Abstract
Rhodococcus sp. strain HA01, isolated through its ability to utilize dibenzofuran (DBF) as the sole carbon and energy source, was also capable, albeit with low activity, of transforming dibenzo-p-dioxin (DD). This strain could also transform 3-chlorodibenzofuran (3CDBF), mainly by angular oxygenation at the ether bond-carrying carbon (the angular position) and an adjacent carbon atom, to 4-chlorosalicylate as the end product. Similarly, 2-chlorodibenzofuran (2CDBF) was transformed to 5-chlorosalicylate. However, lateral oxygenation at the 3,4-positions was also observed and yielded the novel product 2-chloro-3,4-dihydro-3,4-dihydroxydibenzofuran. Two gene clusters encoding enzymes for angular oxygenation (dfdA1A2A3A4 and dbfA1A2) were isolated, and expression of both was observed during growth on DBF. Heterologous expression revealed that both oxygenase systems catalyze angular oxygenation of DBF and DD but exhibited complementary substrate specificity with respect to CDBF transformation. While DfdA1A2A3A4 oxygenase, with high similarity to DfdA1A2A3A4 oxygenase from Terrabacter sp. strain YK3, transforms 3CDBF by angular dioxygenation at a rate of 29% ± 4% that of DBF, 2CDBF was not transformed. In contrast, DbfA1A2 oxygenase, with high similarity to the DbfA1A2 oxygenase from Terrabacter sp. strain DBF63, exhibited complementary activity with angular oxygenase activity against 2CDBF but negligible activity against 3CDBF. Thus, Rhodococcus sp. strain HA01 constitutes the first described example of a bacterial strain where coexpression of two angular dioxygenases was observed. Such complementary activity allows for the efficient transformation of chlorinated DBFs.
Biarylethers, comprising dibenzo-p-dioxin (DD), dibenzofuran (DBF), diphenyl ether, and their halogenated derivatives, are widespread environmental pollutants. Polychlorinated DDs and DBFs, the contaminating by-products formed during the manufacture of pesticides or the incineration of industrial and domestic wastes, can cause a wide range of serious health effects (1, 4, 13).
Microorganisms play important roles in the degradation and mineralization of xenobiotic and aromatic compounds in natural environments. Their aerobic degradation is frequently initiated by Rieske nonheme iron oxygenases which catalyze the incorporation of two oxygen atoms into the aromatic ring to form arene cis-diols (15). This is then followed by a dehydrogenation reaction catalyzed by a cis-dihydrodiol dehydrogenase to give catechol or substituted catechols, which serve as substrates for oxygenolytic aromatic ring cleavage. Rieske nonheme iron oxygenases are typically composed of a terminal oxygenase (iron-sulfur protein) and different electron transport proteins (8). The catalytic iron-sulfur proteins are homo- or heteromultimers with the α-subunit containing a Rieske-type [2Fe-2S] cluster, a mononuclear nonheme iron oxygen activation center, and a substrate-binding site (8) that is responsible for substrate specificity (15). Comparison of the amino acid sequences of the α-subunits has revealed that they form a family of diverse but evolutionarily related sequences. Distinct major lineages have been identified (15), and although none of the enzymes are completely specific, a broad correlation between the grouping in toluene/biphenyl, naphthalene, benzoate, or phthalate subfamilies and their native substrates has been observed.
Naphthalene- or biphenyl-degrading Pseudomonas or Sphingomonas strains transform DBF, DD, and chlorinated derivatives into dead-end products (9, 30, 31). These substrates are attacked at the lateral 1,2- and 2,3-positions to give dihydrodiols, which are subsequently dehydrogenated to dihydroxy compounds and in some cases undergo ring cleavage in a manner that is analogous to biphenyl or naphthalene transformation. While such lateral dioxygenation is appropriate to initiate degradation of biphenyl and naphthalene, various authors have shown that it is inappropriate for the degradation of biarylethers (5, 45). It was thus argued that cleavage of the ether bridge is critical for the degradation of biarylethers (12), and analysis of Sphingomonas wittichii RW1 indicated that DBF degradation proceeds via initial dioxygenation at the ether bond-carrying carbon (the angular position) and an adjacent carbon atom (angular dioxygenation), resulting in the formation of a highly unstable hemiacetal, which after spontaneous cleavage and rearomatization gives rise to 2,2′,3-trihydroxybiphenyl (THB) (7). THB is subject to meta cleavage and subsequent hydrolysis to give salicylate, analogous to the transformation of 2,3-dihydroxybiphenyl (DHB) to benzoate by biphenyl-degrading bacteria. Angular dioxygenation was also shown to be the initial basic step in the degradation of other biarylether compounds (11).
To date, a large number of bacterial strains capable of degrading biarylethers have been isolated and characterized. Phylogenetically, almost all of these belong to the phyla Proteobacteria and Actinobacteria (predominantly Janibacter strains) (23, 37, 60), and in all strains so far analyzed, degradation of DBF is initiated by angular dioxygenation. Interestingly, like the α-subunit of dioxin dioxygenase from S. wittichii RW1, the DbfA1 α-subunit of DBF dioxygenases from Terrabacter sp. strain DBF63 (28) and Paenibacillus sp. strain YK5 (24), as well as the DfdA1 DBF dioxygenase from Terrabacter sp. strain YK3 (22) and carbazole 1,9a-dioxygenase from Pseudomonas resinovorans strain CA10 (42), all represent novel branches in the phylogeny of α-subunits of Rieske nonheme iron oxygenases.
In contrast to the immense amount of information available for transformation of polychlorinated biphenyl congeners by biphenyl dioxygenases, information on the substrate range of angular dioxygenases regarding the transformation of chlorinated substrates remains scarce. S. wittichii RW1 is capable of transforming several mono- and dichlorinated DDs and DBFs (55), whereas highly chlorinated congeners are rather recalcitrant. Currently, all strains harboring angular dioxygenases share the capability of transforming 2-chlorodibenzofuran (2CDBF) and 2-chlorodibenzo-p-dioxin (14, 16, 55), although differences are observed both in the regioselectivity of attack and substrate specificity. Additionally, Sphingomonas sp. strain HH69, S. wittichii RW1, and Sphingomonas sp. strain RW16 are capable of transforming 3-chlorodibenzofuran (3CDBF) (19, 55, 58); however, such a transformation by angular dioxygenases of gram-positive bacteria has yet to be described. In this study, we identify two angular dioxygenases in Rhodococcus sp. strain HA01 and characterize their regioselectivity of attack on chlorinated DBFs and substrate specificity.
MATERIALS AND METHODS
Bacterial strains and isolates and culture conditions.
Escherichia coli cells were cultured in LB medium at 37°C containing the appropriate selection markers, and Rhodococcus sp. strain ATCC 12674 was cultured in GYM Streptomyces medium (4 g glucose, 4 g yeast extract, and 10 g malt extract per liter) at 30°C. Rhodococcus sp. strain HA01 was cultured at 30°C in mineral medium supplemented with the appropriate carbon sources at concentrations of 2 mM. DBF was added from a 100 mM filter-sterilized stock solution in dimethyl sulfoxide (DMSO), and incubation was performed in sealed, screw-capped Erlenmeyer flasks to avoid evaporation. Growth was monitored by measuring turbidity (A600).
DBF-degrading bacteria were isolated from soil samples collected in areas surrounding chemical-, insecticide-, and pesticide-producing factories in Kafr El Ziat, Egypt. Soil (1 g) was incubated in a 1-liter Erlenmeyer flask containing 100 ml of mineral medium (10) with DBF (2 mM) as the sole carbon and energy source. Following 1 month of cultivation at 30°C, 10% of the culture was transferred to fresh medium and cultured for a further month. Dilutions of the culture were spread onto minimal medium agar plates supplemented with DBF as crystals in the lid of the agar plates and incubated for 7 days after which colonies were sprayed with filter-sterilized DHB (10 mM). Yellow colonies due to extradiol cleavage of DHB were purified on minimal medium agar plates with DBF as the sole carbon source. One predominant colony morphotype, which grew rapidly at 30°C and exhibited a slightly red color, was selected for further studies.
Bacteria, vectors, media, and culture conditions.
