Abstract
Antibiotic-sensitive bacteria have been found to coexist with antibiotic-producing bacteria in biofilms, but little is known about how the former develop in such an environment. Here we isolated pyocyanin-sensitive bacteria belonging to the genus Brevibacillus from a biofilm derived from soil extract and based on the preestablished biofilm of a pyocyanin producer, Pseudomonas aeruginosa strain P1. In addition, pyocyanin-resistant strains belonging to the genus Raoultella were isolated from the same biofilm. Microbial relationships within biofilms were examined by using three strains, strain P1, Brevibacillus strain S1, and Raoultella strain R1, each of which individually formed a biofilm within 2 days in a flow cell. Strain S1 did not fully develop on the preestablished biofilm of strain P1 during 4 days of cultivation, whereas a mutant of strain P1 which was deficient in pyocyanin production allowed strain S1 to cocolonize within a biofilm. On the other hand, strain R1 developed on the biofilm of strain P1 regardless of pyocyanin production. When mixed 1:1 inocula of strains S1 and R1 were introduced into the strain P1 biofilm, all three species were found in the 4-day biofilm. In the mixed biofilm, strain S1 was surrounded by the layer of strain R1 and seemed to be separated from strain P1 and the outflow solution. However, strain S1 did not survive in a three-species mixed culture under planktonic conditions. These results indicate that the survival of sensitive bacteria in biofilm with a pyocyanin producer is achieved by covering them with a layer of resistant bacteria. We also evaluated the influence of antibiotic production on the producer.
In natural and engineered environments, most microorganisms are organized on surface-attached biofilms in which multiple species of microorganisms coexist. Because of spatial and nutrient limitations, biofilms are both highly competitive and highly cooperative environments (5, 43, 56). Previous studies have shown that a high percentage of microorganisms associated with surfaces produce antimicrobial metabolites (14, 52, 56). In general, antibiotics play crucial roles in determining bacterial population and distribution in natural habitats (9, 41). For example, Norman et al. (33) reported that antibiotics contribute to their producers' domination of a crude-oil-degrading environment by eliminating other bacteria. However, in numerous biofilms in nature (e.g., marine, oral, and clinical microbial ecosystems), antibiotic-sensitive bacteria have been found to coexist with antibiotic-producing bacteria (25, 38, 39, 45, 47). Still, little is known about how antibiotic-sensitive bacterial species colonize and develop in biofilm with antibiotic-producing bacteria. In this study, we investigated the mechanism underlying the coexistence of an antibiotic-sensitive bacterium with an antibiotic-producing bacterium in a multispecies biofilm.
In general, cells in biofilms are surrounded by a self-produced matrix, and they grow slowly because of the insufficiency of nutrients and space (46, 53). These characteristics probably provide the component species of the biofilm with an increased tolerance of antibiotics. In fact, some reports have shown that bacteria in biofilms acquire tolerance to antibiotics or to invasion by antibiotic-producing bacteria (6, 38, 45). Furthermore, biofilms have heterogeneous and spatially stratified structures (46, 53). Chemical gradients in the concentrations of nutrients and/or metabolic products determine microbes' individual niches (8, 32, 35, 46, 53), resulting in the coexistence of multiple species in biofilms (27, 38, 44, 45). Therefore, a heterogeneous distribution of antibiotics may lead to a spatial niche for surviving and for developing sensitive bacteria in biofilms.
A previous report indicated that biofilms consisting of more than two species acquire higher tolerance to antibiotics than those consisting of only one species (6). That report suggested that synergistic interactions among component species play important roles in the increased antibiotic resistance of sensitive bacterium. Interspecies relationships may affect sensitivity to antibiotics. Furthermore, it is a well-known phenomenon that the interaction between two bacterial species is influenced by the addition of a third species (18, 21).
In this study, we determined the behavior of antibiotic-sensitive bacteria within biofilms containing antibiotic-producing bacteria and evaluated the effect of adding a third microorganism. We selected soil as a bacterial source for the construction of a model system of multispecies biofilm because soil includes a high diversity of microorganisms (13, 48) and is frequently used as a source of antibiotic-producing bacteria (4, 19). It is also a rich supply of surfaces on which biofilms form vigorously (7, 42, 50).
MATERIALS AND METHODS
Bacterial strains, culture conditions, and isolation.
Tryptic soy broth (TSB) (Becton Dickinson, Rutherford, NJ) was used to cultivate biofilms in a flow cell system and to isolate bacteria. Biofilms directly collected from the flow cell were serially diluted in sterilized distilled water and spread on TSB containing 1.5% agar. The samples were incubated overnight at 37°C. Colonies were purified at least three times.
The strains isolated in this study, as well as the test strains used to detect antibiotic activity (Pseudomonas aeruginosa PAO1, Escherichia coli, Bacillus subtilis, and Staphylococcus aureus), were routinely maintained in TSB at 37°C. E. coli JM109 was used for cloning and plasmid amplification and was grown in Luria-Bertani broth or on 1.5% agar at 37°C. When required, tetracycline (150 μg/ml), ampicillin (50 μg/ml), carbenicillin (200 μg/ml), kanamycin (20 μg/ml), or polymyxin (15 μg/ml) was added to the medium.
Collection and preparation of a soil microbial community.
Soil was collected at a depth of 3 to 5 cm on the campus of the Faculty of Agriculture, University of Tokyo. An aliquot (0.1 g) of soil was suspended in 10 ml of sterile water and mixed vigorously. After the soil suspension was allowed to settle, the supernatant was taken and used as a soil extract. The soil extract was stored at 4°C until use and was used within 7 days.
Identification and quantification of antibiotics.
To identify the antibiotic produced by an isolated bacterium, active fractions of cultures were measured by time-of-flight mass spectrometry (QSTAR XL; Applied Biosystems, Tokyo, Japan).
The pyocyanin concentration was determined by a previously described method (11). An aliquot (5 ml) of culture solution or filtrate was extracted with 3 ml of chloroform and then reextracted into 1 ml of 0.2 N HCl. The absorbance of this solution was measured at 520 nm. Concentrations, expressed as micrograms of pyocyanin produced per milliliter of culture supernatant, were determined by multiplying the absorbance at 520 nm by 17.072 (26).
Experimental biofilm design.
