Abstract
Multilocus sequence typing (MLST), an accurate and phylogenetically robust characterization method for population studies of Campylobacter, was applied to Campylobacter jejuni isolates (n = 297) from the fecal samples of cattle from five dairy farms in Cheshire, United Kingdom, collected throughout 2003. The population dynamics of the C. jejuni strains, as identified by the occurrence of sequence types and clonal complexes, demonstrated variations within and between cattle populations over time. Three clonal lineages have emerged to predominate among the cattle isolates, namely, the ST-61 complex (24.2%), ST-21 complex (23.6%), and ST-42 complex (20.5%). This provided further evidence that the ST-61 clonal complex may present a cattle-adapted C. jejuni genotype. In addition, the ST-42 clonal complex may also represent an important cattle-associated genotype. Strong geographical associations for these genotypes were also found among the farms. This is the first longitudinal study and the largest study to date for C. jejuni involving cattle populations using MLST for accurate strain characterization. This study shows the important associations between cattle and C. jejuni clonal complexes ST-61, ST-21, and ST-42, and it suggests that cattle and/or dairy products are likely to be a source of the human Campylobacter gastroenteritis caused by such genotypes. The reported findings have significant implications for the design of effective intervention strategies for disease control and prevention.
Campylobacter jejuni is the primary cause of human bacterial gastroenteritis worldwide and is a major zoonotic disease. The number of campylobacter infection cases reported to the Health Protection Agency Communicable Disease Surveillance Centre in England and Wales has been increasing from 32,907 cases in 1991 to 57,674 cases in 2000, although an unexplained decrease has been observed in recent years, with 46,236 cases in 2006 (http://www.hpa.org.uk/infections). In the United States, the number of human campylobacteriosis cases per year is estimated to be around 2.1 to 2.5 million, and 2,000 deaths are attributable to the infection (2). Further, these reported figures are often an underestimate of the true incidence, due mainly to underreporting (35).
C. jejuni is a common commensal in the gastrointestinal tracts of wild and farm animals and is ubiquitous in the natural environment. The routes of zoonotic transmission to humans are complex but are believed to be primarily food borne, with various food-producing animals including poultry (6) and cattle and sheep (31) implicated as important reservoirs. Disease outbreaks are rare and cases are often sporadic with the source of infection unidentified (24). However, when outbreaks occur, unpasteurized or contaminated cow's milk is a well-documented source (10, 14, 18, 25, 26). In a survey of infectious intestinal diseases in England and Wales, 26% of milk-borne outbreaks were attributable to campylobacters during an 8-year period (13). Fecal contamination (27), direct udder excretion (15, 23), and wild birds pecking milk bottle tops (30) are among the reported mechanisms by which milk becomes contaminated with campylobacters.
The epidemiology of C. jejuni in animal populations or their products which enter the food chain remains poorly understood, due in part to problems such as the difficulties of interlaboratory comparisons, inadequate discrimination, and poor reproducibility associated with strain characterization methods. Multilocus sequence typing (MLST), a robust and accurate molecular characterization method, has been shown to be a valuable tool for investigating the epidemiology and the population structure of C. jejuni (7), and several studies have delineated the C. jejuni populations of isolates from various food-producing animals (5, 19).
Hence, the purpose of the present study was to investigate the role and the contribution of cattle on dairy farms as a source for zoonotic C. jejuni infections in humans. A longitudinal study of five farms in the northwest of England over an 11-month period was undertaken to explore several aspects of C. jejuni epidemiology within dairy farm populations. We first estimated the prevalence of Campylobacter spp. in the cattle population and their variations between farms and/or over time. With MLST, the diversity and prevalence of C. jejuni genotypes from the cattle were investigated. Further, the C. jejuni clonal complexes identified were analyzed for possible between-group differences in their distributions among the farms, seasons, and cattle management groups, to aid the understanding of potential underlying relationships for the elucidation of C. jejuni epidemiology in cattle on dairy farms.
MATERIALS AND METHODS
Study site and sample collection.
Cattle fecal samples (n = 1,208) were collected by the DEFRA Epidemiology Fellowship Unit from five commercial dairy farms located in Cheshire, United Kingdom, over an 11-month period in 2003. Animals were observed defecating, and freshly voided feces were sampled immediately into sterile containers. Each sample consisted of a composite of three or more sites of each fecal pat. No animal was sampled more than once within any given month, and each animal was identified by unique ear tags. Equipment on each farm was sterilized between sampling events.
Information on cattle management groups was recorded for each sample. The definitions for the different management groups were as follows: unweaned, any calf having access to milk; weaned, any calf not having access to milk but of prebreeding age; heifer, any cow that was of breeding age but not yet calved; lactating, any cow producing milk; and dry, any previously calved cow that was not producing milk. Each farm was situated less than 3 km away from the University of Liverpool field station in Leahurst. Farms 1 and 4 were immediately adjacent to each other, with farm 2 approximately 1 km away. Farms 3 and 5 were 1 km apart from each other and 5 km away from farms 1, 2, and 4. In terms of cattle herd sizes, farms 1 and 3 were the largest, with roughly 100 and 200 cattle, respectively. Farms 2, 4, and 5 had smaller herd sizes, with roughly 50 cattle each. Subsequently, approximately 50 and 25 cattle fecal samples were collected in each sampling month from farms 1 and 3 and from farms 2, 4, and 5, respectively. The months of sampling were January through July, September, and November in 2003, except for farm 4 (on which sampling ceased in April due to the sale of the farm) and farm 5 (on which sampling was not performed for September and November).
Bacterial growth conditions.