Most chemicals used in this study were obtained from Sigma and Aldrich and were of the highest grade available. DHB was obtained from Wako Chemicals GmbH. THB and 2,2′,3-trihydroxybiphenyl ether (THBE) were available from previous syntheses (2). 3CDBF was kindly provided by Stefan Schmidt, Biocenter Klein Flottbek, University of Hamburg. 2CDBF was obtained from the Sigma-Aldrich library of rare chemicals, and DD was from Promochem GmbH. Restriction enzymes and reagents for molecular experiments were purchased from New England Biolabs, Boehringer Mannheim, Promega, Qiagen, Q-Biogene, United States Biochemicals, and Sigma. Bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Sphingomonas wittichii RW1 | Dibenzofuran-utilizing bacterium | DSMZa |
| Rhodococcus sp. strain ATCC 12674 | Bacterium incapable of utilizing dibenzofuran | DSMZb |
| Rhodococcus sp. strain HA01 | Dibenzofuran-utilizing bacterium | This study |
| E. coli BL21(DE3)(pT7-5RW) | pT7-5 with 1.4-kb PstI/SalI insert carrying dbfB gene of S. wittichii RW1 | 18 |
| E. coli JM109 | recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) e14− (mcrA) supE44 relA1 Δ(lac-proAB)/F′ [traD36 proAB+ lacIqlacZΔM15] | Stratagene |
| Plasmids | ||
| pGEM-T Easy | Cloning vector | Promega |
| pUC119 | AprlacZ; pMB9 replicon | Takara Shuzo Co. |
| pRSG43 | E. coli-Rhodococcus shuttle vector; Rhodococcus rhodochrous cryptic plasmid pRC4 integrated into the AflIII site of pHSG299 | Masahiro Takeo |
| pDFDE | Apr; pUC119 with 4.4-kb EcoRI/PstI insert carrying dfdA1A2A3A4 genes of strain HA01 | This study |
| pDFDR | Kmr; pRSG43 with 4.4-kb EcoRI/PstI insert carrying dfdA1A2A3A4 genes of strain HA01 | This study |
| pDBFA12a | Apr; pUC119 with 3-kb HindIII/EcoRI insert carrying dbfA1A2 genes of strain HA01 | This study |
| pDBFA12 | Kmr; pRSG43 with 3-kb XbaI/EcoRI insert carrying dbfA1A2 genes of strain HA01 | This study |
DSMZ, Deutsche Sammlung von Mikoorganismen und Zellkulturen, Braunschweig, Germany. This strain was DSMZ 6014 in the DSMZ culture collection.
This strain was DSMZ 43287 in the DSMZ culture collection.
Transformation of substrates.
To quantify growth rate and substrate uptake, Rhodococcus sp. strain HA01 was grown as described above. Harvested cells were inoculated (A600 of 0.1) into 10-ml glass tubes containing 2 ml mineral medium and DBF as the sole carbon source. To estimate the number of CFU, aliquots were serially diluted onto solid LB medium and incubated for 48 h. To measure substrate depletion, 1 ml of culture was supplemented with 8 ml methanol for extraction of residual DBF from cell walls and to achieve complete dissolution. Samples were centrifuged, and the supernatant was analyzed by reverse-phase high-performance liquid chromatography (HPLC). Noninoculated tubes and tubes without substrate (with DMSO only) served as controls.
To quantify the transformation rates and identify the metabolites, Rhodococcus sp. strain HA01 was grown in minimal medium with DBF (2 mM) or fructose (2 mM) as the sole carbon sources as described above, whereas Rhodococcus sp. strain ATCC 12674 derivatives were grown in GYM Streptomyces medium supplemented with kanamycin (100 μg ml−1). E. coli JM109 derivatives were cultured in LB medium supplemented with the appropriate antibiotic. If necessary, isopropyl-thio-β-d-galactopyranoside (IPTG) (0.5 mM) was added when cultures reached an A600 of 0.6. Cells were harvested during late exponential growth by centrifugation, washed twice with 50 mM phosphate buffer (pH 7.4), and resuspended to an A600 of 5 to 20. Cells were harvested as described above for Rhodococcus sp. strain HA01 when the culture reached an A600 of 1.
Harvested cells were incubated with DBF, DD, 2CDBF, or 3CDBF supplied at a concentration of 0.2 to 2 mM in aliquots of 1 ml in 10-ml reagent tubes closed with Teflon-coated screw caps and incubated at 30°C on an overhead shaker. In the case of transformation by E. coli JM109(pDBF12), transformation mixtures were supplemented with glucose (10 mM). At appropriate time intervals, tubes were analyzed for residual substrate as described above. For quantification of transformation products, cell-free supernatants were directly analyzed by HPLC. Rates were calculated by quantifying substrate depletion or product accumulation, in case authentic standards were available. Product formation rate from 2CDBF by E. coli JM109(pDBF12) was calculated assuming that the product exhibited absorption characteristics similar to those of THB.
Specific activities are expressed as micromoles of substrate depleted or product formed per minute and gram of cell weight (dry weight) (U/g [dry weight]). Dry weight was determined for exponentially growing cells, and it was calculated that a turbidity of A600 of 1 corresponds to 0.208 ± 0.010 g/liter of Rhodococcus sp. strain HA01, 0.214 ± 0.011 g/liter of Rhodococcus sp. strain ATCC 12674, and 0.295 ± 0.015 g (dry weight) per liter of E. coli.
To inactivate extradiol dioxygenases and thus accumulate and identify ring cleavage intermediates during transformation of biarylethers, resting cells of Rhodococcus sp. strain HA01 were incubated with the respective substrates as described above in the presence of 3-chlorocatechol (0.1 mM) (56).
To characterize the product formed after transformation of 3CDBF by resting cells, the reaction mixture was supplemented with an aliquot of cell extract of E. coli BL21(DE3)(pT7-5RW) overexpressing DbfB, a 2,2′,3-trihydroxybiphenyl dioxygenase from S. wittichii RW1 (50 mU/ml [activity determined with DHB]) (18).
Preparation of cell extracts.
Cell extracts were prepared as previously described (39). Protein concentrations were determined by the Bradford method (6). Supernatant fluids were subjected to sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis (35), and gels were stained in 1% (wt/vol) Coomassie brilliant blue G-250 (47). Dilutions were designed so that the amount of protein loaded ranged from 1 to 30 μg. PageRuler unstained protein ladder (Fermentas) was used to evaluate the molecular weights of the denatured protein subunits.
Analytical methods.
Depletion of biarylethers was monitored by HPLC analysis as previously described (59) using an aqueous solvent system (flow rate, 1 ml/min) containing 0.01% (vol/vol) H3PO4 (87%) and 80% (vol/vol) methanol. Product accumulation was quantified using 60% (vol/vol) methanol. HPLC-mass spectrometry (HPLC-MS) was performed as previously described (59).
The one-dimensional 1H nuclear magnetic resonance (NMR) spectra were recorded as previously described (59). For respective transformation experiments, the substrates were added from a 100 mM stock solution in deuterated DMSO in order to avoid interference from protonated DMSO-derived signals.
DNA isolation.
Genomic DNA of Rhodococcus sp. was extracted with the FastDNA spin kit for soil (Q-Biogene). DNA from recombinant E. coli strains was obtained by dissolving colonies in 50 μl water followed by boiling for 10 min (27). Supernatant obtained after centrifugation was used as template for PCRs.
PCR amplification of 16S rRNA genes.
The phylogenetic relationship of the isolate was derived using the nearly complete sequence of the 16S rRNA genes (corresponding to positions 15 to 1480 in the Escherichia coli numbering system) determined using primers and conditions described by Lane (36).
PCR amplification of Rieske nonheme iron oxygenase encoding genes.
Part of the dfdA1 gene of Rhodococcus sp. strain HA01 was amplified with RieskeF and RieskeR primers (28). A 2.5-kb fragment was obtained by PCR with the DfdDOF1 forward primer (5′-AGGTCTGCCGCGCCGACTGG-3′) designed based on the obtained sequence and the DfdDOR1 reverse primer (5′-GAYAGMGGBGGKCGYTSRTASGG-3′) designed based on conserved amino acid sequences of ferredoxin reductases of strain RW1, Rhodococcus sp. strain M5, Rhodococcus erythropolis TA421, and Rhodococcus globerulus P6. Specific forward and reverse primers (DfdDOF2 [5′-TCCACGACAGCTCGGTCCTGC-3′] and DfdDOR2 [5′-CCGGTGCGCAAGTTGAACTTA-3′], respectively) annealing at the borders of the obtained sequence were designed in order to better resolve the inner sequence, and additional primers were successively designed to obtain the complete sequence of the fragment on both strands. Amplification of the complete dfdA1A2A3A4 gene cluster of Rhodococcus sp. strain HA01 (4.4 kb) was achieved using primers DfdF546 (5′-GGGGGGATATTTGGCCTCACCC-3′) and DfdR4900 (5′-TGCGGGTGGACCGGGCGACG-3′). Several oligonucleotides were designed to perform the primer walking and sequence assemblage.