Biofilms were grown in continuous-culture flow cells (channel dimensions, 1 by 4 by 40 mm; Stovall Life Science, Greensboro, NC) at 37°C. Preestablished biofilms of pyocyanin-producing strain P1 or of its derivative strain PHZ201 (see below) were constructed as follows. Flow cell channels were inoculated with preculture of strain P1 or ΔphzM. After 3 h of incubation at 37°C with no flow, TSB was allowed to flow at a 1.5-ml/h rate for 2 days with a peristaltic pump. The preestablished biofilms were inoculated with 500 μl of soil extract or culture of the isolated strain(s). After 3 h of incubation with no flow, the medium was recirculated.
Minimal inhibitory activity (MIC) assay.
The MIC of a planktonic culture was determined according to standard techniques (31). Pyocyanin derived from an isolate was used for the MIC assay. Overnight cultures of bacteria were added at a concentration of 5 × 107 CFU/ml to wells of a microtiter plate with various concentrations of pyocyanin, and the microtiter plate was incubated aerobically at 37°C. Cell viability was determined by reading the turbidity at 590 nm in a 96-well plate reader (Molecular Devices, Sunnyvale, CA). The MIC was defined as the lowest concentration of antibiotic in which there was no visible growth after overnight incubation.
DNA extraction.
DNA from soil, biofilm, and isolated bacterial cells was extracted according to the benzyl chloride method (57). The isolated DNA was used as templates for the PCR experiments described below.
Sequence analysis.
The 16S rRNA sequence of each isolate was determined by direct sequencing of the purified PCR-amplified 16S rRNA gene fragment as described previously (20). Primers 27F and 1512R were used to amplify the 16S rRNA genes of the isolates (10). The PCR products of the isolates were purified with the QIAquick PCR purification kit (Qiagen, Tokyo, Japan). The purified 16S rRNA gene was sequenced directly with the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) and an ABI PRISM model 3130/3130xl genetic analyzer (Applied Biosystems). The sequence obtained was aligned with those from the DDBJ nucleotide sequence database with the BLAST program.
PCR amplification of 16S rRNA fragment for denaturing gradient gel electrophoresis (DGGE).
The PCR products were amplified with AmpliTaq Gold (Applied Biosystems) according to the manufacturer's instructions. For DGGE analysis, the V3 region of the 16S rRNA was amplified with primers 357F-GC and 517R (30). Reaction and cycling conditions were the same as described previously (15). The PCR products were examined by electrophoresis on 1.5% agarose gels before being subjected to DGGE.
PCR-DGGE.
The purified PCR products were analyzed by DGGE with the DCode system (Bio-Rad Laboratories, Hercules, CA) as described previously (15). The protocol described by Haruta et al. (15) was used with the modification of a denaturant gradient of 15 to 55% (where 100% is defined as 7 M urea with 40% formamide). After electrophoresis, the gels were stained with SYBR green I (Molecular Probes, Eugene, OR) and photographed. After each band was reamplified under the reaction conditions described above, the resulting PCR products were checked to confirm that they showed single bands at the same positions on the DGGE gel. Following three rounds of band excision and DGGE analysis, the bands were excised and amplified with the primers described above, except that primer 357F, which lacks the 50-bp GC clamp of 357F-GC, was used. The sequence analysis of the PCR products was performed as described above.
Specific PCR detection of Pseudomonas spp. and fungi.
A PCR assay to detect Pseudomonas spp. was performed with primers Ps-for (5′-GGTCTGAGAGGATGATCAGT-3′) and Ps-rev (5′-TTAGCTCCACCTCGCGGC-3′) (55). The reaction and cycling conditions used were described previously (36).
The 18S rRNA genes of the fungi were amplified with primers NS3 (5′-GCAAGTCTGGTGCCAGCAGCC-3′) and YM951r (5′-TTGGCAAATGCTTTCGC-3′) (54). The reaction and cycling conditions used were described previously (16). The products were examined by electrophoresis on 1.5% agarose gel.
Construction of an antibiotic-deficient mutant.
A phzM-deficient mutant of pyocyanin-producing strain P1 was constructed by insertion of the tetracycline resistance gene (tet) by homologous recombination with plasmid pPHZ, which carries the phzM::tet construct. The homologous-recombination method used was described previously (3). A 2.5-kb fragment carrying phzM was amplified by PCR from the chromosomal DNA of strain P1 with oligonucleotides pf2-for (5′-CAGCGAGCACCGAAGAATTCGGCGC-3′) and pr4-rev (5′-GGTCTCGAAGCTTCGCACGTTGTG-3′). The amplified fragment was digested with SacI and HindIII and inserted into the respective sites of pUC19. The phzM gene on the resultant plasmid was disrupted by digestion with SphI and ligation with the tet fragment, resulting in pPHZ. After strain P1 was transformed with pPHZ, a tetracycline-resistant and carbenicillin-sensitive strain whose phzM gene on the chromosome had been replaced with the plasmid-derived disrupted gene by double-crossover recombination was selected and designated strain PHZ201. The mutation was confirmed by PCR.
Fluorescence in situ hybridization (FISH) analysis.
Flow cell biofilms were injected with 100% ethanol for 15 min at 34°C. Biofilms were fixed with 4% paraformaldehyde in phosphate-buffered saline (1.7 mM KH2PO4, 5 mM Na2HPO4, 0.15 M sodium chloride, pH 7.2) for 18 h at 4°C. After fixation, the biofilms were washed with phosphate-buffered saline and then exposed to a lysozyme solution (10 mg/ml in 0.1 M Tris-HCl-0.05 M EDTA, pH 7.2) for 10 min at room temperature to permeabilize the cells. After permeabilization, the biofilms were dehydrated by a series of 3-min washes with 50, 80, and 95% ethanol. DNA probes labeled at the 5′ end with Cy3, Cy5, or fluorescein isothiocyanate (FITC) were synthesized. Pae997-Cy3 (2), LGC-Cy5 (28), and EUB338-FITC (1) were chosen as probes for the detection of P. aeruginosa, gram-positive bacteria, and all other bacteria, respectively. Furthermore, we selected ENT-FITC (23) as a probe for the detection of Raoultella species. Biofilms were incubated with hybridization buffer (0.9 M NaCl, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl [pH 7.2], 15% formamide) containing the fluorescently labeled probes (0.5 pmol/μl). Biofilms exposed to probes were incubated at 46°C for 8 h. After probe hybridization, the biofilms were washed with a wash buffer containing 200 mM NaCl, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl (pH 7.2), and 5 mM EDTA.
Evaluation of the spatial distribution of bacteria within biofilm.