Fecal samples (1 g) were inoculated into campylobacter enrichment broth (LAB135; Lab M, Bury, United Kingdom) containing selective supplement (X131; Lab M) and incubated at 37°C for 24 h followed by a further 24 h at 42°C. A loopful of enrichment culture was then inoculated onto campylobacter blood-free selective agar (CM0739, SR0155; Oxoid, Basingstoke, United Kingdom) and incubated for 24 to 48 h at 42°C under microaerobic conditions using an atmosphere generation system (CN0025; Oxoid). Growth on the selective agar was subcultured onto Columbia blood agar (CBA) (CM0331; Oxoid) and incubated for 24 h under the same conditions for the selective agar. On the basis of growth and colony morphology, oxidase reaction, and Gram stain, Campylobacter spp. were presumptively identified and inoculated into 20% (vol/vol) glycerol brain heart infusion broths (BHIB) for storage at −80°C until required for DNA extraction.
Preparation of chromosomal DNA.
Isolates were revived by culturing on CBA. A loopful of the glycerol brain heart infusion broths was inoculated onto CBA plates for discrete colonies and incubated under the conditions described above. Two to three colonies of growth were removed from the pure culture and emulsified by vortexing in 250 μl of molecular biology-grade water in a 1.5-ml microcentrifuge tube. Bacterial cells were killed and lysed by heating the suspension at 100°C for 25 min in a heat block followed by a freezing treatment at −20°C for 5 min. The centrifugation of the suspension at 13,000 × g for 10 min results in bacterial cell debris forming a pellet and DNA being suspended in the supernatant.
Differentiation of Campylobacter species.
A real-time PCR assay for the differentiation of C. jejuni and Campylobacter coli using TaqMan PCR (PE Applied Biosystems, Foster City, CA) was used to confirm the presumptively identified Campylobacter spp. isolates as described by Best and colleagues (4). Subsequently, only C. jejuni isolates were included in the study.
MLST.
Internal fragments of seven gene targets were amplified by PCR and their nucleotide sequences determined with primers and reaction conditions in accordance with the published MLST scheme for C. jejuni (9). Briefly, the internal fragments of the seven target gene loci (aspA [aspartase A], glnA [glutamine synthase], gltA [citrate synthase], glyA [serine hydroxymethyltransferase], pgm [phosphoglucomutase], tkt [transketolase], and uncA [ATP synthase alpha subunit]) were amplified by PCR and purified with a MultiScreen PCR filter plate and a MultiScreen vacuum manifold (Millipore Corporation, Billerica, MA) according to the manufacturer's instructions. For each of the seven target gene loci, the dideoxy-termination sequencing reaction was performed at least once on each of the amplified forward and reverse DNA strands with BigDye reaction mix version 3.1 and 5× sequencing buffer (PE Applied Biosystems). Reaction products were purified and unincorporated dye terminators were removed by precipitation with 95% and 70% ethanol and sequenced using an ABI Prism 377 automated DNA sequencer. Chromatograms were imported into the Sequencher software version 4.0.5 (Gene Codes Corporation) for sequence editing and assembly. Allele identification and sequence type (ST) and clonal complex assignments for each isolate were done by interrogating the Campylobacter MLST database (http://pubmlst.org/campylobacter).
Statistical analysis.
The associations between C. jejuni genotypes and the respective farms, seasons, and cattle management groups were analyzed with statistical tests using SPSS software version 14.0. A multivariate model was built to adjust for the possible confounding effects between variables using logistic regression, and where appropriate, a risk statistic (adjusted odds ratio) was included to indicate the strength of the relationship. P values of ≤0.05 were the criteria chosen for statistical significance and, where appropriate, more-stringent criteria for statistical significance were used (Bonferroni correction) to adjust for multiple comparisons. Paired data (isolates which were sampled from the same cattle from different months) as well as data from farms 4 and 5 were excluded from these analyses due to the absence of data in certain months, in order to avoid the introduction of bias to the model.
The genotypic diversity for each farm, season, and management group, as evaluated by the number of clonal complexes present in each group, along with the number of isolates representing each genotype, was normalized and quantified by calculating the Simpson's index of diversity (D) (28), where D is given by the following formula: D = 1 − 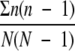 , where n is the number of isolates belonging to a particular clonal complex and N is the total number of isolates in a given group. The value of the index ranges between 0 and 1, where 0 represents no diversity and 1 represents infinite diversity. The index also represents the probability that two individual isolates taken randomly from a given group will have a different clonal complex.
, where n is the number of isolates belonging to a particular clonal complex and N is the total number of isolates in a given group. The value of the index ranges between 0 and 1, where 0 represents no diversity and 1 represents infinite diversity. The index also represents the probability that two individual isolates taken randomly from a given group will have a different clonal complex.
RESULTS
Prevalence of Campylobacter spp.
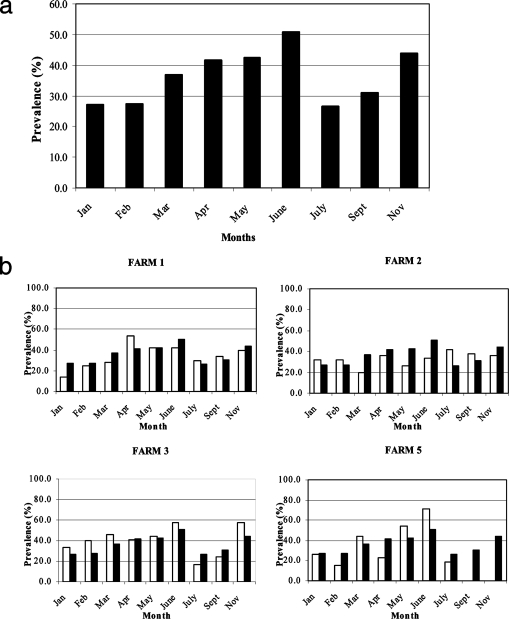
The overall prevalence of Campylobacter spp. in cattle during the study was 35.9% (434/1,208), and from the monthly data, a temporal pattern was observed (Fig. 1a). The prevalence was relatively low from the beginning of the year and gradually increased from March to May and peaked in June (50.8%), followed by a sharp decline in July (26.4%), and it reached a second peak in November (43.9%).
FIG. 1.
(a) Overall monthly prevalence of Campylobacter spp. in cattle during the study (January, 27.0%; February, 27.4%; March, 6.8%; April, 41.5%; May, 42.3%; June, 50.8%; July, 26.4%; September, 30.9%; and November, 43.9%). (b) Monthly prevalence of Campylobacter spp. in cattle on individual farms (white bars) compared with the overall prevalence in panel a (black bars).