To localize dbfA genes, primers Dxn1F (5′-TGYASNTAYCAYGGVTGG-3′) and Dxn2R (5′-TGYASNTAYCAYGGVTGG-3′) were designed to detect and amplify a 700-bp fragment encoding the α-subunit of the dioxin dioxygenase of S. wittichii RW1 and were modified with degenerate positions to integrate the codon usage of the relatively scarce amino acid sequence stretches this sequence has in common with its closest relatives Rhodococcus sp. strain M5 bpdC1, Rhodococcus erythropolis TA421 bphA, and Rhodococcus globerulus P6 bphA. Amplification and sequencing primers were designed based on the angular dioxygenase encoding gene region from Terrabacter sp. strain DBF63.
The PCR products were cleaned using the Qiaquick PCR cleaning kit (Qiagen) and cloned in the pGEM-T system (Promega), and the ligation products were transformed in E. coli JM109 competent cells (Promega).
DNA sequencing and homology research.
Purified PCR products were sequenced using an ABI PRISM BigDye terminator v1.1 ready reaction cycle sequencing kit (Applied Biosystems) and an ABI PRISM 3100 genetic analyzer (Applied Biosystems). Primers used for sequencing reactions were the same as those used in the original PCR, except in the case of inserts in the pGEM-T easy vector (Promega), which were amplified and sequenced with vector-specific M13 forward and M13 reverse primers (47). Raw sequence chromatograms from both strands were assembled with Sequencher software version 4.0.5 (Gene Codes Corporation). Assembled contigs were used for DNA and protein similarity searches on GenBank databases performed with BlastN and BlastP programs of the National Center for Biotechnology Information website. 16S rRNA gene nucleotide sequences were aligned using ClustalW implemented in MEGA software version 3.1 (33). Nucleotide sequences of aromatic dioxygenases were translated and aligned with the program functions available in the same software package. Phylogenetic trees were constructed using the neighbor-joining algorithm. Distances were generated using the Kimura matrix, and tree stability was supported through bootstrap analysis (100 replicates). Bootstrapped consensus of neighbor-joining trees was visualized with MEGA software.
Cloning and transformation in E. coli and Rhodococcus sp.
The complete gene cluster comprising the dfdA1A2A3A4 genes was amplified from DNA from Rhodococcus sp. strain HA01 using the primers DfdPstF (5′-CACGACGACTGCAGCGGTGTGAT-3′) and DfdEcoR (5′-CTACTCTTCGAATTCCTGCGGCATG-3′), which include artificial PstI and EcoRI restriction sites (shown underlined) and which annealed 240 bp upstream and 137 bp downstream, respectively, of the gene cluster. A 3-kb DNA fragment containing the gene region comprising the dbfA1A2 genes from Rhodococcus sp. strain HA01 was amplified using the forward primer DbfHindF (5′-TGACAGCGAAGCTTCAGTGATACC-3′) and reverse primer DbfEcoR (5′-GTACCGGGAATTCTCCACCAGT-3′), which includes artificial HindIII and EcoRI restriction sites (shown underlined) for cloning in pUC119. For cloning in pRSG43, amplification was performed with forward primer DbfXbaF (5′-TGACAGCGTCTAGACAGTGATACC-3′) instead of DbfHindF to introduce the necessary XbaI restriction site (shown underlined). After confirmation of the sequences, the fragments were cloned into the respective restriction sites of pUC119, giving pDFDE (containing the dfdA1A2A3A4 genes) and pDF32 (containing the dbfA1A2 genes), or cloned into respective restriction sites of pRSG43, giving pDFDR (containing the dfdA1A2A3A4 genes) and pDBFA12 (containing the dbfA1A2 genes). Vectors were introduced into E. coli JM109 competent cells (Promega) or electrocompetent cells of Rhodococcus sp. strain ATCC 12674.
Extraction of mRNA, cDNA synthesis, and RT-PCR.
For gene expression studies, Rhodococcus sp. strain HA01 was grown on DBF (2 mM) as the sole carbon source. To assess constitutive expression, the strain was grown in parallel on fructose (2 mM). Cultures were harvested during exponential growth by centrifugation. Total RNA was isolated from 3 ml of DBF- or fructose-grown cells. In the case of DBF-grown cells, residual crystals of DBF were removed by filtration (595 1/2 filter paper; Schleicher & Schuell) before harvesting. Harvested cells were resuspended in 100 μl water and immediately processed as previously described (59). The RNA was subsequently purified using the RNeasy kit (Qiagen), and 2 μl of the eluted RNA (60 μl) was separated in 1% agarose gels and stained with ethidium bromide. The ratio between the total RNA content of DBF and fructose-grown cells was quantified by ImageQuant software (Molecular Dynamics) as 0.7:1.0. cDNA was synthesized from 1 μl of total RNA using a first-strand cDNA synthesis kit for reverse transcriptase PCR (RT-PCR) (Roche, Mannheim, Germany). The reverse transcription reaction mixtures were serially diluted (3.2-fold) with nuclease-free water (Qiagen), and 1 μl of each dilution was subjected to amplification by PCR using the primer set DfdRNAF (5′-CAACGTGTTCCCCAACTTCT-3′) and DfdRNAR (5′-GGTCGATTACCTCGGTCGTA-3′) to amplify a dfdA gene fragment and primer set DbfRNAF (5′-TCTACCGCAAGGAATTGGAC-3′) and DbfRNAR (5′-ATCCCGACGTCGTTCTGATA-3′) to amplify a dbfA gene fragment. Amplification products were separated on 1% agarose gels and stained with ethidium bromide. Product bands were purified from agarose gels using a QIAquick PCR purification kit (Qiagen) and sequenced to verify their identity.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this study are available under GenBank accession numbers EU622789 to EU622791.
RESULTS
Characterization of strain HA01.
Analysis of the nearly complete 16S rRNA gene sequence (corresponding to positions 15 to 1494 according to the E. coli numbering system) revealed that the novel DBF-degrading isolate designated HA01 belongs to the genus Rhodococcus (Fig. 1) and is the most similar to the type strains of Rhodococcus rhodochrous, Rhodococcus pyridinivorans, and Rhodococcus sp. strain T104, which has previously been reported to grow on biphenyl as well as on terpenoids as the sole carbon and energy source (21).
FIG. 1.
Dendrogram showing the relationship of the nearly complete 16S rRNA gene sequence of Rhodococcus sp. strain HA01 with those sequences reported for several Rhodococcus sp. type strains. The tree was constructed using the neighbor-joining method. The reliability of the inferred trees was tested by bootstrap analysis using 100 resamplings. Bootstrap values beyond 50% are denoted at the branching points. The scale bar indicates 0.01 nucleotide substitution per position. GenBank/EMBL/DDBJ accession numbers used for phylogenetic analysis are shown after the bacterial strain name.
Growth of Rhodococcus sp. strain HA01 on DBF and transformation of substrate analogues.
Rhodococcus sp. strain HA01 can utilize DBF as the sole carbon and energy source with a doubling time of 3 h, while neither DD, 2CDBF, nor 3CDBF could be used as a carbon source. Cells pregrown on DBF and having a turnover rate of 20.5 ± U/g (dry weight) were able to remove DD, 2CDBF, and 3CDBF at rates of 1.8 ± 0.1, 2.3 ± 0.1, and 5.8 ± 1.0 U/g (dry weight), respectively. Depletion of DBF by fructose-grown cells was negligible (<0.4 U/g [dry weight]), indicating that the enzymes involved in DBF degradation by strain HA01 are inducible.
The removal of both DBF and DD occurred without significant accumulation of intermediates. In the presence of 3-chlorocatechol (0.1 mM), which is known to be a strong inhibitor of extradiol dioxygenases (3, 54), the intermediate accumulation of up to 20% of applied DBF (1 mM) as THB and up to 40% of applied DD as THBE indicated that the degradation is initiated by angular dioxygenation. However, 3-chlorocatechol-mediated inactivation was not quantitative, and trihydroxybiphenyl metabolites were slowly transformed further.
