Abstract
To obtain an efficient ethanologenic Escherichia coli strain, we reduced the functional space of the central metabolic network, with eight gene knockout mutations, from over 15,000 pathway possibilities to 6 pathway options that support cell function. The remaining pathways, identified by elementary mode analysis, consist of four pathways with non-growth-associated conversion of pentoses and hexoses into ethanol at theoretical yields and two pathways with tight coupling of anaerobic cell growth with ethanol formation at high yields. Elimination of three additional genes resulted in a strain that selectively grows only on pentoses, even in the presence of glucose, with a high ethanol yield. We showed that the ethanol yields of strains with minimized metabolic functionality closely matched the theoretical predictions. Remarkably, catabolite repression was completely absent during anaerobic growth, resulting in the simultaneous utilization of pentoses and hexoses for ethanol production.
Microorganisms that are specialized to convert sugars into ethanol in the most efficient way are expected to be cells with minimal functionality, equipped only with the specialized catalytic capability for the formation of the desired product and for the replication and renewal of this catalytic function. The creation of a minimal cell that is able to self-assemble and to self-replicate based on a limited number of genes is an intriguing goal in biology. Several approaches have been proposed to realize this goal, using comparative genomic, genetic, or biochemical tools (14-16, 18, 27). It is reasoned that knowledge of the entire parts list of a cell should enable one to combine a minimal set of parts to assemble a functional unit. However, it is unclear how this can be accomplished without solid theoretical principles that define cell function and guide the experimental assembly of parts.
A metabolic network can function according to many different pathway options. Such redundancy of pathways enables cells to compete efficiently and to survive under changing environmental conditions (39). Elementary mode (EM) analysis has emerged as a powerful systems biological tool that rigorously dissects a metabolic network into its basic building blocks (35, 36). The metabolism of a functioning cell has to be viewed as a weighted average of the fluxes through all fundamental pathways (EMs) that its metabolic network supports (40, 43). These EMs therefore represent the inherent building blocks of the metabolic structure. The set of EMs is in fact the parts list for cell function encoded at a higher level in the hierarchy of biological complexity. The quest for the minimal cell can be conducted, therefore, at this functional level. DNA replication and protein synthesis appear to be only supporting functions that ensure the self renewal of cell functionality.
Ethanol has emerged as an important renewable and sustainable energy source that can reduce our reliance on fossil resources. It can be produced from inexpensive, abundant, and renewable feedstocks, including cellulosic and lignocellulosic biomass, by fermentation using microorganisms, and significant advances have already been made (6, 17, 20, 30, 44, 45). The key to this technology is developing efficient and robust microorganisms to convert the biomass-derived hexoses and pentoses to ethanol at the best possible yields and high productivities (33).
To construct a minimal cell that is dedicated to produce ethanol in the most efficient way, we took a top-down approach and started with a complex, functioning Escherichia coli cell that has at its disposition all the numerous metabolic pathway options characteristic of a wild-type cell. We chose E. coli as a model organism to demonstrate this approach because it can degrade a variety of pentoses and hexoses and its genetics can be easily engineered according to rational strain design with well-established molecular techniques.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Table 1 shows a list of bacterial strains and plasmids used in this study. E. coli MG1655 was used as the wild type. All mutants with single deleted genes were obtained from the single-gene-knockout library, the Keio collection (4). These mutants were derived from BW25113, a derivative of MG1655, and constructed by using the technique of one-step disruption of chromosomal genes (9). To construct mutants with multiple deleted genes, all single deleted genes whose parent strains are BW25113 were first transferred into the wild type by generalized P1 transduction. Then mutants with multiple deleted genes were created by multiple steps of P1 transduction from strains with a single deleted gene (41). At each step, the kanamycin cassette was removed from the recipient strain that contained one or more deleted genes by using the temperature-sensitive helper plasmid pFT-A (31). Donor strains used to prepare P1 lysates had a single deleted gene with an intact kanamycin cassette. The PCR test was designed to detect a complete gene disruption by using primers located outside of the undeleted portion of the structural gene (see Table S1 in the supplemental material).
TABLE 1.