All five farms were positive for Campylobacter spp., and the overall prevalence for the individual farms covered a narrow range, from 32.7% to 39.1%. However, distinct temporal dynamics were observed within each farm during the study period (Fig. 1b), with peaks in prevalence seen in March (farm 3 and farm 5), April (farm 1), June (farm 3 and farm 5), July (farm 2), and November (farm 1 and farm 3). Conversely, reduced prevalence was observed in January (farm 1 and farm 3), February (farm 5), March (farm 2), April (farm 5), May (farm 2), and July (farm 1, farm 3, and farm 5). The smallest and largest changes in prevalence observed between adjacent months in a farm were recorded, respectively, between January and February in farm 2 with no change in prevalence and between June and July in farm 5 with a 73.7% reduction in prevalence. Farm 4 was withdrawn from the study due to the sale of cattle after April, and hence a trend was not established.
C. jejuni genotypes.
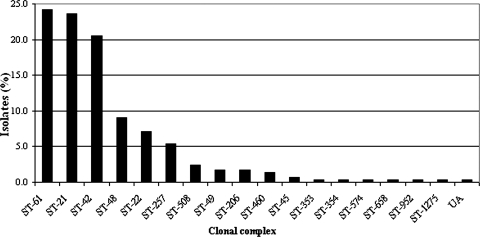
From a total of 297 C. jejuni isolates, 41 STs were identified, of which 22 appeared only once in the data set (Table 1). The most-common STs in the study were ST-61 (23.2%), ST-42 (18.2%), ST-21 (8.8%), ST-48 (8.4%), ST-22 (6.4%), and ST-257 (4.4%), which collectively represented 69% of the data set.
TABLE 1.
C. jejuni STs and clonal complexes among 297 cattle isolates
| Clonal complex (% of isolates) | ST | No. (%) of isolates |
|---|---|---|
| MST-61 (24.2) | ST-61 | 69 (23.2) |
| ST-618 | 1 (0.3) | |
| ST-2369a | 1 (0.3) | |
| ST-2370a | 1 (0.3) | |
| ST-21 (23.6) | ST-21 | 26 (8.8) |
| ST-19 | 21 (7.1) | |
| ST-50 | 15 (5.1) | |
| ST-917 | 3 (1.0) | |
| ST-376 | 2 (0.7) | |
| ST-104 | 1 (0.3) | |
| ST-518 | 1 (0.3) | |
| ST-577 | 1 (0.3) | |
| ST-42 (20.5) | ST-42 | 54 (18.2) |
| ST-758 | 2 (0.7) | |
| ST-2371a | 2 (0.7) | |
| ST-2372a | 1 (0.3) | |
| ST-447 | 1 (0.3) | |
| ST-517 | 1 (0.3) | |
| ST-48 (9.1) | ST-48 | 25 (8.4) |
| ST-38 | 1 (0.3) | |
| ST-205 | 1 (0.3) | |
| ST-22 (7.1) | ST-22 | 19 (6.4) |
| ST-545 | 1 (0.3) | |
| ST-1047 | 1 (0.3) | |
| ST-257 (5.4) | ST-257 | 13 (4.4) |
| ST-286 | 2 (0.7) | |
| ST-776 | 1 (0.3) | |
| ST-508 (2.3) | ST-508 | 6 (2.0) |
| ST-2373a | 1 (0.3) | |
| ST-206 (1.7) | ST-206 | 3 (1.0) |
| ST-227 | 2 (0.7) | |
| ST-49 | ST-49 | 5 (1.7) |
| ST-460 | ST-606 | 4 (1.3) |
| ST-45 | ST-137 | 2 (0.7) |
| ST-353 | ST-356 | 1 (0.3) |
| ST-354 | ST-354 | 1 (0.3) |
| ST-574 | ST-574 | 1 (0.3) |
| ST-658 | ST-311 | 1 (0.3) |
| ST-952 | ST-799 | 1 (0.3) |
| ST-1275 | ST-1231 | 1 (0.3) |
| Unassigned | ST-1673 | 1 (0.3) |
Newly identified ST.
With the exception of ST-1673, represented by a single isolate, all STs identified were grouped into 17 clonal complexes, with 90% of isolates belonging to the six most-prevalent lineages (ST-61, ST-21, ST-42, ST-48, ST-22, and ST-257 clonal complexes). Just over two-thirds (68%) of the isolates belonged to one of three clonal complexes which dominated the data set (Fig. 2). Only eight clonal complexes included more than one ST, and six clonal complexes occurred only once in the data set. From six isolates, five STs (ST-2369, ST-2370, ST-2371, ST-2372, and ST-2373) were described for the first time and assigned to three previously established clonal complexes.
FIG. 2.
Distribution of C. jejuni clonal complexes among all 297 cattle isolates in the study. UA, unassigned.
ST-61, ST-21, and ST-42 clonal complexes.
Three clonal complexes (ST-61, ST-21, and ST-42) have emerged to predominate among the cattle isolates, where each complex individually represented over 20% of all isolates. The ST-61 complex, the most-common clonal complex in the study, represented 24.2% of all isolates (n = 72) and comprised four different STs. Similarly, the ST-21 complex represented 23.6% of all isolates (n = 70) and comprised eight STs, while the ST-42 complex represented 20.5% of all isolates (n = 61) and comprised six STs (Table 1). In all three cases, the central genotype of the clonal complex was also the most-frequent ST present in each clonal complex, with ST-61 and ST-42 representing 95.8% and 88.5% of isolates belonging to the ST-61 complex and the ST-42 complex, respectively. However, ST-21, despite being the most-frequent ST in the lineage, represented only 37% of isolates belonging to the ST-21 complex, as the lineage was codominated by two other STs, ST-19 (30.0%) and ST-50 (21.4%).
Genotypic diversity and distribution of genotypes among farms, seasons, and management groups.