Accumulation of intermediates was observed during the transformation of both 2CDBF and 3CDBF. HPLC analysis revealed the accumulation of one major intermediate concomitant with the depletion of 3CDBF, which showed a retention behavior and an absorption spectrum identical with an authentic standard of 4-chlorosalicylate. The identity of the metabolite was further confirmed by HPLC-MS analysis (negative ionization mode), which showed a metabolite with a molecular ion at m/z 171 and 173 (ratio 3:1). Transformation to 4-chlorosalicylate was nearly stoichiometric (95% ± 5% of applied 3CDBF accumulated as 4-chlorosalicylate). However, the culture supernatant during 3-CDBF depletion was yellow, which is indicative of the accumulation of an extradiol ring cleavage product. The yellow color was not depleted during further incubation, indicating that this compound might be a dead-end metabolite, probably formed by lateral dioxygenation of 3CDBF followed by dehydrogenation and ring cleavage. Spectrophotometric analysis of the culture supernatant after complete removal of 2 mM 3CDBF showed that the yellow product exhibited an absorption maximum at 455 nm (A455 of <1) similar to the reported absorption maximum of the meta cleavage product of 1,2-dihydroxydibenzofuran (464 to 470 nm [49, 50]).
HPLC analysis of the supernatants during the transformation of 2CDBF revealed the accumulation of two major metabolites. One of the metabolites was identified as 5-chlorosalicylate based on the identical retention behavior and UV spectrum compared with an authentic standard. Moreover, HPLC-MS analysis confirmed the identity of one metabolite as 5-chlorosalicylate. Quantification with an authentic standard revealed that 40% ± 5% of the applied 2CDBF was transformed into 5-chlorosalicylate. HPLC-MS analysis indicated that the second metabolite has a negative molecular ion at m/z 235/237 corresponding to a parent compound, C12H9ClO3, that is either a chlorosubstituted trihydroxybiphenyl or a dihydrodiol of 2CDBF.
The structures of both metabolites were elucidated using in situ 1H NMR spectroscopic analysis of a freshly prepared product mixture (Fig. 2). This analysis showed the presence of two major metabolites, one of which was 5-chlorosalicylate. The three aromatic protons of this compound showed chemical shifts, δ, at 6.96 (H-3), 7.45 (H-4), and 7.83 (H-6) ppm, respectively, with characteristic coupling constants of J (H-3/H-4) = 9.0 Hz and J (H-4/H-6) = 2.2 Hz.
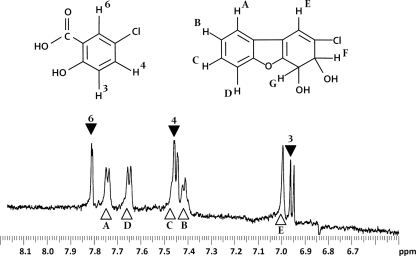
FIG. 2.
The 1H NMR spectrum of the metabolite mixture formed from 2CDBF indicates the presence of two major metabolites in approximately equal amounts. One metabolite is identical to 5-chlorosalicylate (▾). The NMR spectrum of the second metabolite (▵) indicates its identity to 2-chloro-3,4-dihydro-3,4-dihydroxydibenzofuran. The structures of the metabolites are shown at the top of the figure. The signals at the bottom of the figure are marked with numbers and letters according to the assignments of the respective protons.
The second metabolite possessed five aromatic or vinylic protons, exhibiting chemical shifts at δ = 7.75 (H-A), 7.42 (H-B), 7.45 (H-C), 7.67 (H-D), and 6.99 (H-E) ppm, respectively. Although the signals of H-4 of 5-chlorosalicylate and of H-C of the second metabolite could not be separated, the integral clearly showed two distinct protons at 7.45 ppm. The presence of five aromatic or vinylic protons excludes the chlorosubstituted trihydroxybiphenyls (either 2,2′3-trihydroxy-5-chlorobiphenyl or 2,2′3-trihydroxy-5-chlorobiphenyl can be formed after angular dioxygenation), which harbor six aromatic protons and indicates the presence of a chlorosubstituted DBF dihydrodiol. As the metabolite contained at least four aromatic protons showing vicinal couplings (J = 8 to 10 Hz) with at least one other proton, dioxygenolytic attack has occurred in the chlorosubstituted ring. Thus, the observed NMR data are compatible only with the structure of the metabolite as 2-chloro-3,4-dihydro-3,4-dihydroxydibenzofuran, with a singlet vinylic proton resonating at δ = 6.99 ppm (H-E). Protons F and G (see the chemical structures in Fig. 2), are expected to resonate at δ = 4 to 4.5 ppm and are under the suppressed water signal. Comparison of the integrals of the resonance lines showed that in different preparations, the ratio in which these products have been formed (5-chlorosalicylate to 2-chloro-3,4-dihydro-3,4-dihydroxydibenzofuran) was always 0.8 to 0.9:1. There was no significant accumulation of any other metabolites.
PCR amplification and characterization of genes encoding a Rieske nonheme iron oxygenase in Rhodococcus sp. strain HA01.
As described above, strain HA01 showed a regioselectivity of attack on 2CDBF not previously reported. Hence, as only a few genes involved in the metabolism of DBF via angular dioxygenation have been described, we have attempted to identify the genes responsible for angular dioxygenation in strain HA01. Amplification using previously described RieskeF and RieskeR primers (28) resulted in the amplification of an 78-bp fragment. Sequence analysis revealed the presence of two different dioxygenase gene fragments. One of these fragments showed the highest nucleotide sequence identity (92%) to gene fragments encoding the Rieske clusters of benzoate dioxygenases. The second fragment was highly similar (99% identity) to a gene fragment encoding part of the α-subunit of the DfdA angular dioxygenase of Terrabacter sp. strain YK3 (GenBank/EMBL/DDBJ accession number AB075242).
As described in Materials and Methods, a 4.4-kb gene fragment was obtained which showed high similarity over the entire sequence with the dfdA1A2A3A4 gene cluster of Terrabacter sp. strain YK3 (22). The predicted dfdA1 α-subunit sequence differed from that of Terrabacter sp. strain YK3 by two amino acids, whereas the predicted dfdA2 β-subunit sequences of strains YK3 and HA01 were identical. Three amino acid differences were observed between the DfdA3 ferredoxins, while the DfdA4 ferredoxin reductase of strain HA01 was five amino acids longer than that of strain YK3.
To investigate whether the identified dfdA genes are expressed in response to DBF, RT-PCR experiments were performed with total RNA extracted from Rhodococcus sp. strain HA01 growing on DBF or fructose. Amplification products of the expected size were observed in DBF-grown cells using the DfdRNAF and DfdRNAR primers targeting the α-subunit (DfdA1), whereas no product was detected with RNA extracted from fructose-grown cells (Fig. 3). In addition, no amplification products were observed in controls devoid of reverse transcriptase or template cDNA (Fig. 3C). Sequencing of the approximately 435-bp product confirmed that it was identical with the corresponding dfdA gene fragment from the genomic DNA of Rhodococcus sp. strain HA01 (Fig. 3C, lane 6). The detection of transcripts in DBF-grown cells was possible in as little as 0.8 picogram of total RNA, whereas transcription of the gene was virtually absent in fructose-grown cells, indicating that gene transcripts are induced at least 106-fold.
FIG. 3.
RT-PCR amplification of dfdA mRNA from Rhodococcus sp. strain HA01 grown on DBF (A) or fructose (B). M lanes contain the molecular size marker Hyperladder 1 (Bioline). cDNA generated from 1 μg template RNA was serially diluted (3.2-fold) with nuclease-free water, and 1 μl of each dilution was subjected to amplification by PCR (lanes 1 to 15). Negative controls included undiluted RT-PCR mixtures devoid of reverse transcriptase (lanes 1 to 4 in panel C) or template cDNA (lane 5 in panel C). PCR mixtures containing 1 ng of genomic DNA as a template were used as positive control (lane 6 in panel C) in the same experiment.
The complete gene cluster comprising the dfdA1A2A3A4 genes was amplified from DNA from Rhodococcus sp. strain HA01 by using primers DfdPstF and DfdEcoR, and the gene cluster was initially cloned into pUC119 to give pDFDE and introduced into E. coli JM109. However, as DBF transformation was absent in E. coli (data not shown), the dfdA1A2A3A4 genes were cloned into the pRSG43 shuttle vector to give pDFDR, and introduced into Rhodococcus sp. strain ATCC 12674, which was shown to be incapable of degrading DBF. Analysis of cell extracts by sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed the presence of a prominent band with a molecular mass of 51 ± 2 kDa in cell extracts of Rhodococcus sp. strain ATCC 12674(pDFDR) independent of whether the strain was grown in the presence or absence of IPTG. This is in agreement with the expected molecular mass of the dfdA1 gene product (53 kDa).