List of strains and plasmids
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| MG1655 | Wild type | 5 |
| JW0855 | BW25113, poxB::Kan+ | 4 |
| JW2294 | BW25113, pta::Kan+ | 4 |
| JW1087 | BW25113, ptsG::Kan+ | 4 |
| JW2385 | BW25113, glk::Kan+ | 4 |
| JW1806 | BW25113, manX::Kan+ | 4 |
| TCS062 | MG1655, Δzwf Δndh ΔsfcA ΔmaeB ΔldhA ΔfrdA::Kan− | 4 |
| TCS083 | MG1655, Δzwf Δndh ΔsfcA ΔmaeB ΔldhA ΔfrdA ΔpoxB Δpta::Kan− | This study |
| CT1101 | MG1655, Δzwf Δndh ΔsfcA ΔmaeB ΔldhA ΔfrdA ΔpoxB Δpta ΔptsG Δglk ΔmanX::Kan− | This study |
| pFT-A | Ampr | 31 |
| pLOI297 | pUC18 backbone vector containing pdcZM and adhBZM from Z. mobilis under lac promoter with Ampr and Tetr | ATCC 68239 |
Growth medium.
All controlled batch bioreactor studies used Lauria Bertani (LB) rich medium containing 5 g/liter NaCl, 5 g/liter yeast extract, 10 g/liter trypone, 80 g/liter total sugars (unless otherwise specified), and 10 μg/ml tetracycline. The LB components were autoclaved. The sugars and tetracycline were sterile filtered and added into bioreactors. Growth experiments conducted in baffled shake flasks used defined medium containing 12.8 g/liter Na2HPO4.7H2O, 3 g/liter KH2PO4, 0.5 g/liter NaCl, 1 g/liter NH4Cl, 0.2% (vol/vol) 1 M MgSO4, 0.01 g/liter CaCl2, 0.1% (vol/vol) stock trace metals solution, 1 mg/liter thiamine, 2 g/liter xylose, and 2 g/liter glucose. The stock trace metal solutions consisted of 0.15 g/liter H3BO4, 0.065 g/liter CoSO4, 0.05 g/liter ZnSO4.7H2O, 0.015 g/liter MnCl2.4H2O, 0.015 g/liter NaMo4.2H2O, 0.01 g/liter NiCl2.6H2O, 0.005 g/liter CuSO4.5H2O, and 3 g/liter Fe(NH4) citrate.
Growth in batch bioreactors.
Batch bioreactor experiments were conducted in 10-liter Braun bioreactors (Biostat MD; B. Braun Biotech International, Melsungen, Germany) with a working volume of 6 liters under anaerobic conditions. The temperature and agitation rate were set at 37°C and 200 rpm, respectively. Single colonies were picked from freshly streaked plates and grown overnight in 15-ml tubes containing 5 ml of rich medium. The cultures were then transferred to 250-ml capped shake flasks containing 100 ml of rich medium. Exponential cultures grown in shake flasks (37°C, 225 rpm) were then used for inoculation. The media used for inoculation and for the bioreactors were identical. The initial optical density measured at a 600-nm wavelength (OD600) after inoculation in all batch bioreactors was 0.05. To maintain anaerobic growth conditions, nitrogen was sparged into bioreactors through a 0.2-μm filter at a volumetric flow rate of 100 ml/min at least 4 h before inoculation and throughout the fermentation. The exhaust gas was first passed through an exhaust gas condenser and then a 0.2-μm filter and a pressure regulator and finally into a Prima δ-B mass spectrometer (ThermoOnix, Houston, TX) to analyze gas composition. The reactor gauge pressure was set at 1 lb/in2 to minimize air diffusion into the bioreactors and hence maintain anaerobic growth conditions. pH was controlled at 6.5 by using 6 M NaOH and 40% H3PO4. The anaerobic growth conditions could be confirmed by the absence of oxygen signals from mass spectroscopy. Fermentation was completed when H3PO4 started being added to the bioreactors.
For growth conducted in baffled shake flasks, the procedures used were as previously reported (41). For growth in anaerobic shake tubes, anaerobic 30-ml glass tubes were filled with 20 ml of defined medium, sparged with N2, and sealed with aluminum crimples on a rubber stopper. The initial OD600 after inoculation was set at 0.01. Measurement of the optical density of the anaerobic tubes was carried out directly by using the Spectronic 20D+ (model no. 333182000; Houston, TX) that has a tube adapter.
Analytical techniques.