The number of distinct C. jejuni STs or clonal complexes present on each farm ranged from 2 on farm 5 to 19 on farm 3. The genotypic diversity of each farm, as quantified by the Simpson's index of diversity (28), based on the number of clonal complexes present, highlighted the diversity of farms 1 to 4, while farm 5 was the least diverse (Table 2).
TABLE 2.
Genotypic diversity of C. jejuni isolates among farms, seasons, and management groups
| Isolate group | No. of:
|
Index of diversitya | ||
|---|---|---|---|---|
| Isolates | STs | Clonal complexes | ||
| All isolates | 297 | 41 | 17 | 0.829 |
| Farms | ||||
| 2 | 46 | 18 | 12 | 0.823 |
| 4 | 32 | 12 | 7 | 0.786 |
| 3 | 103 | 19 | 10 | 0.785 |
| 1 | 87 | 16 | 9 | 0.752 |
| 5 | 29 | 4 | 2 | 0.123 |
| Seasons | ||||
| Summer | 88 | 23 | 11 | 0.837 |
| Winter | 63 | 17 | 10 | 0.832 |
| Autumn | 49 | 19 | 11 | 0.827 |
| Spring | 97 | 20 | 13 | 0.820 |
| Management groups | ||||
| Dry | 25 | 11 | 8 | 0.887 |
| Lactating | 47 | 15 | 11 | 0.862 |
| Heifer | 63 | 22 | 11 | 0.839 |
| Unweaned | 123 | 14 | 8 | 0.825 |
| Weaned | 36 | 17 | 8 | 0.803 |
Simpson's index of diversity for isolates of each farm, season, and management group based on clonal complexes, in order of descending diversity.
There was a distinct variation in the distribution of C. jejuni clonal complexes on individual farms (Table 3). The ST-21 complex was the only genotype that was present on all five farms, while complexes ST-61, ST-42, ST-22, and ST-257 were present on four farms. Complexes ST-21 and ST-42 constituted a higher proportion of isolates on farms 1, 2, and 4 than on the other farms. Similarly, the ST-61 complex appeared to be relatively more common on farm 3 (35.9%) and farm 5 (93.6%) than on other farms, while the ST-48 complex appeared to be common only on farm 3 (23.3%).
TABLE 3.
Prevalence of major C. jejuni clonal complexes found among farms, seasons, and cattle management groups from 297 cattle isolates
| Clonal complex | Prevalence (%)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Farm
|
Season
|
Management group
|
||||||||||||
| 1 | 2 | 3 | 4 | 5 | Winter | Spring | Summer | Autumn | Unweaned | Weaned | Heifer | Lactating | Dry | |
| ST-61 | 5.7 | 6.5 | 35.9 | 0.0 | 93.5 | 22.2 | 28.9 | 22.7 | 20.4 | 30.6 | 22.8 | 27.0 | 23.4 | 20.0 |
| ST-21 | 29.9 | 32.6 | 16.5 | 31.3 | 6.5 | 27.0 | 21.6 | 19.3 | 30.6 | 25.0 | 26.8 | 20.6 | 23.4 | 16.0 |
| ST-42 | 37.9 | 26.1 | 5.8 | 31.3 | 0.0 | 19.0 | 20.6 | 21.6 | 20.4 | 13.9 | 23.6 | 20.6 | 14.9 | 16.0 |
| ST-48 | 0.0 | 4.3 | 23.3 | 3.1 | 0.0 | 4.8 | 5.2 | 17.0 | 8.2 | 8.3 | 13.0 | 7.9 | 6.4 | 0.0 |
| ST-22 | 13.8 | 4.3 | 1.9 | 15.6 | 0.0 | 7.9 | 10.3 | 5.7 | 2.0 | 8.3 | 7.3 | 3.2 | 6.4 | 16.0 |
| ST-257 | 4.6 | 6.5 | 4.9 | 12.5 | 0.0 | 11.1 | 3.1 | 5.7 | 2.0 | 2.8 | 3.3 | 4.8 | 10.6 | 12.0 |
| Others | 8.1 | 19.6 | 11.7 | 6.3 | 0.0 | 7.9 | 10.3 | 8.0 | 16.3 | 11.1 | 3.3 | 15.9 | 14.9 | 20.0 |
The numbers of STs and clonal complexes present in each season were relatively stable and ranged from 17 STs in winter to 23 in summer and from 10 clonal complexes in winter to 13 in spring. The Simpson's index of diversity determined for each season, with a range of 0.820 to 0.837, indicated high levels of genotypic diversity in C. jejuni isolates across seasons, with summer being the most diverse and spring being the least diverse (Table 2).
The distribution of different C. jejuni clonal complexes in each of the four seasons demonstrated slight variations (Table 3). Clonal complexes ST-61, ST-21, ST-42, ST-48, ST-22, ST-257, and ST-508 were the genotypes present in all four seasons. The ST-61 complex was more prevalent in spring (28.9%), whereas the ST-21 complex appeared to be more prevalent in winter (27.0%) and autumn (30.6%) and the ST-42 complex demonstrated a relatively consistent prevalence throughout. Clonal complexes ST-48, ST-22, and ST-257 were most prevalent in summer, spring, and winter, respectively.
Similarly, the number of STs presented in the cattle management or age groups ranged from 11 (8 clonal complexes) in the dry group to 22 (11 clonal complexes) in heifers. The Simpson's index of diversity, with a range of 0.803 to 0.887, also indicated a consistently high diversity among the management groups, although C. jejuni isolates originating from older cattle appeared to be the most diverse (Table 2).
Clonal complexes ST-61, ST-21, ST-42, ST-22, and ST-257 were present in C. jejuni isolates from cattle from all five management groups, and a progressive change in the distribution of C. jejuni clonal complexes in each of the five cattle groups was observed (Table 3). Collectively, the ST-61, ST-42, and ST-21 complexes appeared to be more prevalent in younger cattle. Conversely, older cattle appeared to harbor uncommon genotypes more frequently.