While cells of Rhodococcus sp. strain ATCC 12674(pRSG43) were not capable of DBF depletion and no metabolites were observed by HPLC analysis, cells of Rhodococcus sp. strain ATCC 12674(pDFDR) were capable of DBF removal at a rate of 0.49 ± 0.05 U/g (dry weight). A single metabolite was observed, which cochromatographed with authentic THB and showed an identical UV absorption spectrum. Transformation of DBF into THB was stoichiometric, confirming that the dfdA1A2A3A4 genes from Rhodococcus sp. strain HA01 encode a functional angular dioxygenase.
Resting cells of Rhodococcus sp. strain ATCC 12674 (pDFDR) also converted 3CDBF into one product. As shown by HPLC analysis, 3CDBF was depleted at a rate of 0.14 ± 0.02 U/g (dry weight). HPLC-MS indicated a chlorosubstituted metabolite, C12H9ClO3, from the negative molecular ion at m/z 235/237. To exclude its identity to a dihydrodiol derivative and validate that it is a chlorosubstituted trihydroxybiphenyl, the reaction mixture was supplemented with DbfB, a 2,2′,3-trihydroxybiphenyl dioxygenase from S. wittichii RW1. The metabolite was transformed at rates similar in magnitude to that with THB as the substrate; hence, it can be assumed that it is 4-chloro-2,2′,3-trihydroxybiphenyl.
Two metabolites were formed from DD, which was removed at a rate of 0.09 ± 0.015 U/g (dry weight). One of the metabolites was identified as THBE by comparing its retention behavior and spectral characteristics with those of an authentic standard. THBE was the major metabolite formed, accounting for approximately 90% of transformed DD. The second metabolite, like THBE, displayed a negative molecular ion at m/z 217 and is probably identical to a DD dihydrodiol. In contrast, no conversions were observed when Rhodococcus sp. strain ATCC 12674(pRSG43) was used in experiments with DBF, 3CDBF, or DD as a substrate. Similarly, resting cells of Rhodococcus sp. strain ATCC 12674(pDFDR) did not transform 2CDBF.
PCR amplification and detection of a second angular dioxygenase in Rhodococcus sp. strain HA01.
The above cloned and characterized DfdA DBF dioxygenase showed a substrate spectrum and regioselectivity, which does not explain the observed transformation by the wild-type HA01. To localize a proposed second angular dioxygenase in strain HA01, new degenerate primers Dxn1F and Dxn2R were designed (see Materials and Methods). PCRs using these primers with genomic DNA of Rhodococcus sp. strain HA01 amplified an approximately 700-bp fragment, the nucleotide sequence of which showed high similarity (98%) to the dbfA gene encoding the α-subunit of dibenzofuran dioxygenase of Terrabacter sp. strain DBF63 (GenBank/EMBL/DDBJ accession number AB054975) (28). Using the strategy described in Materials and Methods, a 4,863-bp fragment was obtained and sequence analysis revealed the presence of five open reading frames. The deduced 443-amino-acid sequence encoded by dbfA1 showed high similarity (98.6% identity) with the α-subunit DbfA1 of the angular DBF dioxygenase from Terrabacter sp. strain DBF63 (accession number BAC75993). Similar identities were observed with the α-subunit of DBF dioxygenases of Rhodococcus sp. strain YK2 (accession number BAC00802; 98.9% identity) and Rhodococcus sp. strain DFA3 (accession number BAD51811; 98.3% identity); however, both of these sequences were incomplete and do not cover the entire α-subunits. The deduced amino acid sequence of the 167-amino-acid protein encoded by dbfA2, located downstream of dbfA1, showed 98.2% identity with both the β-subunit of the dibenzofuran dioxygenase (DbfA2) from Terrabacter sp. strain DBF63 (accession number BAC75994) and that from Rhodococcus sp. strain YK2 (accession number BAC00803). Similarly, the deduced amino acid sequence of the 319-amino-acid protein encoded by ORF3 located downstream of dbfA2 showed high similarity (98.4% identity) to putative meta cleavage compound hydrolases encoded downstream of the α- and β-subunits of DBF dioxygenase of Rhodococcus sp. strain YK2 and Terrabacter sp. strain DBF63. A similar protein was also reported to be encoded by the genome of Rhodococcus rhodochrous K37 (97.6% identity in a stretch of 126 amino acids analyzed) (51). As with Rhodococcus sp. strain YK2, Terrabacter sp. strain DBF63, and Rhodococcus rhodochrous K37, the gene encoding a putative meta cleavage compound hydrolase (ORF3) was followed in Rhodococcus sp. strain HA01 by a gene encoding a putative extradiol dioxygenase (ORF4). This protein, however, does not belong to the type I (vicinal oxygen chelate superfamily) extradiol dioxygenases typically involved in the degradation of biphenyl and related compounds but to the type II extradiol dioxygenases, which include enzymes such as protocatechuate 4,5-dioxygenase (LigAB) from Pseudomonas paucimobilis (43). The deduced amino acid sequence showed 99.3% identity with the respective proteins from Rhodococcus sp. strain YK2 and Rhodococcus rhodochrous K37 but only 94.3% identity with that from Terrabacter sp. strain DBF63. The deduced amino acid sequence of the 81-amino-acid protein encoded by ORF5 located downstream of ORF4 showed high similarity (98.7% identity) to the ferredoxin proteins of Rhodococcus sp. strain YK2 and Rhodococcus rhodochrous K37 but relatively low similarity (90%) with the ferredoxin gene of Rhodococcus sp. strain DFA3.
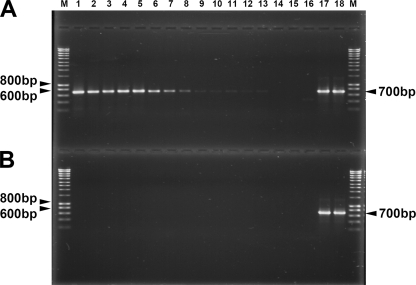
To verify whether the dbf genes are also expressed in response to DBF, RT-PCR experiments were performed with total RNA extracted from Rhodococcus sp. strain HA01 growing on DBF and fructose. RT-PCR amplification products of the expected 700-bp size were observed with 2.5 pg or more of RNA extracted from the culture grown on DBF (Fig. 4A, lanes 1 to 13), whereas no product was detected with RNA extracted from the culture grown on fructose (Fig. 4B, lanes 1 to 13), indicating that gene transcripts are induced at least 105-fold. In addition, no amplification products were observed in controls devoid of reverse transcriptase (Fig. 4A and B, lanes 14 and 15) or template cDNA (Fig. 4A and B, lanes 16). Sequencing of the approximately 700-bp product confirmed that it was identical to the corresponding dbfA1 gene fragment from genomic DNA of Rhodococcus sp. strain HA01. These results showed that the DbfA dioxygenase from Rhodococcus sp. strain HA01 is also specifically induced in the presence of DBF, and thus, at least two angular dioxygenases are simultaneously expressed when strain HA01 is confronted with DBF.
FIG. 4.
RT-PCR amplification of dbfA mRNA from Rhodococcus sp. strain HA01 grown on DBF (A) or fructose (B). M lanes contain the molecular size marker Hyperladder 1(Bioline). cDNA generated from 1 μg template RNA was serially diluted (3.2-fold) with nuclease-free water, and 1 μl of each dilution was subjected to amplification by PCR (lanes 1 to 13). Negative controls included undiluted RT-PCR mixtures devoid of reverse transcriptase (lanes 14 and 15) or template cDNA (lane 16). PCR mixtures containing 1 ng of genomic DNA as a template were used as a positive control (lanes 17 and 18).
As dbfA1A2 from Terrabacter sp. strain DBF63 was previously cloned using pUC119 (pDF32) in E. coli JM109 and some activity against DBF was observed using this host (28), a 3-kb DNA fragment containing the gene region comprising the dbfA1A2 genes from Rhodococcus sp. strain HA01 was amplified, cloned into pUC119, giving plasmid pDBFA12a, and transformed into E. coli JM109. Resting cells of E. coli (pDBFA12a) (A600 of 10) showed a low activity against DBF where after 18 h of incubation, approximately 2 μM of THB was produced. Activity against all other substrates was below the detection limit.
Thus, the 3-kb DNA fragment was cloned into pRSG43, giving pDBFA12, and transformed into E. coli JM109. A prominent band at a molecular mass of approximately 49 kDa was observed in cell extracts of E. coli JM109(pDBFA12), which was absent in cell extracts of E. coli JM109(pRSG43). This is in agreement with the expected molecular mass of the dbfA1 gene product (49.5 kDa).