Biofilms were visualized by FISH and 4,6-diamino-2-phenylindole (DAPI) staining with a Fluoview FV500 confocal laser scanning microscope (Olympus, Tokyo, Japan). The biofilm structure was analyzed by using a series of horizontal optodigital sections, each 0.62 μm thick, with the intervening gaps between the horizontal sections ranging over the entire height (z axis) of the biofilm. In addition, we analyzed the vertical sections which were recorded from each biofilm to determine spatial distribution. Biofilm cultivation under the same conditions was conducted three times, and six fluorescence images were randomly acquired for each culture at each time point. The percentage of the substratum coverage of each strain and the shortest distance between bacteria were evaluated from at least 18 vertical sections.
Planktonic culture system.
Isolates were individually precultivated to the stationary phase. One microliter of each preculture solution was inoculated into 10 ml of TSB medium with either 2.5 μg/ml or no pyocyanin. These cultures were incubated while being shaken (200 rpm) at 37°C. During 6 days of cultivation, the cells were counted daily after plating on agar plates. A TSB agar plate containing polymyxin (15 μg/ml) was used for counting the cells of gram-positive bacteria. A TSB agar plate containing kanamycin (20 μg/ml) was used for counting the cells of gram-negative bacteria.
Catalase and superoxide dismutase (SOD) activities.
The catalase and SOD activities of the isolates were measured by adapting a previously described method (33). Briefly, the growth of the isolates was monitored spectrophotometrically and 2.5 μg/ml pyocyanin was added to each culture at the onset of the log phase. At the stationary phase, cells were harvested by centrifugation. After the cells were disrupted by sonication on ice, lysate was taken and stored at −80°C. The protein concentration in the cell lysate was measured with a bicinchoninic acid protein assay kit (Pierce, Rockford, IL). Catalase activity was measured spectrophotometrically at 240 nm (22). One unit of catalase activity catalyzes 1 μM H2O2/mg of protein/min at 25°C. SOD activity was determined by the SOD Assay Kit-WST (Dojindo Molecular Technology, Tokyo, Japan). Units of SOD activity were calculated according to the manufacturer's protocol.
Nucleotide sequence accession numbers.
The nucleotide sequences of the seven isolates and three DGGE bands in this study were deposited in the GenBank database under accession numbers AB364957 to AB364963 and AB364947 to AB364949, respectively.
RESULTS
Isolation and characterization of an antibiotic-producing bacterium.
To isolate an antibiotic-producing bacterium capable of biofilm formation, we first introduced the soil extract in a flow cell. After 5 days of cultivation, a biofilm approximately 50 μm thick was directly collected from the flow cell. The bacteria within the biofilm were harvested and isolated by a conventional cultivation method (see Materials and Methods). The antibacterial activity in the supernatant of each isolate was evaluated by measuring the inhibitory zones around test strains on agar plates. Among the isolates, strain P1 exhibited significant inhibitory activity against gram-positive bacteria (B. subtilis and S. aureus) but not against gram-negative bacteria (E. coli and P. aeruginosa) (data not shown). Strain P1 was identified as P. aeruginosa by the 16S rRNA sequence (Table 1). Most P. aeruginosa strains are known to produce a low-molecular-weight antibiotic that is a chloroform-soluble phenazine agent active against gram-positive bacteria and fungi. The chloroform extract fraction of the strain P1 culture supernatant inhibited the growth of gram-positive bacteria as well as the whole culture supernatant did (data not shown). Mass spectroscopy of the chloroform fraction revealed a protonated molecular ion cluster at m/z 211, which was identical to that of pyocyanin reported previously (51).
TABLE 1.
Identification and characteristics of bacteria isolated from a multispecies biofilm
| Strain | GenBank accession no. | Closest relative | 16S rRNA gene identification (%) | MIC of pyocyanin (μg/ml)a | Activity (U/mg protein)b
|
Corresponding DGGE band(s) | |
|---|---|---|---|---|---|---|---|
| Catalase | SOD | ||||||
| P1 | AB364957 | Pseudomonas aeruginosa | 99.8 | a | |||
| R1 | AB364958 | Raoultella ornithinolytica | 99.7 | >20 | 355 ± 12 | 140 ± 7 | b, c |
| R2 | AB364959 | Raoultella planticola | 99.8 | >20 | 289 ± 7 | 93 ± 3 | NDc |
| S1 | AB364960 | Brevibacillus borstelensis | 100 | 0.92 | 22 ± 2 | 18 ± 1 | d |
| S2 | AB364961 | Bacillus licheniformis | 99.7 | 1.01 | 45 ± 5 | 19 ± 5 | ND |
| S3 | AB364962 | Bacillus lentus | 97.0 | 1.31 | 87 ± 6 | 28 ± 3 | ND |
| S4 | AB364963 | Bacillus subtilis | 99.6 | 1.08 | 43 ± 2 | 21 ± 2 | ND |
Each value was obtained by measuring turbidity at 590 nm on a 96-well plate reader.
The unit of activity is defined in Materials and Methods.
ND, not detected.
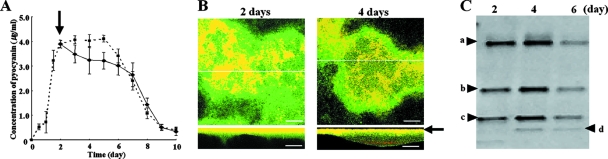
We examined pyocyanin productivity in a single-species biofilm of strain P1 in a flow cell. Approximately 2.5 to 4.0 μg/ml pyocyanin was detected in the outflow solution of 6-day biofilm (Fig. 1A); this concentration range effectively inhibited the growth of gram-positive bacteria (data not shown). During this cultivation period, a formation about 50 μm thick was confirmed by confocal laser scanning microscopy (CLSM) (Fig. 2a and b).
FIG. 1.