Associations between ST-61, ST-21, and ST-42 clonal complexes and the respective farms, seasons, and cattle management groups.
The apparent nonrandom associations in the distributions of three predominant clonal complexes were explored with a multivariate logistic regression model with isolates from farms 1 to 3 (Table 4). Data from farms 4 and 5 were omitted from these analyses due to the absence of data in certain months, in order to avoid the introduction of bias to the model.
TABLE 4.
Pairwise comparison of the probability of isolating C. jejuni clonal complexes ST-61, ST-21, and ST-42 among farms and seasons
| Effects by group | OR [95% CI (lower/upper)]a
|
P | P*b | |
|---|---|---|---|---|
| Unadjusted | Adjusted | |||
| Farm effects | ||||
| ST-61 complex | ||||
| Farm 3c | 1 | 1 | ||
| Farm 1 | 0.11 (0.04/0.33) | 0.08 (0.03/0.48) | <0.001 | 0.003 |
| Farm 2 | 0.15 (0.04/0.51) | 0.12 (0.02/0.48) | 0.003 | 0.009 |
| ST-21 complex | ||||
| Farm 3c | 1 | 1 | ||
| Farm 1 | 1.98 (0.95/4.13) | 2.32 (0.93/4.20) | 0.075 | |
| Farm 2 | 2.26 (0.98/5.21) | 2.32 (0.96/5.59) | 0.061 | |
| ST-42 complex | ||||
| Farm 3c | 1 | 1 | ||
| Farm 1 | 9.62 (3.75/25) | 9.60 (3.66/25.21) | <0.001 | 0.003 |
| Farm 2 | 4.88 (1.66/14.29) | 4.45 (1.42/13.99) | 0.011 | 0.033 |
| Temporal effects (quarter) | ||||
| ST-61 complex | ||||
| Summerc | 1 | 1 | ||
| Autumn | 0.50 (0.17/1.46) | 0.40 (0.12/1.31) | 0.126 | |
| Winter | 1.69 (0.52/5.46) | 3.48 (0.87/13.95) | 0.079 | |
| Spring | 3.13 (1.19/8.26) | 5.05 (1.64/15.56) | 0.005 | 0.03 |
OR, odds ratio.
P*, P values of significance adjusted for pairwise comparisons using a Bonferroni correction for three comparisons (farm effects) or six comparisons (temporal effects).
Reference group.
For farm effects, the adjusted odds ratios indicated a significantly increased chance of isolating the ST-61 complex from fecal samples on farm 3 compared to the chances on farm 1 (P = 0.003) and farm 2 (P = 0.009). Similarly, there was a significantly increased probability of isolating the ST-42 complex from fecal samples on farm 1 (P = 0.003) and farm 2 (P = 0.033) compared to the probability on farm 3. In addition, although not statistically significant, a positive association for the ST-42 complex was seen in farm 1 compared to farm 2 (P = 0.099), as well as for the ST-21 complex on farm 1 (P = 0.075) and farm 2 (P = 0.061) compared to farm 3.
The adjusted odds ratio for temporal effects indicated that there was a significantly increased probability of isolating the ST-61 complex in feces collected in spring compared to summer (P = 0.03). A nonsignificant positive statistical association was seen for the ST-61 complex in feces collected in winter compared to summer (P = 0.079). No other significant temporal effects were seen for isolating the ST-21 and ST-42 complexes under this model.
There were also no statistically significant differences in the distributions of individual clonal complexes among the five cattle management groups under the multivariate model (data not shown).
Changes in genotype carriage.
Thirty animals were sampled on two or more occasions at different months during the study and were characterized by MLST (Table 5). Of the 30 cows, 25 were sampled twice and 5 were sampled three times. Sixteen cows carried a different genotype on successive sampling occasions (animals 1 to 16), including one which was sampled three times (animal 13). Of these, 13 animals (animals 1 to 13) carried a different clonal complex on each occasion, while three animals carried a different ST of the same clonal complex (animals 14 to 16). Twelve cows possessed the same ST on successive sampling occasions (animals 19 to 30), including two which were sampled on three occasions (animals 19 and 20). The two remaining cows (animals 17 and 18) were sampled three times and carried an identical ST in the first two samples with a change in clonal complex for the third sample. ST-61, ST-22, ST-48, ST-42, ST-19, and ST-49 were the only STs which were carried on successive occasions.
TABLE 5.
Analysis of the dynamics of C. jejuni genotype carriage from 30 animals on successive sampling occasionsa
| Type of carriage change (n) | Animal no. | Initial sample
|
Successive sample
|
||
|---|---|---|---|---|---|
| ST | CC | ST | CC | ||
| Change in both ST and CC (15) | 1 | ST-42 | ST-42 | ST-21 | ST-21 |
| 2 | ST-42 | ST-42 | ST-21 | ST-21 | |
| 3 | ST-42 | ST-42 | ST-50 | ST-21 | |
| 4 | ST-48 | ST-48 | ST-21 | ST-21 | |
| 5 | ST-206 | ST-206 | ST-19 | ST-21 | |
| 6 | ST-21 | ST-21 | ST-42 | ST-42 | |
| 7 | ST-19 | ST-21 | ST-42 | ST-42 | |
| 8 | ST-22 | ST-22 | ST-42 | ST-42 | |
| 9 | ST-257 | ST-257 | ST-42 | ST-42 | |
| 10 | ST-22 | ST-22 | ST-1231 | ST-1275 | |
| 11 | ST-42 | ST-42 | ST-61 | ST-61 | |
| 12 | ST-618 | ST-61 | ST-508 | ST-508 | |
| 13* | ST-22 | ST-22 | ST-61 | ST-61 | |
| 13* | ST-61 | ST-61 | ST-38 | ST-48 | |
| 17* | ST-21 | ST-21 | ST-42 | ST-42 | |
| 18* | ST-61 | ST-61 | ST-48 | ST-48 | |
| Change in ST only (3) | 14 | ST-758 | ST-42 | ST-2372 | ST-42 |
| 15 | ST-21 | ST-21 | ST-917 | ST-21 | |
| 16 | ST-21 | ST-21 | ST-50 | ST-21 | |
| No change (14) | 17* | ST-21 | ST-21 | ST-21 | ST-21 |
| 18* | ST-61 | ST-61 | ST-61 | ST-61 | |
| 19* | ST-61 | ST-61 | ST-61 | ST-61 | |
| 19* | ST-61 | ST-61 | ST-61 | ST-61 | |
| 20* | ST-61 | ST-61 | ST-61 | ST-61 | |
| 20* | ST-61 | ST-61 | ST-61 | ST-61 | |
| 21 | ST-61 | ST-61 | ST-61 | ST-61 | |
| 22 | ST-61 | ST-61 | ST-61 | ST-61 | |
| 23 | ST-61 | ST-61 | ST-61 | ST-61 | |
| 24 | ST-22 | ST-22 | ST-22 | ST-22 | |
| 25 | ST-22 | ST-22 | ST-22 | ST-22 | |
| 26 | ST-48 | ST-48 | ST-48 | ST-48 | |
| 27 | ST-48 | ST-48 | ST-48 | ST-48 | |
| 28 | ST-19 | ST-21 | ST-19 | ST-21 | |
| 29 | ST-42 | ST-42 | ST-42 | ST-42 | |
| 30 | ST-49 | ST-49 | ST-49 | ST-49 | |
CC, clonal complex; *, animal sampled on three occasions.