Resting cells of E. coli JM109(pDBFA12) transformed DBF and DD into single metabolites, and HPLC analysis revealed that these metabolites cochromatographed with authentic THB and THBE standards and possessed UV absorption spectra identical to those of the respective standards, indicating DBF and DD are dioxygenated exclusively at the angular positions. The metabolites were formed at rates of 0.5 ± 0.1 mU/g (dry weight) and 0.25 ± 0.07 mU/g (dry weight), respectively, which were significantly lower than those observed in experiments using Rhodococcus sp. strain ATCC 12674 (pDFDR). 2CDBF was transformed into a single product, which was not identical to either 5-chlorosalicylate or 2-chloro-3,4-dihydro-3,4-dihydroxydibenzofuran formed by Rhodococcus sp. strain RHA01. It is thus possible that the observed metabolite has the 5′-chloro-2,2′,3-trihydroxybiphenyl structure, which is further transformed into 5-chlorosalicylate in the HA01 strain. Product accumulation occurred at a rate of 0.14 ± 0.04 mU/g (dry weight). No significant activity as evidenced by the absence of metabolite accumulation was observed with 3CDBF, even after extended incubation for 12 h. To evaluate whether higher activities could be obtained by expression in Rhodococcus, pDBFA12 was introduced into Rhodococcus sp. strain ATCC 12674. However, resting cells of Rhodococcus sp. strain ATCC 12674(pDBFA12) (A600 of up to 20) did not show any detectable depletion of DBF, 3CDBF, 2CDBF or DD or any detectable product formation with incubations of up to 24 h.
DISCUSSION
Although the involvement of angular dioxygenases in the degradation pathways of DBF and its structural analogues has been studied for over 2 decades, there is only limited information on the diversity and distribution of such dioxygenases, their substrate range, and the regioselectivity of attack. In the present study, two angular dioxygenases were found in Rhodococcus sp. strain HA01, one of which (DbfAHA01) exhibits high similarity to DBF dioxygenase from Terrabacter sp. strain DBF63 (DbfADBF63) (28), while the other (DfdAHA01) exhibits high similarity with dibenzofuran dioxygenase DfDA from Terrabacter sp. strain YK3 (DfdAYK3) (22). It has been previously suggested that enzymes similar to DfdAYK3 are distributed among DBF-degrading actinobacteria (22), while enzymes similar to DbfADBF63 have been recently reported in Rhodococcus strains, such as Rhodococcus sp. strain YK2 or DFA3 (44).
The existence of multiple dioxygenase genes in a single bacterium has been reported previously and seems to be common in environmental strains. For example, the genome sequence of Rhodococcus sp. strain RHA1 revealed the presence of six ring-hydroxylating dioxygenases (40). The presence of multiple Rieske nonheme iron oxygenases in Sphingomonas (2, 46) and Rhodococcus species may, at least in part, explain why these organisms are able to efficiently degrade a wide range of aromatic hydrocarbons. However, to our knowledge, the presence of two distinct angular dioxygenases in a single bacterium has not previously been reported. Both gene clusters were expressed by strain HA01 in response to DBF and encode functional enzymes, as revealed by their successful heterologous expression.
The capability of DfdAYK3 to transform DBF by angular attack has previously been reported (22), although transformation of chlorinated derivatives was not assessed. Analysis of the substrate specificity and regioselectivity of attack of DfdAHA01 revealed that this enzyme, like DfdAYK3, catalyzes exclusively an angular dioxygenation of DBF. Also, like DfdAYK3, DD was preferentially subject to angular dioxygenation, although some lateral dioxygenation was also observed. Interestingly, DfdAHA01 was capable of rapidly transforming 3CDBF into 4-chloro-2,2′,3-trihydroxybiphenyl (Fig. 5). Previously, Harms et al. (19) showed that a mutant of Sphingomonas sp. strain HH69, defective in 2,3-dihydroxybiphenyl 1,2-dioxygenase, transforms 3CDBF into equal amounts of 4′-chloro-2,2′,3-trihydroxybiphenyl and 4-chloro-2,2′,3-trihydroxybiphenyl, indicating a nonselective attack of both the substituted and nonsubstituted aromatic nuclei of 3CDBF. Similarly, Wilkes et al. (55) reported that S. wittichii RW1 attacks both the substituted and nonsubstituted aromatic nuclei of 3CDBF that form after ring cleavage and hydrolysis, salicylate and 4-chlorosalicylate, respectively, a situation similar to that observed in Sphingomonas sp. strain RW16 (58). Evidently, the transformation of 3CDBF by DfdAHA01 analyzed here is different (Fig. 5), as it preferentially attacks the nonsubstituted aromatic nucleus of 3CDBF.
FIG. 5.
Metabolism of 2-chlorodibenzofuran and 3-chlorodibenzofuran by Rhodococcus sp. strain HA01 (black arrows) and by DbfAHA01 (gray arrow) and DfdAHA01 (broken black arrow) dioxygenases. Lateral dioxygenation of 3-chlorodibenzofuran by strain HA01 was observed to be a minor reaction, and the sites of dioxygenation are tentative.
Strain HA01 transforms 3CDBF almost stoichiometrically into 4-chlorosalicylate, as well as minor amounts of a yellow ring cleavage intermediate, which is presumably generated after lateral dioxygenation (Fig. 5). The stability of the yellow intermediate accounts only for a minor portion of the formed products in strain HA01. This excludes its identity with a ring cleavage product formed from 4-chloro-2,2′,3-trihydroxybiphenyl or 4′-chloro-2,2′,3-trihydroxybiphenyl, as such compounds, like the situation in the THB ring cleavage product, are presumably highly unstable and should spontaneously rearrange to form 3-(chroman-4-on-2-yl)-pyruvates (19). Evidently, DfdAHA01 is responsible for 3CDBF transformation by DBF-grown cells, although the lateral oxygenation indicates that a third oxygenase, besides DfdAHA01 and DbfAHA01, is expressed during growth on DBF.
Heterologous expression of DbfAHA01 indicated that this enzyme is capable of 2CDBF transformation, whereas no detectable transformation of 3CDBF was observed, suggesting that both angular oxygenase systems have complementary substrate specificities. As described previously for DbfADBF63 and CARDO angular dioxygenases of Pseudomonas resinovorans CA10 (16), angular dioxygenation occurred on the nonsubstituted aromatic nucleus, giving rise to 5′-chloro-2,2′,3-trihydroxybiphenyl, which contrasts with the situation in S. wittichii RW1 and Sphingomonas sp. strain RW16 where angular attack occurred both at the substituted and unsubstituted aromatic nuclei of 2CDBF, giving rise to the accumulation of both 5-chlorosalicylate and salicylate (55, 58).
In contrast to the single product formed by DbfAHA01, wild-type Rhodococcus sp. strain HA01 forms two distinct products originating from angular and lateral oxygenation which, as described above, is evidence for the expression of a third lateral oxygenase system during growth of the strain on DBF. Lateral oxygenation is observed quite often in a number of DBF-degrading organisms (26, 32, 41, 60), but it is not observed when cloned angular dioxygenases are confronted with DBF (22, 28, 48, 53). This suggests fortuitous expression of lateral oxygenases, as naphthalene dioxygenase in Rhodococcus opacus SAO101 (29) is common in DBF-degrading organisms. However, the extent to which substrates are misrouted by lateral dioxygenation will depend on the relative expression levels and affinities of the expressed enzymes toward their respective substrates. In Rhodococcus sp. strain HA01, the expression of two angular dioxygenase systems during growth on DBF evidently prevents lateral oxygenation of this substrate. In contrast, chlorinated derivatives are subject to lateral oxygenation. The current observation of two specifically expressed angular oxygenases in a single host also shows that studies using wild-type organisms to reveal substrate specificities of angular dioxygenases have to be considered with care.
The presence and expression of two different angular dioxygenases in Rhodococcus sp. strain HA01 prevent significant substrate misrouting from DBF, although they do not allow mineralization of DD. Interestingly, various DBF-degrading strains, such as Janibacter sp. strain YY-1 (60), Janibacter sp. strain YA (25), Terrabacter sp. strain DBF63 (41), and Sphingomonas sp. strain HH69 (20), are also not able to mineralize DD, although it is believed that both DBF and DD are degraded by similar pathways (57). Information on the reasons for the failure of DBF-mineralizing strains to mineralize DD is scarce. Harms et al. (20) reported that Sphingomonas sp. strain HH69 transforms DD into a 2-phenoxy derivative of muconate, indicating that extradiol ring cleavage of intermediate THBE is a crucial step. Rhodococcus sp. strain HA01 cannot grow on DD as a carbon source, but DBF-grown cells do transform DD, at a rate <10% that observed for DBF as a substrate, without significant accumulation of intermediates. This indicates that the initial angular attack is one limiting factor in DD degradation by this strain. However, the possibility that additional factors are responsible for the failure of the strain to degrade DD cannot be excluded.