Analysis of the multispecies biofilm derived from soil developed on strain P1. (A) Change in pyocyanin concentration in outflow solution. Cultivation was performed for 10 days. Solid and dotted lines indicate pyocyanin concentrations of multispecies biofilm and single-species P1 biofilm, respectively. An arrow indicates the time of inoculation of soil extract into strain P1 biofilm. The data are means of triplicate experiments. Error bars indicate standard deviations. (B) FISH analysis of the mixed-species biofilm in horizontal and vertical sections at 2 and 4 days of cultivation after inoculation. Vertical CLSM images show a cross-section through the x-z dimension at the positions marked by the lines on the horizontal panels. An arrow indicates an abiotic surface. P. aeruginosa strain P1 appears yellow on a green background with the Pae997-Cy3 probe, gram-positive bacteria appear red with LGC-Cy5, and all other bacteria appear green with the EUB338-FITC universal probe. Representative fields of vision are shown. Scale bars = 50 μm. (C) DGGE analysis of the multispecies biofilm at 2, 4, and 6 days of cultivation after soil inoculation. Bands a to d were excised and sequenced. The sequence identities are as follows: a, P. aeruginosa (accession no. AB364949, 100% identical); b and c, R. ornithinolytica (accession no. AB364948, 100% identical); d, B. borstelensis (accession no. AB364947, 100% identical).
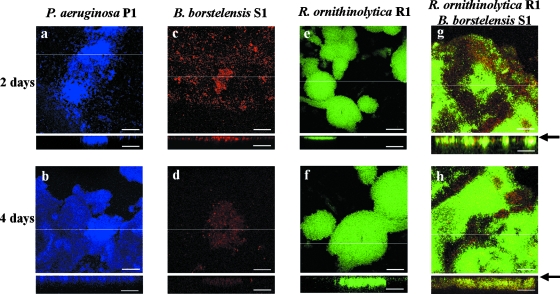
FIG. 2.
FISH analysis of single- and dual-species biofilms on an abiotic surface in horizontal and vertical sections. An arrow indicates an abiotic surface. P. aeruginosa strain P1 (a and b), B. borstelensis strain S1 (c and d), and R. ornithinolytica strain R1 (e and f) were hybridized by the EUB338 probe labeled with Cy5. For dual-species biofilm (g and h), B. borstelensis S1 and R. ornithinolytica R1 were concurrently introduced as 1:1 inocula into a flow cell. B. borstelensis appears red with LGC-Cy5, and R. ornithinolytica R1 appears green with the ENT183-FITC probe. Biofilms were cultivated for 2 or 4 days in a flow cell. Vertical CLSM images show a cross-section through the x-z dimension at the positions marked by the lines on the horizontal panels. Representative fields of vision are shown. Scale bars = 50 μm.
Composition and spatial distribution of multispecies biofilm derived from soil on a pyocyanin-producing strain.
To determine the bacterial species that could coexist with strain P1 within biofilm, a soil extract was inoculated into the preestablished strain P1 biofilm in a flow cell. To exclude the effects of other pyocyanin producers, we selected a soil extract that was different from the source of strain P1. The soil extract did not contain any Pseudomonas species, as confirmed by PCR analysis with the Pseudomonas-specific primers. Furthermore, without the addition of strain P1, pyocyanin was not detected in the flow cell solution after cultivation of the soil extract for 8 days in the flow cell (data not shown).
The pyocyanin concentration in the outflow solution gradually decreased after inoculation of the soil extract at 2 days of cultivation, indicating that the soil extract affected the strain P1 biofilm. However, the concentration was kept high enough to inhibit the growth of gram-positive bacteria until 6 days (Fig. 1A). After 2 or 4 days of cultivation following inoculation of the soil extract, the biofilm was analyzed by FISH with probes Pae997-Cy3 for strain P1, LGC-Cy5 for gram-positive bacteria, and EUB338-FITC for all bacteria (Fig. 1B). P. aeruginosa, appearing in yellow with the Pae997-Cy3 and EUB 338-FITC probes, dominated in the 2-day biofilm, where there was no obvious signal from the LGC-Cy5 probe. However, after 4 days of cultivation, a layer of gram-positive bacteria, shown in red, appeared separately from strain P1 on the abiotic surface. Here, a greenish layer derived from neither P. aeruginosa nor gram-positive bacteria was found between the yellow and red layers. In addition, a red portion was covered by a thin greenish layer. A red signal upon FISH analysis indicated the existence of gram-positive bacteria in the biofilm. The biofilm thickness observed by FISH analysis was the same as that observed by DAPI staining, suggesting that the pretreatment for FISH (e.g., lysozyme, dehydration, and hybridization) had no effect on the biofilm structure. To confirm the bacteria's sensitivity to pyocyanin, bacteria were isolated from the 6-day biofilm with TSB agar plates. In addition to strain P1, six strains were isolated. Designated R1, R2, and S1 to S4, these strains were identified as gram-negative Enterobacteriales and gram-positive Bacillales, respectively, by 16S rRNA analysis (Table 1). The sensitivity of the isolates against pyocyanin derived from strain P1 was determined by MIC analysis (Table 1). The MICs for the isolated Bacillales bacteria (strains S1 to S4) were lower than the pyocyanin concentration in the outflow solution (Fig. 1A). Strains R1 and R2 did not show any growth inhibition at the pyocyanin concentrations tested. Pyocyanin's wide range of antibiotic activity is attributed to its ability to catalyze the generation of toxic radicals such as superoxide and hydrogen peroxide (17). Therefore, the activities of the antioxidation enzymes catalase and SOD were determined (Table 1). Strains R1 and R2 showed high activity levels of both the catalase and SOD enzymes, suggesting that these antioxidant enzymatic activities contributed to the pyocyanin tolerance observed.
DGGE was used to analyze the biofilm after 2, 4, or 6 days of cultivation following soil inoculation on preestablished strain P1 biofilm. The DGGE profile of the 2-day biofilm showed three observable bands (Fig. 1C, bands a to c). After 4 days, a new band (Fig. 1C, band d) appeared and the banding profile at 6 days was identical to that at 4 days. Sequencing analysis of each band indicated that band a was derived from strain P1. Both bands b and c were shown to be derived from a gram-negative bacterium, Raoultella ornithinolytica. As observed by FISH analysis, band d, appearing after 4 days of cultivation, was related to a gram-positive bacterium, Brevibacillus borstelensis. Isolated strains R1 and S1, which were identified as R. ornithinolytica and B. borstelensis, corresponded to DGGE bands b, c, and d, respectively. Fungi were not detected by a PCR analysis with specific primers (data not shown).
Relationships among the three strains and their spatial distribution within biofilm.