DISCUSSION
Cattle play a significant role in C. jejuni epidemiology as an important host to campylobacter strains that are capable of causing disease in humans (31). Based on data sets comprised of sample collections of C. jejuni isolates from disparate animal sources, a number of MLST studies have provided a growing body of evidence for host specificity among C. jejuni genotypes, including distinct pathogenic isolates associated with cattle (5, 8, 12, 19). Here, we present findings from the first longitudinal study and the largest to date MLST survey with respect to C. jejuni populations in cattle, based on a set of epidemiologically linked isolates from dairy cattle farms within a defined geographical region over a temporally continuous period, with the primary objective of investigating the importance of cattle as a reservoir for human campylobacter infections.
Depending on a range of factors, including sampling type and size, recovery methods, herd type, season, and geography, wide discrepancies in the overall prevalence of C. jejuni in cattle have been reported in past studies. Nonetheless, the prevalence rates of 39.5% (34), 38.6% (3), and 23.0% (22) for thermophilic campylobacters described in several studies involving fecal samples of dairy cattle were generally in concordance with our findings (35.9%). Temporal fluctuations in prevalence were observed with a distinct peak in June (50.8%; Fig. 1a), coinciding with the seasonal peak traditionally seen in human infections in England and Wales (http://www.hpa.org.uk/infections) and consistent with the pronounced late spring peak observed in the shedding numbers of thermophilic campylobacters in cattle in a previous study (32). Although it is a widely held view that poultry is the predominant source of campylobacter infections, the findings from this study suggest that cattle may play a role in the seasonal peak in human infections or that the exposure to a common source of contamination may exist. Furthermore, a number of recent studies, including a 6-year survey, have shown evidence against a direct causal link between the seasonality of campylobacter prevalence in chickens and human infections (20, 36).
Several studies have delineated the C. jejuni populations of isolates from various food-producing animals, which provided increasing evidence that members of the ST-61 complex are more restricted to cattle/bovine sources (8, 12, 19). Our study further confirms the hypothesis by clearly demonstrating the dominance of the ST-61 complex in a large cattle data set. Interestingly, clonal complexes ST-21 and ST-42 were also frequently isolated, and clonal complexes ST-48, ST-22, and ST-257 were moderately common. Whereas the ST-42 complex was found to be strongly associated with ovine sources in previous studies (5, 8, 19), this study provided the first description of a high prevalence in cattle. This finding raises the possibility that there may be similarities in the physiological features of the gastrointestinal tracts of cattle and sheep that cause ruminants to be more likely to carry similar C. jejuni genotypes. Additionally, the previous report of cattle and sheep isolates sharing similar pulsed-field gel electrophoresis and flagellin genotypes fits well with this speculation (11). Further investigation involving more ruminant isolates may well provide the key to explain certain aspects of the host specificity for C. jejuni ST-61 and ST-42 clonal complexes. Indeed, there is evidence that the guts of cattle may provide a stable environment for bacterial strains, as there were instances where organisms were found to be genetically stable, dominant cattle strains. For example, certain pathogenic Salmonella STs have been found to be associated exclusively with bovine sources (1). In addition, C. coli strains isolated from cattle were found to be highly clonal in a previous study, where more than 80% of the strains belonged to a single ST (21). Incidentally, this group of C. coli strains possessed the uncA allele 17, which is found in the C. jejuni genotypes belonging to the ST-61 clonal complex found in high numbers in this study.
To put into a public health context the contribution of the C. jejuni genotypes identified in this study, we interrogated the Campylobacter MLST database, which revealed that all of the C. jejuni clonal complexes and at least 66% (27/41) of the STs identified in this study have been isolated from human disease in the past. In addition, all of the six most-common clonal complexes (ST-21, ST-45, ST-206, ST-61, ST-48, and ST-257) identified in the largest human data set to date (8) have all been isolated from cattle in this study. Further, 13 of the clonal complexes reported here were present in an investigation of human campylobacter isolates conducted in the northwest of England in 2003, where the ST-21 complex was clearly the dominant genotype (29). However, the genotypes identified in the present study are distinct from those recovered from environmental sources, where isolates belonging to the ST-45 complex and various uncommon genotypes were more frequently isolated (12). In addition, it has been found that new and unassigned genotypes in humans were predominantly associated with swimming in natural bodies of water (17). This suggests that the transmission pathway of campylobacters from cattle to humans may not necessarily include environmental sources or routes.