While pDBFR, used in the current study for heterologous expression, encodes the electron transport chain (dfdA3A4) of DfdAHA01 in addition to the terminal oxygenase component (dfdA1A2), pDBFA12 encodes only the terminal oxygenase (dbfA1A2) of DbfAHA01. Despite this lack of a primary electron transport chain, E. coli cells harboring pDBFA12 were capable of angular dioxygenation of DBF, indicating the recruitment of host components for electron transport. Thus far, there are various examples showing that electron transport to terminal oxygenases can be achieved by components derived from E. coli host cells (28, 34, 38). However, as previously reported for DbfADBF63 (28), the observed activity was rather weak and significantly lower than that of heterologously expressed DfdAHA01. Expression of DbfAHA01 in Rhodococcus sp. strain ATCC 12674 failed, suggesting that the electron transport components of this strain could not be recruited. It has been reported that a ferredoxin encoded downstream of the dbfA1A2 genes of Terrabacter sp. strain DBF63 can transfer electrons to the terminal oxygenase and that ferredoxin reductase of phthalate dioxygenase, encoded upstream of dbfA1A2, can complement an effective electron transport chain (17, 52). As the respective gene organization in Rhodococcus sp. strain HA01 is similar to that in Terrabacter sp. strain DBF63, a functional DbfA angular oxygenase may also be formed in strain HA01. However, the possibility that DfdA3A4 fulfills such a function in strain HA01 remains to be elucidated.
Acknowledgments
We thank Iris Plumeier, Christel Kakoschke, and Beate Jaschok-Kentner for excellent technical assistance and Melissa Wos for critically reading the manuscript. Masahiro Takeo kindly provided plasmid pRSG43.
H.A.H.A. was supported by a scholarship from the Egyptian Government, Ministry of Higher Education, and N.B.H. was supported by a DAAD scholarship.
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Aoki, Y. 2001. Polychlorinated biphenyls, polychlorinated dibenzo-p-dioxins, and polychlorinated dibenzofurans as endocrine disrupters: what we have learned from Yusho disease. Environ. Res. 86:2-11. [DOI] [PubMed] [Google Scholar]
- 2.Armengaud, J., B. Happe, and K. N. Timmis. 1998. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J. Bacteriol. 180:3954-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartels, I., H.-J. Knackmuss, and W. Reineke. 1984. Suicide inactivation of catechol 2,3-dioxygenase from Pseudomonas putida mt-2 by 3-halocatechols. Appl. Environ. Microbiol. 47:500-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellin, J. S., and D. G. Barnes. 1985. Health hazard assessment for chlorinated dioxins and dibenzofurans other than 2,3,7,8-TCDD. Toxicol. Ind. Health 1:25-248. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi, D., A. Bosetti, D. Cidaria, A. Bernardi, I. Gagliardi, and P. D'Amico. 1997. Oxidation of polycyclic aromatic heterocycles by Pseudomonas fluorescens TTC1. Appl. Microbiol. Biotechnol. 47:596-599. [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Bünz, P. V., and A. M. Cook. 1993. Dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1: angular dioxygenation by a three-component enzyme system. J. Bacteriol. 175:6467-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler, C. S., and J. R. Mason. 1997. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv. Microb. Physiol. 38:47-84. [DOI] [PubMed] [Google Scholar]
- 9.Cerniglia, C. E., J. C. Morgan, and D. T. Gibson. 1979. Bacterial and fungal oxidation of dibenzofuran. Biochem. J. 180:175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorn, E., M. Hellwig, W. Reineke, and H.-J. Knackmuss. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. [DOI] [PubMed] [Google Scholar]
- 11.Engesser, K. H., W. Fietz, P. Fischer, P. Schulte, and H.-J. Knackmuss. 1990. Dioxygenolytic cleavage of aryl ether bonds: 1,2-dihydroxy-1,2-dihydroxy-4-carboxybenzophenone as evidence for initial 1,2-dioxygenation in 3-and 4-carboxy biphenyl ether degradation. FEMS Microbiol. Lett. 69:317-322. [DOI] [PubMed] [Google Scholar]
- 12.Engesser, K. H., V. Strubel, K. Christoglou, P. Fischer, and H. G. Rast. 1989. Dioxygenolytic cleavage of aryl ether bonds: 1,10-dihydro-1,10-dihydroxyfluoren-9-one, a novel arene dihydrodiol as evidence for angular dioxygenation of dibenzofuran. FEMS Microbiol. Lett. 65:205-210. [DOI] [PubMed] [Google Scholar]
- 13.Ferre-Huguet, N., M. Nadal, M. Schuhmacher, and J. L. Domingo. 2006. Environmental impact and human health risks of polychlorinated dibenzo-p-dioxins and dibenzofurans in the vicinity of a new hazardous waste incinerator: a case study. Environ. Sci. Technol. 40:61-66. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda, K., S. Nagata, and H. Taniguchi. 2002. Isolation and characterization of dibenzofuran-degrading bacteria. FEMS Microbiol. Lett. 208:179-185. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, D. T., and R. E. Parales. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11:236-243. [DOI] [PubMed] [Google Scholar]
- 16.Habe, H., J. S. Chung, J. H. Lee, K. Kasuga, T. Yoshida, H. Nojiri, and T. Omori. 2001. Degradation of chlorinated dibenzofurans and dibenzo-p- dioxins by two types of bacteria having angular dioxygenases with different features. Appl. Environ. Microbiol. 67:3610-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habe, H., M. Miyakoshi, J. Chung, K. Kasuga, T. Yoshida, H. Nojiri, and T. Omori. 2003. Phthalate catabolic gene cluster is linked to the angular dioxygenase gene in Terrabacter sp. strain DBF63. Appl. Microbiol. Biotechnol. 61:44-54. [DOI] [PubMed] [Google Scholar]
- 18.Happe, B., L. Eltis, H. Poth, R. Hedderich, and K. Timmis. 1993. Characterization of 2,2′,3-trihydroxybiphenyl dioxygenase, an extradiol dioxygenase from the dibenzofuran- and dibenzo-p-dioxin-degrading bacterium Sphingomonas sp. strain RW1. J. Bacteriol. 175:7313-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harms, H., H. Wilkes, V. Sinnwell, R.-M. Wittich, K. Figge, W. Francke, and P. Fortnagel. 1991. Transformation of 3-chlorodibenzofuran by Pseudomonas sp. HH69. FEMS Microbiol. Lett. 81:25-30. [DOI] [PubMed] [Google Scholar]
- 20.Harms, H., R.-M. Wittich, V. Sinnwell, H. Meyer, P. Fortnagel, and W. Francke. 1990. Transformation of dibenzo-p-dioxin by Pseudomonas sp. strain HH69. Appl. Environ. Microbiol. 56:1157-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez, B. S., S. C. Koh, M. Chial, and D. D. Focht. 1997. Terpene-utilizing isolates and their relevance to enhanced biotransformation of polychlorinated biphenyls in soil. Biodegradation 8:153-158. [Google Scholar]
- 22.Iida, T., Y. Mukouzaka, K. Nakamura, and T. Kudo. 2002. Plasmid-borne genes code for an angular dioxygenase involved in dibenzofuran degradation by Terrabacter sp. strain YK3. Appl. Environ. Microbiol. 68:3716-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iida, T., Y. Mukouzaka, K. Nakamura, I. Yamaguchi, and T. Kudo. 2002. Isolation and characterization of dibenzofuran-degrading actinomycetes: analysis of multiple extradiol dioxygenase genes in dibenzofuran-degrading Rhodococcus species. Biosci. Biotechnol. Biochem. 66:1462-1472. [DOI] [PubMed] [Google Scholar]
- 24.Iida, T., K. Nakamura, A. Izumi, Y. Mukouzaka, and T. Kudo. 2006. Isolation and characterization of a gene cluster for dibenzofuran degradation in a new dibenzofuran-utilizing bacterium, Paenibacillus sp. strain YK5. Arch. Microbiol. 184:305-315. [DOI] [PubMed] [Google Scholar]
- 25.Iwai, S., A. Yamazoe, R. Takahashi, F. Kurisu, and O. Yagi. 2005. Degradation of mono-chlorinated dibenzo-p-dioxins by Janibacter sp. strain YA isolated from river sediment. Curr. Microbiol. 51:353-358. [DOI] [PubMed] [Google Scholar]
- 26.Jin, S., T. Zhu, X. Xu, and Y. Xu. 2006. Biodegradation of dibenzofuran by Janibacter terrae strain XJ-1. Curr. Microbiol. 53:30-36. [DOI] [PubMed] [Google Scholar]
- 27.Kanakaraj, R., D. L. Harris, J. G. Songer, and B. Bosworth. 1998. Multiplex PCR assay for detection of Clostridium perfringens in feces and intestinal contents of pigs and in swine feed. Vet. Microbiol. 63:29-38. [DOI] [PubMed] [Google Scholar]
- 28.Kasuga, K., H. Habe, J. S. Chung, T. Yoshida, H. Nojiri, H. Yamane, and T. Omori. 2001. Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the Gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochem. Biophys. Res. Commun. 283:195-204. [DOI] [PubMed] [Google Scholar]
- 29.Kimura, N., W. Kitagawa, T. Mori, N. Nakashima, T. Tamura, and Y. Kamagata. 2006. Genetic and biochemical characterization of the dioxygenase involved in lateral dioxygenation of dibenzofuran from Rhodococcus opacus strain SAO101. Appl. Microbiol. Biotechnol. 73:474-484. [DOI] [PubMed] [Google Scholar]
- 30.Klečka, G. M., and D. T. Gibson. 1980. Metabolism of dibenzo-p-dioxin and chlorinated dibenzo-p-dioxins by a Beijerinckia species. Appl. Environ. Microbiol. 39:288-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klecka, G. M., and D. T. Gibson. 1979. Metabolism of dibenzo[1,4]dioxan by a Pseudomonas species. Biochem. J. 180:639-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubota, M., K. Kawahara, K. Sekiya, T. Uchida, Y. Hattori, H. Futamata, and A. Hiraishi. 2005. Nocardioides aromaticivorans sp. nov., a dibenzofuran-degrading bacterium isolated from dioxin-polluted environments. Syst. Appl. Microbiol. 28:165-174. [DOI] [PubMed] [Google Scholar]
- 33.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 34.Kurkela, S., H. Lehväslaiho, E. T. Palva, and T. H. Teeri. 1988. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB9816. Gene 73:355-362. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, United Kingdom.