The optical densities of the cultures were measured at a wavelength of 600 nm in 1-cm cuvettes using a Hewlett Packard 8452A diode array spectrophotometer (Palo Alto, CA). Ten milliliters of a culture was withdrawn periodically from a bioreactor and immediately processed to determine the cell dry weight and the levels of secreted metabolites in the supernatant. First, the sample was spun at 7,000 × g at 4°C for 10 min. Then its supernatant was stored at −20°C for later analysis, and the cell pellet was washed once with deionized water, vacuum filtered, and weighed in a weighing dish after being dried in a 65°C oven for at least 1 day. The formula for weight conversion of optical density is 1 OD600 = 0.259 g/liter (R2 = 0.942). Metabolite concentrations were determined by using a high-pressure liquid chromatography system (Shimadz10A; Shimadzu, Columbia, MD) equipped with an autosampler (SIL-10AF), a cation exchange column (HPX-87H; Bio-Rad, Hercules, CA), and two detectors in series consisting of a UV-visual spectroscopy detector (SPD-10A) and a refractive index detector (RID-10A). Samples from cell supernatants were first filtered through a 0.22-μm filter unit. Then the samples were loaded into the column operated at 65°C. A 5-mM H2SO4 solution was used as the mobile phase and run isocratically at a flow rate of 0.5 ml/min.
Yield calculation and carbon balance.
Ethanol yields on sugars (YETOH/sugars) were determined by the formula YETOH/sugars = rETOH/rsugars (g of ethanol/g of sugars), where rETOH (g of ethanol/liter/h) and rsugars (g of sugars/liter/h) represent the ethanol production rate and sugar consumption rate, respectively. In all experiments, the YETOH/sugars appeared to be constant, since the linear regression of ethanol produced (g/liter) and sugars consumed (g/liter) yielded a perfect fit with R2 > 0.99. Yields of other by-products on sugars were computed in the same way.
The percent of carbon recovery was calculated by the following equation: 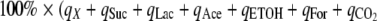 ), where qX, qSuc, qLac, qAce, qETOH, qFor, and
), where qX, qSuc, qLac, qAce, qETOH, qFor, and  (Cmol of product/Cmol of sugars/h) are the specific rates of biomass, succinic acid, lactic acid, acetic acid, ethanol, formic acid, and carbon dioxide, respectively. The specific rates of CO2 were not directly measured but were estimated from the formation of ethanol, succinic acid, and acetic acid. Specifically, the formation of ethanol and acetic acid leads to gains of equimolar amounts of CO2, but the formation of succinic acid under anaerobic growth conditions leads to losses of equimolar amounts of CO2. The carbon recovery rate was close to 100%, as shown in Table S2 in the supplemental material.
(Cmol of product/Cmol of sugars/h) are the specific rates of biomass, succinic acid, lactic acid, acetic acid, ethanol, formic acid, and carbon dioxide, respectively. The specific rates of CO2 were not directly measured but were estimated from the formation of ethanol, succinic acid, and acetic acid. Specifically, the formation of ethanol and acetic acid leads to gains of equimolar amounts of CO2, but the formation of succinic acid under anaerobic growth conditions leads to losses of equimolar amounts of CO2. The carbon recovery rate was close to 100%, as shown in Table S2 in the supplemental material.