This is the first study to outline and compare the genotypic diversity of C. jejuni in cattle herds with a set of spatially and temporally related isolates using MLST. We have demonstrated that C. jejuni genotypes were highly diverse in cattle, with 90% of this data set represented by only 6 clonal complexes while the remaining 10% were represented by 11 complexes. These observations are in agreement with the notion that C. jejuni has a weakly clonal population structure (9, 33). The similarities in genotypic diversity observed between different seasons according the Simpson's index of diversity (Table 2) were somewhat surprising, as these similarities were in contrast to the hypothesis that the characteristic seasonal variation, which is the hallmark of C. jejuni epidemiology, would be reflected to a certain extent by variations in the diversity of the genotypes. On the other hand, the apparent differences in the genotypic diversity of C. jejuni between farms were consistent with the fact that there were geographical differences recorded for C. jejuni clonal complexes, as discussed below. The evaluation of the differences in the genotypic diversity of C. jejuni within and between various sources, regions, and time frames may provide additional insights into the ecological aspects of C. jejuni populations and may, for example, assist in the identification of specific animals or environments that are potential drivers for generating novel genotypes.
To better define the C. jejuni epidemiology in cattle on dairy farms, C. jejuni clonal complexes were analyzed for differences in their distributions among farms, seasons, and cattle management groups.
We have demonstrated that the distribution of C. jejuni genotypes was not random among farms (Tables 3 and 4), suggesting a strong geographical association among genotypes. Consequently, these findings led to the observation that two groups of farms which were 5 km apart (farms 3 and 5 and farms 1, 2, and 4) appeared to have maintained different genotypes, while farms within the same group (no more than 1 km apart) appeared to possess similar C. jejuni genotypes. This observation suggests that campylobacters may be readily transmitted between distances of approximately 1 km, but transmission may be limited on a larger geographical scale. This is consistent with the previous report that C. jejuni isolates from distances of less than 1 km were genetically more similar than isolates separated by greater distances, independent of sample or host types (12).
Certain dynamics were also noted among clonal complexes between farms, in particular, the presence of the ST-61 complex and ST-42 complex genotypes were almost mutually exclusive; these strains were not present on any of the farms with comparable prevalence. This may be explained by a potential ecological competition between strains within the bovine gut, although further studies are required to corroborate this observation. In addition, although the results of this study and past studies have suggested that the ST-61 complex may be a cattle-adapted C. jejuni genotype, the apparent geographical associations shown in this study have revealed that there are indeed cattle farms with only a few ST-61 complex isolates. More importantly, this raises the question of how a strain that is well adapted to cattle can also exist in very low numbers in nearby groups of cattle within a defined geographic area. It is possible that the variations of genotypes observed were due to farm management factors, which may possibly be explained only by scrutinizing the particular differences in farming practices or the diets of animals on individual farms.
Surprisingly, a temporal pattern was detected only for one of the clonal complexes in the logistic regression model (Table 4), where the probability of recovering ST-61 complex isolates was marginally increased in winter and significantly increased in spring, compared to summer. Consequently, this may suggest that no particular genotypes are exclusively responsible for the seasonal peak seen in summer. Nonetheless, the elucidation of the temporal characteristics of genotypes from different sources may help clarify the role of particular genotypes and their attribution to the rise of human infections during specific time frames.
Although there were no significant differences in the distributions of individual genotypes among cattle management groups, it appeared that the dominant genotypes (ST-61, ST-21, and ST-42 complexes) were more prevalent in younger cattle and gradually became relatively less prevalent in older cattle with higher proportions of uncommon genotypes. This observation agrees with a previous report which showed that adult animals carry a broader range of C. jejuni serotypes than calves (22).
We examined strain carriage over time in a small number of animals and observed no striking patterns, except for ST-61, which appeared to be more persistent in cattle and was carried on two sampling occasions in four animals, as well as on all three occasions from two animals. However, the number of animals involved was too small for this observation to be considered significant. Further investigations into this aspect of C. jejuni epidemiology could provide insights into the ecological dynamics of different genotypes within animal hosts.
This is the first report of a longitudinal study of C. jejuni in dairy cattle herds using MLST, and the results have provided important advances in knowledge in several key aspects of C. jejuni epidemiology, indicating that dairy cattle and their products may have a significant role as sources or transmission routes for human campylobacter infections.
Acknowledgments
The funding of this work was supported by the Health Protection Agency North West.
We thank the following people for their assistance: Chris Roberts for his valuable advice on statistical analyses; Steve Keeney and Pam Watson for allowing access to their sequencing facilities; and all the farmers who participated in this study.
This study made use of the Campylobacter jejuni Multi Locus Sequence Typing website (http://pubmlst.org/campylobacter/) developed by Keith Jolley and Man-Suen Chan and hosted by the University of Oxford (16). The development of this site has been funded by the Wellcome Trust.
Footnotes
Published ahead of print on 18 April 2008.