- 37.Lang, E., R. M. Kroppenstedt, J. Swiderski, P. Schumann, W. Ludwig, A. Schmid, and N. Weiss. 2003. Emended description of Janibacter terrae, including ten dibenzofuran-degrading strains and Janibacter brevis as its later heterotypic synonym. Int. J. Syst. Evol. Microbiol. 53:1999-2005. [DOI] [PubMed] [Google Scholar]
- 38.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKay, D. B., M. Prucha, W. Reineke, K. N. Timmis, and D. H. Pieper. 2003. Substrate specificity and expression of three 2,3-dihydroxybiphenyl 1,2-dioxygenases from Rhodococcus globerulus strain P6. J. Bacteriol. 185:2944-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLeod, M. P., R. L. Warren, W. W. Hsiao, N. Araki, M. Myhre, C. Fernandes, D. Miyazawa, W. Wong, A. L. Lillquist, D. Wang, M. Dosanjh, H. Hara, A. Petrescu, R. D. Morin, G. Yang, J. M. Stott, J. E. Schein, H. Shin, D. Smailus, A. S. Siddiqui, M. A. Marra, S. J. Jones, R. Holt, F. S. Brinkman, K. Miyauchi, M. Fukuda, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 103:15582-15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monna, L., T. Omori, and T. Kodama. 1993. Microbial degradation of dibenzofuran, fluorene, and dibenzo-p-dioxin by Staphylococcus auriculans DBF63. Appl. Environ. Microbiol. 59:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nam, J. W., Z. Fujimoto, H. Mizuno, H. Yamane, T. Yoshida, H. Habe, H. Nojiri, and T. Omori. 2002. Crystallization and preliminary crystallographic analysis of the terminal oxygenase component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10. Acta Crystallogr. Sect. D 58:1350-1352. [DOI] [PubMed] [Google Scholar]
- 43.Noda, Y., S. Nishikawa, K.-I. Shiozuka, H. Kadokuda, H. Nakajima, K. Yoda, Y. Katayama, N. Morohoshi, T. Haraguchi, and M. Yamasaki. 1990. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J. Bacteriol. 172:2704-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noumura, T., H. Habe, J. Widada, J. S. Chung, T. Yoshida, H. Nojiri, and T. Omori. 2004. Genetic characterization of the dibenzofuran-degrading actinobacteria carrying the dbfA1A2 gene homologues isolated from activated sludge. FEMS Microbiol. Lett. 239:147-155. [DOI] [PubMed] [Google Scholar]
- 45.Resnick, S. M., and D. T. Gibson. 1996. Regio- and stereospecific oxidation of fluorene, dibenzofuran, and dibenzothiophene by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. Appl. Environ. Microbiol. 62:4073-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Sato, S. I., J. W. Nam, K. Kasuga, H. Nojiri, H. Yamane, and T. Omori. 1997. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J. Bacteriol. 179:4850-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seeger, M., B. Camara, and B. Hofer. 2001. Dehalogenation, denitration, dehydroxylation, and angular attack on substituted biphenyls and related compounds by a biphenyl dioxygenase. J. Bacteriol. 183:3548-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selifonov, S. A., A. V. Slepenkin, V. M. Adanin, M. Nefedova, and I. I. Starovoitov. 1991. Oxidation of dibenzofuran by Pseudomonas strains harboring plasmids of naphthalene degradation. Mikrobiologiya 60:67-71. (In Russian.) [PubMed] [Google Scholar]
- 51.Taguchi, K., M. Motoyama, and T. Kudo. 2004. Multiplicity of 2,3-dihydroxybiphenyl dioxygenase genes in the Gram-positive polychlorinated biphenyl degrading bacterium Rhodococcus rhodochrous K37. Biosci. Biotechnol. Biochem. 68:787-795. [DOI] [PubMed] [Google Scholar]
- 52.Takagi, T., H. Habe, T. Yoshida, H. Yamane, T. Omori, and H. Nojiri. 2005. Characterization of [3Fe-4S] ferredoxin DbfA3, which functions in the angular dioxygenase system of Terrabacter sp. strain DBF63. Appl. Microbiol. Biotechnol. 68:336-345. [DOI] [PubMed] [Google Scholar]
- 53.Takagi, T., H. Nojiri, T. Yoshida, H. Habe, and T. Omori. 2002. Detailed comparison between the substrate specificities of two angular dioxygenases, dibenzofuran 4,4a-dioxygenase from Terrabacter sp. and carbazole 1,9a-dioxygenase from Pseudomonas resinovorans. Biotechnol. Lett. 24:2099-2106. [Google Scholar]
- 54.Vaillancourt, F. H., G. Labbe, N. M. Drouin, P. D. Fortin, and L. D. Eltis. 2002. The mechanism-based inactivation of 2,3-dihydroxybiphenyl 1,2-dioxygenase by catecholic substrates. J. Biol. Chem. 277:2019-2027. [DOI] [PubMed] [Google Scholar]
- 55.Wilkes, H., R. M. Wittich, K. N. Timmis, P. Fortnagel, and W. Francke. 1996. Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 62:367-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wittich, R.-M., H. Wilkes, V. Sinnwell, W. Francke, and P. Fortnagel. 1992. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 58:1005-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wittich, R. M. 1998. Degradation of dioxin-like compounds by microorganisms. Appl. Microbiol. Biotechnol. 49:489-499. [DOI] [PubMed] [Google Scholar]
- 58.Wittich, R. M., C. Strompl, E. R. B. Moore, R. Blasco, and K. N. Timmis. 1999. Interaction of Sphingomonas and Pseudomonas strains in the degradation of chlorinated dibenzofurans. J. Ind. Microbiol. Biotechnol. 23:353-358. [DOI] [PubMed] [Google Scholar]
- 59.Witzig, R., H. A. H. Aly, C. Strömpl, V. Wray, H. Junca, and D. H. Pieper. 2007. Molecular detection and diversity of novel diterpenoid dioxygenase DitA1 genes from proteobacterial strains and soil samples. Environ. Microbiol. 9:1202-1218. [DOI] [PubMed] [Google Scholar]
- 60.Yamazoe, A., O. Yagi, and H. Oyaizu. 2004. Degradation of polycyclic aromatic hydrocarbons by a newly isolated dibenzofuran-utilizing Janibacter sp. strain YY-1. Appl. Microbiol. Biotechnol. 65:211-218. [DOI] [PubMed] [Google Scholar]