Isolated strains P1, S1, and R1 were chosen for an investigation of microbial relationships within biofilm, since the DGGE analysis showed that they were dominant species in the mixed biofilm (Fig. 1C). The three strains were able to individually colonize the abiotic surface during 4 days of cultivation (Fig. 2a to f). First, the effect of the inoculation of each isolate on the preestablished strain P1 biofilm was evaluated by FISH analysis with the specific probe for each strain. Strain R1 colonized on the strain P1 biofilm within 2 days of cultivation (Fig. 3a and b). On the other hand, strain S1 did not develop at all in 4 days of cultivation after inoculation (Fig. 3c and d). To confirm visual inspection of biofilms, we calculated the percentage of the substratum coverage of each strain from visual fields. At 4 days of cultivation following the introduction of strain S1 or R1, the substratum coverage of strain P1 was 100% and 95.2% ± 0.5%, respectively, indicating that the introduction of other bacteria did not disrupt the structure of the preestablished strain P1 biofilm. In addition, the introduction of other bacteria did not affect pyocyanin productivity (data not shown). To confirm pyocyanin's effect on strain S1 behavior, we constructed a pyocyanin-deficient strain by the disruption of one of the pyocyanin biosynthesis genes, phzM, by double-crossover homologous recombination from strain P1 (see Materials and Methods). The constructed mutant strain, designated PHZ201, did not produce pyocyanin, but still its biofilm-forming ability was comparable to that of the wild-type strain (data not shown). Strain S1 was inoculated into the preestablished biofilm of strain PHZ201 (Fig. 3g and h). Strain S1 colonized the biofilm of strain PHZ201 after 2 days of cultivation, suggesting that pyocyanin derived from P. aeruginosa strain P1 inhibited biofilm colonization by B. borstelensis strain S1. Furthermore, the percentage of the substratum coverage of P. aeruginosa strain PHZ201 (61.5% ± 3.8%) was much lower in the biofilm formed by P. aeruginosa strain P1 (100%). On the other hand, pyocyanin production did not affect the growth of R. ornithinolytica strain R1 (Fig. 3e and f).
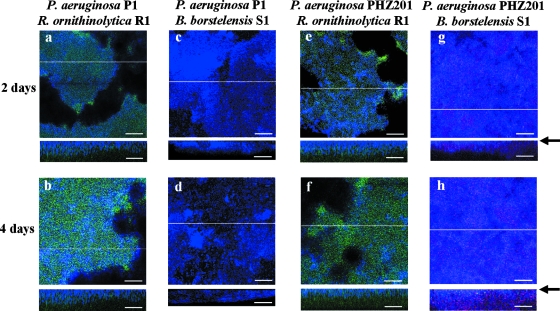
FIG. 3.
FISH analysis of biofilm on P. aeruginosa strain P1 biofilm in horizontal and vertical sections. Strain R. ornithinolytica R1 (panels a and b and panels e and f) or B. borstelensis S1 (panels c and d and panels g and h) was inoculated into a preestablished biofilm of strain P1 (a to d) or PHZ201 (e to h). P. aeruginosa P1 and PHZ201 appear blue with the Pae997-Cy3 probe, B. borstelensis S1 appears red with LGC-Cy5, and R. ornithinolytica R1 appears green with the ENT-FITC probe. Biofilms were cultivated for 2 or 4 days after inoculation into a preestablished biofilm of strain P1 or PHZ201. Vertical CLSM images show a cross-section through the x-z dimension at the positions marked by the lines on the horizontal panels. An arrow indicates an abiotic surface. Representative fields of vision are shown. Scale bars = 50 μm.
Secondly, we evaluated strain R1's effect on the behavior of strain S1. We prepared mixed 1:1 inocula of strains S1 and R1 and introduced them into the preestablished strain P1 biofilm. At 2 days of cultivation following the introduction of the inocula, strain R1 developed on the layer of strain P1, but strain S1 was not clearly detected in the biofilm (Fig. 4a). However, after 4 days of cultivation, the three species were found and the spatial distribution of each was clear (Fig. 4b). The substratum coverage of strain P1 was 89.6% ± 5.7%. Strain S1 was colonized on the layer of strain R1 and did not directly come into contact with strain P1. Strain R1 developed a thick layer and completely covered the strain P1 biofilm. In the mixed-species biofilm containing strain P1, spherical colony formation with strain P1 in the interior and strain R1 on the periphery, which was approximately 200 μm in diameter (Fig. 4), was a consistent feature of the coexistence between pyocyanin-producing and -sensitive bacteria. The average shortest distance between strains P1 and S1 within the biofilm was 30.5 ± 3.5 μm. Strain R1 was also covered with the surface layer of strain S1. This distribution was a consistent feature of the coexistence of strains P1 and S1 and was similar to that seen when the soil sample was used (Fig. 1B). Because the pyocyanin concentration in the outflow solution at 4 days of cultivation was approximately 2.5 μg/ml, which was high enough to inhibit the growth of strain S1, the colonization within the layer of strain R1 was probably necessary for strain S1 to survive in the biofilm. Furthermore, strains S1 and R1 were intermingled in the biofilm without strain P1 at 2 days of cultivation (Fig. 2g and h). In addition, when strains S1 and R1 were inoculated into the preestablished biofilm of strain PHZ201, the surface was incompletely covered with strain PHZ201 (Fig. 4c and d). The substratum coverage of strains PHZ201, R1, and S1 in the biofilm after 4 days was 54.4% ± 4.2%, 18.7% ± 1.2%, and 26.9% ± 3.0%, respectively. These results indicate that pyocyanin produced by a member of the multispecies biofilm determines the spatial distribution of the film's component species.
FIG. 4.
FISH analysis of three-species biofilms in horizontal and vertical sections. P. aeruginosa strains P1 and PHZ201 appear blue with the Pae997-Cy3 probe, B. borstelensis S1 appears red with LGC-Cy5, and R. ornithinolytica R1 appears green with the ENT183-FITC probe. Mixed 1:1 inocula of strains S1 and R1 were inoculated into a preestablished biofilm of strain P1 (a and b) or PHZ201 (c and d). Biofilms were cultivated for 2 or 4 days. Vertical CLSM images show a cross-section through the x-z dimension at the positions marked by the lines on the horizontal panels. An arrow indicates an abiotic surface. Representative fields of vision are shown. Scale bars = 50 μm.