REFERENCES
- 1.Alcaine, S. D., Y. Soyer, L. D. Warnick, W. L. Su, S. Sukhnanand, J. Richards, E. D. Fortes, P. McDonough, T. P. Root, N. B. Dumas, Y. Gröhn, and M. Wiedmann. 2006. Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl. Environ. Microbiol. 72:7575-7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, W., K. N. Kaya, D. D. Hancock, D. R. Call, Y. H. Park, and T. E. Besser. 2005. Prevalence and antimicrobial resistance of thermophilic Campylobacter spp. from cattle farms in Washington State. Appl. Environ. Microbiol. 71:169-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Best, E. L., E. J. Powell, C. Swift, K. A. Grant, and J. A. Frost. 2003. Applicability of a rapid duplex real-time PCR assay for speciation of Campylobacter jejuni and Campylobacter coli directly from culture plates. FEMS Microbiol. Lett. 229:237-241. [DOI] [PubMed] [Google Scholar]
- 5.Colles, F. M., K. Jones, R. M. Harding, and M. C. J. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corry, J. E. L., and H. I. Atabay. 2001. Poultry as a source of Campylobacter and related organisms. J. Appl. Microbiol. 90:96S-114S. [DOI] [PubMed] [Google Scholar]
- 7.Dingle, K. E., F. M. Colles, D. Falush, and M. C. J. Maiden. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. A. Wareing, and M. C. J. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahey, T., D. Morgan, C. Gunneburg, G. K. Adak, F. Majid, and E. Kaczmarski. 1995. An outbreak of Campylobacter jejuni enteritis associated with failed milk pasteurization. J. Infect. 31:137-143. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald, C., K. Stanley, S. Andrew, and K. Jones. 2001. Use of pulsed-field gel electrophoresis and flagellin gene typing in identifying clonal groups of Campylobacter jejuni and Campylobacter coli in farm and clinical environments. Appl. Environ. Microbiol. 67:1429-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French, N., M. Barrigas, P. Brown, P. Ribiero, N. Williams, H. Leatherbarrow, R. Birtles, E. Bolton, P. Fearnhead, and A. Fox. 2005. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7:1116-1126. [DOI] [PubMed] [Google Scholar]
- 13.Gillespie, I. A., G. K. Adak, S. J. O'Brien, and F. J. Bolton. 2003. Milkborne general outbreaks of infectious intestinal disease, England and Wales, 1992-2000. Epidemiol. Infect. 130:461-468. [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington, P. 2002. Outbreak of Campylobacter jejuni infections associated with drinking unpasteurized milk procured through a cow-leasing program—Wisconsin, 2001. MMWR Morb. Mortal. Wkly. Rep. 51:548-549. [PubMed] [Google Scholar]
- 15.Hutchinson, D. N., F. J. Bolton, P. M. Hinchliffe, H. C. Dawkins, S. D. Horsley, E. G. Jessop, P. A. Robertshaw, and D. E. Counter. 1985. Evidence of udder excretion of Campylobacter jejuni as the cause of milk-borne campylobacter outbreak. J. Hyg. (London) 94:205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jolley, K. A., M. S. Chan, and M. C. J. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karenlampi, R., H. Rautelin, D. Schonberg-Norio, L. Paulin, and M. L. Hanninen. 2007. Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl. Environ. Microbiol. 73:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornblatt, A. N., T. Barrett, G. K. Morris, and F. E. Tosh. 1985. Epidemiologic and laboratory investigation of an outbreak of campylobacter enteritis associated with raw-milk. Am. J. Epidemiol. 122:884-889. [DOI] [PubMed] [Google Scholar]
- 19.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meldrum, R. J., J. K. Griffiths, R. M. M. Smith, and M. R. Evans. 2005. The seasonality of human campylobacter infection and Campylobacter isolates from fresh, retail chicken in Wales. Epidemiol. Infect. 133:49-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, W. G., M. D. Englen, S. Kathariou, I. V. Wesley, G. L. Wang, L. Pittenger-Alley, R. M. Siletz, W. Muraoka, P. J. Fedorka-Cray, and R. E. Mandrell. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245-255. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen, E. M. 2002. Occurrence and strain diversity of thermophilic campylobacters in cattle of different age groups in dairy herds. Lett. Appl. Microbiol. 35:85-89. [DOI] [PubMed] [Google Scholar]
- 23.Orr, K. E., N. F. Lightfoot, P. R. Sisson, B. A. Harkis, J. L. Tweddle, P. Boyd, A. Carroll, C. J. Jackson, D. R. A. Wareing, and R. Freeman. 1995. Direct milk excretion of Campylobacter jejuni in a dairy cow causing cases of human enteritis. Epidemiol. Infect. 114:15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pebody, R., M. J. Ryan, and P. G. Wall. 1997. Outbreaks of Campylobacter infection: rare events for a common pathogen. Commun. Dis. Rep. CDR Rev. 7:R33-R37. [PubMed] [Google Scholar]
- 25.Peterson, M. C. 2003. Campylobacter jejuni enteritis associated with consumption of raw milk. J. Environ. Health 65:20-21. [PubMed] [Google Scholar]
- 26.Robinson, D. A., and D. M. Jones. 1981. Milk-borne Campylobacter infection. Br. Med. J. 282:1374-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schildt, M., S. Savolainen, and M. L. Hanninen. 2006. Long-lasting Campylobacter jejuni contamination of milk associated with gastrointestinal illness in a farming family. Epidemiol. Infect. 134:401-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 29.Sopwith, W., A. Birtles, M. Matthews, A. Fox, S. Gee, M. Painter, M. Regan, Q. Syed, and E. Bolton. 2006. Campylobacter jejuni multilocus sequence types in humans, Northwest England, 2003-2004. Emerg. Infect. Dis. 12:1500-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Southern, J. P., R. M. M. Smith, and S. R. Palmer. 1990. Bird attack on milk bottles—possible mode of transmission of Campylobacter jejuni to man. Lancet 336:1425-1427. [DOI] [PubMed] [Google Scholar]
- 31.Stanley, K., and K. Jones. 2003. Cattle and sheep farms as reservoirs of Campylobacter. J. Appl. Microbiol. 94:104S-113S. [DOI] [PubMed] [Google Scholar]
- 32.Stanley, K. N., J. S. Wallace, J. E. Currie, P. J. Diggle, and K. Jones. 1998. The seasonal variation of thermophilic campylobacters in beef cattle, dairy cattle and calves. J. Appl. Microbiol. 85:472-480. [DOI] [PubMed] [Google Scholar]
- 33.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wesley, I. V., S. J. Wells, K. M. Harmon, A. Green, L. Schroeder-Tucker, M. Glover, and I. Siddique. 2000. Fecal shedding of Campylobacter and Arcobacter spp. in dairy cattle. Appl. Environ. Microbiol. 66:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler, J. G., D. Sethi, J. M. Cowden, P. G. Wall, L. C. Rodrigues, D. S. Tompkins, M. J. Hudson, and P. J. Roderick. 1999. Study of infectious intestinal disease in England: rates in the community, presenting to general practice, and reported to national surveillance. Br. Med. J. 318:1046-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson, I. G. 2002. Salmonella and campylobacter contamination of raw retail chickens from different producers: a six year survey. Epidemiol. Infect. 129:635-645. [DOI] [PMC free article] [PubMed] [Google Scholar]