appl. Microbiol. 92:1105-1115. [DOI] [PubMed] [Google Scholar]
- 22.Myers, G. S., D. A. Rasko, J. K. Cheung, J. Ravel, R. Seshadri, R. T. DeBoy, Q. Ren, J. Varga, M. M. Awad, L. M. Brinkac, S. C. Daugherty, D. H. Haft, R. J. Dodson, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. A. Sullivan, H. Khouri, G. I. Dimitrov, K. L. Watkins, S. Mulligan, J. Benton, D. Radune, D. J. Fisher, H. S. Atkins, T. Hiscox, B. H. Jost, S. J. Billington, J. G. Songer, B. A. McClane, R. W. Titball, J. I. Rood, S. B. Melville, and I. T. Paulsen. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ou, L. T., and R. E. Marquis. 1970. Electromechanical interactions in cell walls of gram-positive cocci. J. Bacteriol. 101:92-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paredes-Sabja, D., D. Raju, J. A. Torres, and M. R. Sarker. 2008. Role of small, acid-soluble spore proteins in the resistance of Clostridium perfringens spores to chemicals. Int. J. Food Microbiol. 122:333-335. [DOI] [PubMed] [Google Scholar]
- 27.Paredes-Sabja, D., J. A. Torres, P. Setlow, and M. R. Sarker. 2008. Clostridium perfringens spore germination: characterization of germinants and their receptors. J. Bacteriol. 190:1190-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popham, D. L., M. E. Gilmore, and P. Setlow. 1999. Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J. Bacteriol. 181:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popham, D. L., B. Illades-Aguiar, and P. Setlow. 1995. The Bacillus subtilis dacB gene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration. J. Bacteriol. 177:4721-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popham, D. L., S. Sengupta, and P. Setlow. 1995. Heat, hydrogen peroxide, and UV resistance of Bacillus subtilis spores with increased core water content and with or without major DNA-binding proteins. Appl. Environ. Microbiol. 61:3633-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popham, D. L., and P. Setlow. 1993. The cortical peptidoglycan from spores of Bacillus megaterium and Bacillus subtilis is not highly cross-linked. J. Bacteriol. 175:2767-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raju, D., P. Setlow, and M. R. Sarker. 2007. Antisense-RNA-mediated decreased synthesis of small, acid-soluble spore proteins leads to decreased resistance of Clostridium perfringens spores to moist heat and UV radiation. Appl. Environ. Microbiol. 73:2048-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raju, D., M. Waters, P. Setlow, and M. R. Sarker. 2006. Investigating the role of small, acid-soluble spore proteins (SASPs) in the resistance of Clostridium perfringens spores to heat. BMC Microbiol. 6:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotman, Y., and M. L. Fields. 1967. A modified reagent for dipicolinic acid analysis. Anal. Biochem. 22:168. [DOI] [PubMed] [Google Scholar]
- 35.Sarker, M. R., R. J. Carman, and B. A. McClane. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 33:946-958. [DOI] [PubMed] [Google Scholar]
- 36.Sarker, M. R., R. P. Shivers, S. G. Sparks, V. K. Juneja, and B. A. McClane. 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid genes versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 66:3234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Schuch, R., and P. J. Piggot. 1994. The dacF-spoIIA operon of Bacillus subtilis, encoding sigma F, is autoregulated. J. Bacteriol. 176:4104-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779-786. [DOI] [PubMed] [Google Scholar]
- 39.Setlow, B., C. A. Loshon, P. C. Genest, A. E. Cowan, C. Setlow, and P. Setlow. 2002. Mechanisms of killing spores of Bacillus subtilis by acid, alkali and ethanol. J. Appl. Microbiol. 92:362-375. [DOI] [PubMed] [Google Scholar]
- 40.Setlow, B., and P. Setlow. 1993. Binding of small, acid-soluble spore proteins to DNA plays a significant role in the resistance of Bacillus subtilis spores to hydrogen peroxide. Appl. Environ. Microbiol. 59:3418-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Setlow, P. 2007. I will survive: DNA protection in bacterial spores. Trends Microbiol. 15:172-180. [DOI] [PubMed] [Google Scholar]
- 42.Setlow, P. 2001. Resistance of spores of Bacillus species to ultraviolet light. Environ. Mol. Mutagen. 38:97-104. [DOI] [PubMed] [Google Scholar]
- 43.Setlow, P. 1988. Small, acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function, and degradation. Annu. Rev. Microbiol. 42:319-338. [DOI] [PubMed] [Google Scholar]
- 44.Setlow, P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514-525. [DOI] [PubMed] [Google Scholar]
- 45.Simpson, E. B., T. W. Hancock, and C. E. Buchanan. 1994. Transcriptional control of dacB, which encodes a major sporulation-specific penicillin-binding protein. J. Bacteriol. 176:7767-7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sloan, J., T. A. Warner, P. T. Scott, T. L. Bannam, D. I. Berryman, and J. I. Rood. 1992. Construction of a sequenced Clostridium perfringens-Escherichia coli shuttle plasmid. Plasmid 27:207-219. [DOI] [PubMed] [Google Scholar]
- 47.Sowell, M. O., and C. E. Buchanan. 1983. Changes in penicillin-binding proteins during sporulation of Bacillus subtilis. J. Bacteriol. 153:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spratt, B. G., and K. D. Cromie. 1988. Penicillin-binding proteins of gram-negative bacteria. Rev. Infect. Dis. 10:699-711. [DOI] [PubMed] [Google Scholar]
- 49.Tennen, R., B. Setlow, K. L. Davis, C. A. Loshon, and P. Setlow. 2000. Mechanisms of killing of spores of Bacillus subtilis by iodine, glutaraldehyde and nitrous acid. J. Appl. Microbiol. 89:330-338. [DOI] [PubMed] [Google Scholar]
- 50.Warth, A. D., and J. L. Strominger. 1972. Structure of the peptidoglycan from spores of Bacillus subtilis. Biochemistry 11:1389-1396. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]