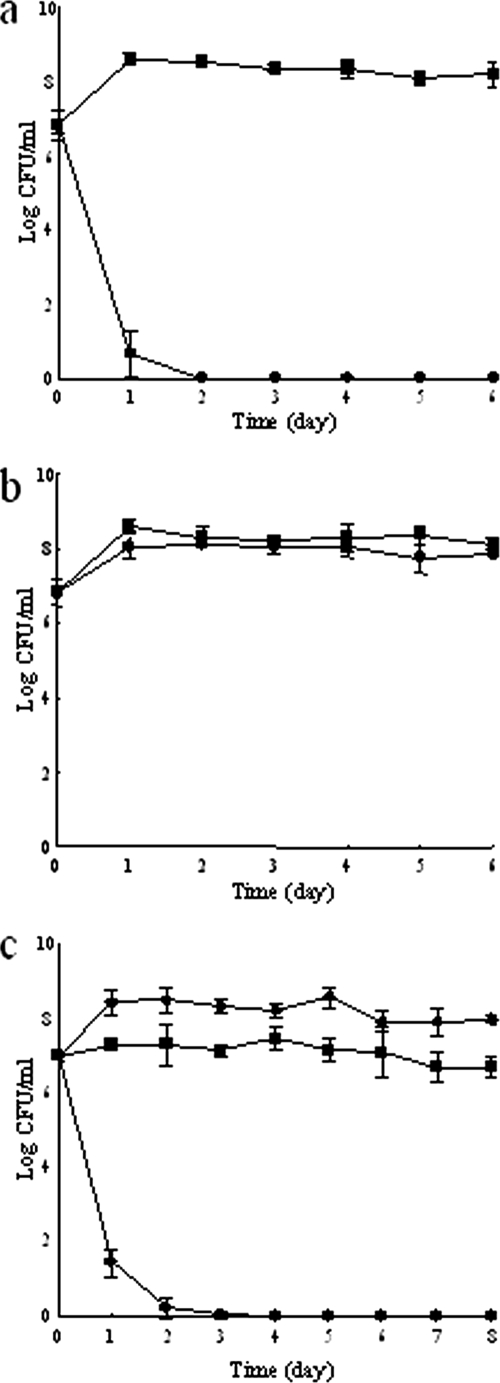
The effect of spatial distribution on the growth of strain S1 was evaluated by a coculture experiment with strains S1 and R1 under planktonic conditions with pyocyanin. Strains S1 and R1 were introduced into TSB medium with 2.5 μg/ml pyocyanin (Fig. 5a) or P. aeruginosa P1 (Fig. 5c). Strain S1 dropped below the detection limit at 2 days of cultivation. This indicates that biofilm development is necessary for the survival of strain S1 in the presence of pyocyanin. Furthermore, the presence of strain R1 did not affect the behavior of strain S1 (Fig. 5b). This indicates that the existence of strain R1 does not help the growth of strain S1 under planktonic conditions with pyocyanin.
FIG. 5.
Community dynamics in a shaken test tube. B. borstelensis strain S1 (•) and R. ornithinolytica strain R1 (▪) were concurrently inoculated into a test tube containing TSB medium with (a) and without (b) pyocyanin. P. aeruginosa strain P1 (⧫), B. borstelensis S1 (•), and R. ornithinolytica R1 (▪) were concurrently inoculated into a test tube containing TSB medium (c). The cells of the two species were counted every day after plating on agar plates. The results are expressed as means ± standard deviations of three independent experiments.
DISCUSSION
Niche differentiation is often the basis of the coexistence of multiple species in plant and animal ecology. Recent experimental studies with laboratory cultures have shown that this is also true for microbial species in spatially restricted environments. For example, three strains of colicinogenic, colicin-sensitive, and colicin-resistant E. coli create a spatial distribution on a petri dish, resulting in the strain's stable coexistence (24). In another example, it has been suggested that the coexistence of epiphytic bacteria on a leaf surface could be achieved through the spatial separation of individual microcolonies (29). It seems likely that the opportunities for interaction among the microcolonies are limited to only a few sites on the leaf surface. Unlike those environments, strong interactions among the component bacterial species are expected in microenvironments inside biofilms. In addition, heterogeneous environments are often observed in a biofilm (46, 53). Chemical gradients of nutrients, oxygen, metabolites, and/or inhibitory substances have been determined (8, 32, 35, 46, 53). This has been considered a major factor in the coexistence of multiple species in a biofilm (27, 38, 44, 45). In this study, we showed that the survival of sensitive bacteria in biofilm was achieved by shielding them from the pyocyanin producer and the pyocyanin-containing medium by inserting a layer of resistant bacteria between them (Fig. 1B and 4b). Thus, this spatial distribution would provide a niche where the concentration of the inhibitory substance was low in biofilm containing the inhibitor producer, resulting in the coexistence of pyocyanin-producing and -sensitive bacterial species in a single microenvironment. Previous research indicates that high densities of cells within microcolonies enhance persistence or resistance against superior competitors (46). However, such a mechanism did not seem applicable to this work, because the sensitive strain was introduced into the preestablished biofilm of the antibiotic-producing strain.
It is believed that pyocyanin's antibiotic activity occurs through the formation of reactive oxygen species (ROS) such as superoxide and hydrogen peroxide. We used hydrorhodamine 123 (Sigma Chemical Co., St. Louis, MO) to test for the presence of ROS in situ. ROS were detected in the three-species biofilm containing P. aeruginosa P1 but not when pyocyanin-deficient strain PHZ201 was used (see Fig. S1 in the supplemental material), indicating that pyocyanin produced by strain P1 caused the generation of ROS within the biofilm. Antioxidant enzymes SOD and catalase are known to be involved in resistance to pyocyanin (17). In fact, the pyocyanin-resistant strains isolated from the multispecies biofilm in this work had relatively high SOD and catalase enzymatic activity levels, while the sensitive strains had low levels (Table 1). The ability of the pyocyanin-resistant strains to coexist with pyocyanin-producing strain P1 might be attributable to those antioxidant activities. Previous studies of dual-species biofilms indicate that antioxidant enzymes derived from antibiotic-resistant bacteria contribute to the enhanced viability of sensitive bacteria in the presence of antibiotics (34). Thus, the scavenging of pyocyanin-elicited oxidative stress by the resistant strains might be a major reason for the survival of the sensitive bacteria that were sequestered into the layer of resistant bacteria. Another possible reason is an increased biofilm matrix viscosity, which would reduce the permeation of antibiotics. Raoultella (formerly Klebsiella) is known to produce high amounts of exopolysaccharide in the biofilm state (40). Another study suggested that interactions between the different matrix polymers result in a more viscous matrix in multispecies biofilm (49). One subject for future research is the matrix viscosity in the multispecies biofilm used in the present study.