E. coli metabolic network.
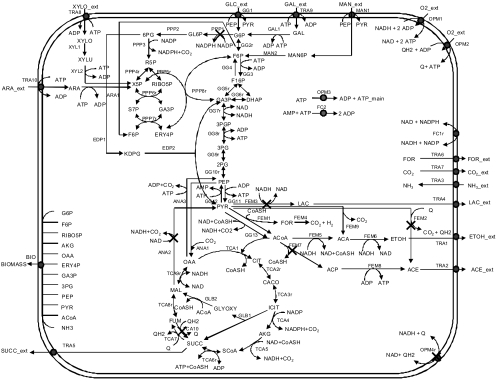
A metabolic network was constructed for E. coli that can grow on pentoses and hexoses, including d-(+)-xylose, l-(+)-arabinose, d-(+)-glucose, d-(+)-galactose, and d-(+)-mannose, by using available public databases (22) and published literature (8, 28), as shown in Fig. 1 and Table S3 in the supplemental material. We considered that the transport of glucose and mannose in the model was facilitated by the phosphoenolpyruvate sugar transferase system (PTS), while the uptake of galactose, xylose, and arabinose was mediated by ABC transporters with high affinity (26). The constructed model represents the intermediary metabolism of E. coli. The validity of the model was experimentally tested in previous studies (41, 43). The pyruvate decarboxylase reaction that converts pyruvate to acetaldehyde was also considered in constructing the model. The pyruvate decarboxylase does not exist in E. coli but was introduced into E. coli through the plasmid pLOI297 (ATCC 68239) (3). The introduction of pyruvate decarboxylase into the strain contributed to the use of the precursor pyruvate for the ethanol production pathway and mimicked the ethanol-producing pathway of native ethanologenic strains such as Zymomonas mobilis and Saccharomyces cerevisiae. It has been reported that pyruvate carboxylase has a higher affinity to pyruvate than pyruvate formate lyase, which converts pyruvate to acetyl coenzyme A (CoA) (19). Furthermore, the conversion of pyruvate into acetaldehyde is not dependent on the cofactor CoA, which can be highly regulated and become limiting. Some reactions in sugar degradation pathways that occur in series without branches have been lumped to simplify the model without affecting the analysis. We calculated all EMs of the E. coli metabolic network using METATOOL 5.0, the currently available, fast, and flexible Matlab-based software package designed to handle complex metabolic networks (42).
FIG. 1.
Metabolic map of E. coli central metabolic network. Deleted reactions in TCS083 are shown next to the symbol X. Listed are glucose-6-phosphate-1-dehydrogenase (PPP1, zwf), NADH dehydrogenase II (OPM4r, ndh), NAD/NADP-dependent malate enzyme (ANA2, sfcA/maeB), d-lactate dehydrogenase (FEM3, ldhA), fumarate reductase (TCA10, frdA), pyruvate oxidase (FEM2, poxB), and phosphate acetyltransferase (FEM7, pta). ETOH, ethanol.
Statistical analysis.
Each case study conducted in batch bioreactor experiments was performed at least in duplicate. For each bioreactor run, three samples were collected and analyzed for each time point during the course of fermentation. The fermentation parameters such as titers and yields are reported as means ± standard deviations of at least six independent measurements from at least two batch bioreactor runs. Each case study conducted in anaerobic shake tubes was performed with at least six replicates.
RESULTS
Pentose utilization.
Xylose and arabinose are the dominant pentoses found in biomass. Therefore, it is important to focus design considerations on these two sugars. Based on the metabolic network considered, we identified 15,185 EMs that E. coli can use to metabolize either xylose or arabinose. Among these, 1,004 EMs can consume either xylose or arabinose under anaerobic conditions. Of these anaerobic EMs, 964 can make ethanol, 443 can synthesize biomass, and 415 can coproduce ethanol and biomass (Table 2). The set of EMs that does not involve biomass production is associated with cell maintenance and/or production of by-products only. These EMs represent the production phase of E. coli when one of the substrates other than the carbon source that is required for growth is depleted. Such a non-growth-associated production phase is possible if functional pathway enzymes are present and the cellular redox balance is met.
TABLE 2.
EMs that utilize different pentoses and hexoses as carbon sources and ranges of ethanol and biomass yields
| No. of reactions deleted | EM characteristica | Value(s) for indicted carbon source
|
|||
|---|---|---|---|---|---|
| Xylose or arabinose | Glucose | Mannose | Galactose | ||
| None | Total EMsb | ||||
| Anaerobic EMs | 1,004 | 5,010 | 2,841 | 1,620 | |
| ETOH-producing anaerobic EMs | 964 | 4,913 | 2,745 | 1,580 | |
| Biomass-producing anaerobic EMs | 443 | 4,157 | 2,134 | 1,297 | |
| Biomass- and ETOH-producing anaerobic EMs | 415 | 4,080 | 2,064 | 1,269 | |
| Ranges of ethanol yield (g ETOH/g sugars)c | 0.00-0.51 | 0.00-0.51 | 0.00-0.51 | 0.00-0.51 | |
| Ranges of biomass yield (g biomass/g sugars)c | 0.00-0.19 | 0.02-0.31 | 0.02-0.31 | 0.02-0.21 | |
| Seven | Total EMsb | 116 | 116 | 116 | 116 |
| Anaerobic EMs | 12 | 12 | 12 | 12 | |
| ETOH-producing anaerobic EMs | 12 | 12 | 12 | 12 | |
| Biomass-producing anaerobic EMs | 4 | 4 | 4 | 4 | |
| Biomass- and ETOH-producing anaerobic EMs | 4 | 4 | 4 | 4 | |
| Ranges of ethanol yield (g ethanol/g sugars)c | 0.45-0.51 | 0.36-0.51 | 0.36-0.51 | 0.44-0.51 | |
| Ranges of biomass yield (g biomass/g sugars)c | 0.12 | 0.30 | 0.3 | 0.15 | |
ETOH, ethanol.