Antibiotics are believed to have helped their producers to dominate their habitats by inhibiting the growth of other sensitive bacteria (for reviews, see references 12 and 37). Pyocyanin production was necessary for the development of the stratified structure shown in Fig. 4. This is the first report that an endogenous antibiotic plays a key role in niche determination of multiple bacterial species in a biofilm. Pyocyanin production also seemed to suppress the dispersion of strain P1 in the three-species mixed cultures (Fig. 4b and d). Thus, strain S1 in the presence of pyocyanin would be effective for maintaining strain P1 on an abiotic surface and for protecting strain P1 from grazing, desiccation, and other stresses. We propose that the provision of such advantages is a novel function of antibiotics in multispecies biofilm.
This work shows that the presence of pyocyanin-resistant bacteria is necessary for the coexistence of pyocyanin-producing and -sensitive bacteria in a multispecies biofilm. The existence of strain P1 had an effect on the layer distribution between strains R1 and S1 because the two strains were intermingled in the dual-species biofilm without strain P1 (Fig. 2g and h). Interestingly, a clear boundary was observed between pyocyanin-producing strain P1 and the resistant strains in multispecies biofilm that included sensitive bacteria (Fig. 1B and 4b), whereas the dual-species biofilm had a fuzzy boundary between strains P1 and R1 (Fig. 3b). These results indicate that the pyocyanin-sensitive bacteria also affect the interaction between pyocyanin-producing and -resistant bacteria. The detailed mechanisms underlying the physical and chemical interactions at work among pyocyanin-producing, -sensitive, and -resistant bacteria in multispecies biofilms remain to be elucidated, and studies to this end are in progress. The spatial distribution observed in this work may represent the notable role of antibiotic production in the maintenance of species diversity under biofilm conditions.
Supplementary Material
Acknowledgments
We thank Naoya Ohmura (Central Research Institute of the Electric Power Industry) for help with the identification of antibiotics.
Footnotes
Published ahead of print on 25 April 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, R. Schulze, S. Spring, E. Moore, and K.-H. Schleifer. 1996. rRNA-targeted oligonucleotide probes for the identification of genuine and former pseudomonads. Syst. Appl. Microbiol. 19:501-509. [Google Scholar]
- 3.Arai, H., M. Hayashi, A. Kuroi, M. Ishii, and Y. Igarashi. 2005. Transcriptional regulation of the flavohemoglobin gene for aerobic nitric oxide detoxification by the second nitric oxide-responsive regulator of Pseudomonas aeruginosa. J. Bacteriol. 187:3960-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram, R., M. Schlicht, K. Mahr, H. Nothaft, M. H. Saier, and F. Titgemeyer. 2004. In silico and transcriptional analysis of carbohydrate uptake systems of Streptomyces coelicolor A3(2). J. Bacteriol. 186:1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgess, J. G., E. M. Jordan, M. Bregu, A. Mearns-Spragg, and K. G. Boyd. 1999. Microbial antagonism: a neglected avenue of natural products research. J. Biotechnol. 70:27-32. [DOI] [PubMed] [Google Scholar]
- 6.Burmølle, M., J. S. Webb, D. Rao, L. H. Hansen, S. J. Sørensen, and S. Kjelleberg. 2006. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 72:3916-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burmølle, M., L. H. Hansen, and S. J. Sørensen. 2007. Establishment and early succession of a multispecies biofilm composed of soil bacteria. Microb. Ecol. 54:352-362. [DOI] [PubMed] [Google Scholar]
- 8.Cowan, S. E., E. Gilbert, D. Liepmann, and J. D. Keasling. 2000. Commensal interactions in a dual-species biofilm exposed to mixed organic compounds. Appl. Environ. Microbiol. 66:4481-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, J. 2006. Are antibiotics naturally antibiotics? J. Ind. Microbiol. Biotechnol. 33:496-499. [DOI] [PubMed] [Google Scholar]
- 10.Devereux, R., and S. S. Wilkinson. 2004. Amplification of ribosomal RNA sequences, p. 509-522. In G. A. Kowalchuk, F. J. de Brujin, I. M. Head, A. D. L. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 11.Essar, D. W., L. Eberly, A. Hadero, and I. P. Crawford. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firn, R. D., and C. G. Jones. 2003. Natural products—a simple model to explain chemical diversity. Nat. Prod. Rep. 20:382-391. [DOI] [PubMed] [Google Scholar]
- 13.Gans, J., M. Wolinsky, and J. Dunbar. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387-1390. [DOI] [PubMed] [Google Scholar]
- 14.Haas, D., and G. Défago. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3:307-319. [DOI] [PubMed] [Google Scholar]
- 15.Haruta, S., Z. Cui, Z. Huang, M. Li, M. Ishii, and Y. Igarashi. 2002. Construction of a stable microbial community with high cellulose-degradation ability. Appl. Microbiol. Biotechnol. 59:529-534. [DOI] [PubMed] [Google Scholar]
- 16.Haruta, S., S. Ueno, I. Egawa, K. Hashiguchi, A. Fujii, M. Nagano, M. Ishii, and Y. Igarashi. 2006. Succession of bacterial and fungal communities during a traditional pot fermentation of rice vinegar assessed by PCR-mediated denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 109:79-87. [DOI] [PubMed] [Google Scholar]
- 17.Hassan, H. M., and I. Fridovich. 1980. Mechanism of the antibiotic action of pyocyanine. J. Bacteriol. 141:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobley, L., J. R. King, and E. Sockett. 2006. Bdellovibrio predation in the presence of decoys: three-way bacterial interactions revealed by mathematical and experimental analyses. Appl. Environ. Microbiol. 72:6757-6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huddleston, A. S., N. Cresswell, M. C. Neves, J. E. Beringer, S. Baumberg, D. I. Thomas, and E. M. Wellington. 1997. Molecular detection of streptomycin-producing streptomycetes in Brazilian soils. Appl. Environ. Microbiol. 63:1288-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, S., S. Haruta, Z. J. Cui, M. Ishii, and Y. Igarashi. 2004. Effective cellulose degradation by a mixed-culture system composed of a cellulolytic Clostridium and aerobic non-cellulolytic bacteria. FEMS Microbiol. Ecol. 51:133-142. [DOI] [PubMed] [Google Scholar]
- 21.Kato, S., S. Haruta, Z. J. Cui, M. Ishii, and Y. Igarashi. 2005. Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl. Environ. Microbiol. 71:7099-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katsuwon, J., and A. J. Anderson. 1989. Response of plant-colonizing pseudomonads to hydrogen peroxide. Appl. Environ. Microbiol. 55:2985-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kempf, V. A., K. Trebesius, and I. B. Autenrieth. 2000. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38:830-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerr, B., M. A. Riley, M. W. Feldman, and B. J. Bohannan. 2002. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171-174. [DOI] [PubMed] [Google Scholar]
- 25.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187:7193-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurachi, M. 1958. Studies on the biosynthesis of pyocyanine. II. Isolation and determination of pyocyanine. Bull. Inst. Chem. Res. Kyoto Univ. 36:174-187. [Google Scholar]
- 27.Leriche, V., R. Briandet, and B. Carpentier. 2003. Ecology of mixed biofilms subjected daily to a chlorinated alkaline solution: spatial distribution of bacterial species suggests a protective effect of one species to another. Environ. Microbiol. 5:64-71. [DOI] [PubMed] [Google Scholar]
- 28.Meier, H., R. Amann, W. Ludwig, and K.-H. Schleifer. 1999. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst. Appl. Microbiol. 22:186-196. [DOI] [PubMed] [Google Scholar]
- 29.Monier, J. M., and S. E. Lindow. 2005. Spatial organization of dual-species bacterial aggregates on leaf surfaces. Appl. Environ. Microbiol. 71:5484-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 32.Nielsen, A. T., T. Tolker-Nielsen, K. B. Barken, and S. Molin. 2000. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ. Microbiol. 2:59-68. [DOI] [PubMed] [Google Scholar]
- 33.Norman, R. S., P. Moeller, T. J. McDonald, and P. J. Morris. 2004. Effect of pyocyanin on a crude-oil-degrading microbial community. Appl. Environ. Microbiol. 70:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connell, H. A., G. S. Kottkamp, J. L. Eppelbaum, B. A. Stubblefield, S. E. Gilbert, and E. S. Gilbert. 2006. Influences of biofilm structure and antibiotic resistance mechanisms on indirect pathogenicity in a model polymicrobial biofilm. Appl. Environ. Microbiol. 72:5013-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okabe, S., T. Ito, K. Sugita, and H. Satoh. 2005. Succession of internal sulfur cycles and sulfur-oxidizing bacterial communities in microaerophilic wastewater biofilms. Appl. Environ. Microbiol. 71:2520-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pesaro, M., and F. Widmer. 2006. Identification and specific detection of a novel Pseudomonadaceae cluster associated with soils from winter wheat plots of a long-term agricultural field experiment. Appl. Environ. Microbiol. 72:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price-Whelan, A., L. E. P. Dietrich, and D. K. Newman. 2006. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2:71-78. [DOI] [PubMed] [Google Scholar]
- 38.Rao, D., J. S. Webb, and S. Kjelleberg. 2005. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 71:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao, D., J. S. Webb, and S. Kjelleberg. 2006. Microbial colonization and competition on the marine alga Ulva australis. Appl. Environ. Microbiol. 72:5547-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rättö, M., R. Verhoef, M. L. Suihko, A. Blanco, H. A. Schols, A. G. Voragen, R. Wilting, M. Siika-Aho, and J. Buchert. 2006. Colanic acid is an exopolysaccharide common to many enterobacteria isolated from paper-machine slimes. J. Ind. Microbiol. Biotechnol. 33:359-367. [DOI] [PubMed] [Google Scholar]
- 41.Riley, M. A., and D. M. Gordon. 1999. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 7:129-133. [DOI] [PubMed] [Google Scholar]
- 42.Roberson, E. B., and M. K. Firestone. 1992. Relationship between desiccation and exopolysaccharide production in a soil Pseudomonas sp. Appl. Environ. Microbiol. 58:1284-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slattery, M., I. Rajbhandari, and K. Wesson. 2001. Competition-mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microb. Ecol. 41:90-96. [DOI] [PubMed] [Google Scholar]
- 44.Stewart, P. S., A. K. Camper, S. D. Handran, C. Huang, and M. Warnecke. 1997. Spatial distribution and coexistence of Raoultella pneumoniae and Pseudomonas aeruginosa in biofilms. Microb. Ecol. 33:2-10. [DOI] [PubMed] [Google Scholar]
- 45.Tait, K., and I. W. Sutherland. 2002. Antagonistic interactions amongst bacteriocin-producing enteric bacteria in dual species biofilms. J. Appl. Microbiol. 93:345-352. [DOI] [PubMed] [Google Scholar]
- 46.Teal, T. K., D. P. Lies, B. J. Wold, and D. K. Newman. 2006. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl. Environ. Microbiol. 72:7324-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tong, H., W. Chen, J. Merritt, F. Qi, W. Shi, and X. Dong. 2007. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol. Microbiol. 63:872-880. [DOI] [PubMed] [Google Scholar]
- 48.Torsvik, V., J. Goksöyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Von Canstein, H., S. Kelly, Y. Li, and I. Wagner-Döbler. 2002. Species diversity improves the efficiency of mercury-reducing biofilms under changing environmental conditions. Appl. Environ. Microbiol. 68:2829-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker, T. S., H. P. Bais, E. Déziel, H. P. Schweizer, L. G. Rahme, R. Fall, and J. M. Vivanco. 2004. Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 134:320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson, D., J. MacDermot, R. Wilson, P. J. Cole, and G. W. Taylor. 1986. Purification and structural analysis of pyocyanin and 1-hydroxyphenazine. Eur. J. Biochem. 159:309-313. [DOI] [PubMed] [Google Scholar]
- 52.Wawrik, B., L. Kerkhof, G. J. Zylstra, and J. J. Kukor. 2005. Identification of unique type II polyketide synthase genes in soil. Appl. Environ. Microbiol. 71:2232-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Werner, E., F. Roe, A. Bugnicourt, M. J. Franklin, A. Heydorn, S. Molin, B. Pitts, and P. S. Stewart. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White, T. J., T. Burns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and application. Academic Press, San Diego, CA.
- 55.Widmer, F., R. J. Seidler, P. M. Gillevet, L. S. Watrud, and G. D. Di Giovanni. 1998. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl. Environ. Microbiol. 64:2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan, L., K. G. Boyd, D. R. Adams, and J. G. Burgess. 2003. Biofilm-specific cross-species induction of antimicrobial compounds in bacilli. Appl. Environ. Microbiol. 69:3719-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, H., F. Qu, and L. H. Zhu. 1993. Isolation of genomic DNAs from plants, fungi and bacteria with benzyl chloride. Nucleic Acids Res. 21:5279-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.