The number of aerobic EMs was calculated as the difference in total EMs and anaerobic EMs.
The data presented is for anaerobic growth conditions.
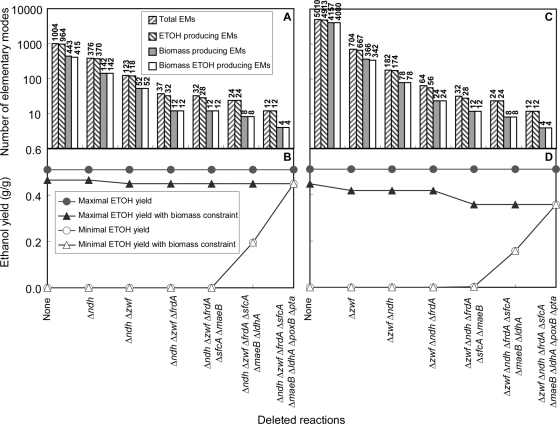
As indicated in Fig. 2A, deletion of a set of seven reactions encoded in eight genes can reduce the total number of anaerobic EMs from 1,004 to 12. All 12 modes produce ethanol, and 4 modes can make biomass; i.e., they support growth. The seven reactions were identified through the application of the following three basic rules to the complete set of EMs. First, the effect of the elimination of individual reactions on the number of remaining EMs was evaluated. For this purpose, the reactions were sorted in increasing-number order of the remaining EMs after the elimination of each reaction. Second, the maximum ethanol and biomass yields were evaluated in the set of remaining pathways. Third, the reaction with the least number of remaining EMs still supporting maximum ethanol and high biomass yields was selected for elimination. Successive application of this algorithm resulted in the set of reactions to be eliminated. Exceptions based on the nature of the identified reaction were made to these selections, since some reactions cannot be easily eliminated.
FIG. 2.
Effect of reaction deletions on the numbers of anaerobic EMs for growth on xylose (A and B) and glucose (C and D). The bars in panels A and C specify the numbers of EMs for strains with deletions of the indicated genes. In each group of bars, the numbers of (i) total modes, (ii) modes that make ethanol, (iii) modes that produce biomass, and (iv) modes that make both biomass and ethanol are listed. The possible maximal and minimal ethanol and biomass yields for xylose (B) and glucose (D) are shown. Note that the minimal yields are pushed toward the upper theoretical limit with increasing numbers of deleted genes. ETOH, ethanol.
The number of functional anaerobic EMs is, in fact, even lower, because 6 of the 12 EMs use the pyruvate dehydrogenase complex, which has been reported to not be active under anaerobic conditions (1). Therefore, deletion of the set of seven reactions results in only six remaining EMs that function under anaerobic conditions. Two of these EMs coproduced biomass and ethanol during the growth phase with an ethanol yield of 0.45 g ethanol/g of sugars (Table 3, Fig. 2B). This finding implies that an engineered strain designed to function only according to this set of pathways always couples cell growth and ethanol production. The other four EMs make only ethanol with or without maintenance energy at the theoretical yield of 0.51 g ethanol/g sugars. Since all remaining pathways generate the same optimum yield, the design criteria were met and it was not necessary to further reduce the number of possible pathways.
TABLE 3.
Stoichiometric equations for the efficient ethanol-producing EMs that use pentoses and hexosesa
| Substrate | Equation | YETOH/sugar (g/g) | No. of EMs |
|---|---|---|---|
| Glucose | 1 glucose = 2 ethanol + 2 CO2 | 0.51 | 3 |
| 1 glucose = 2 ethanol + 2 CO2 + 2 ATP | 0.51 | 1 | |
| 1 glucose + 0.49 NH3 = 1.79 biomass + 1.40 ethanol + 0.11 H2 + 1.41 CO2 | 0.36 | 1 | |
| 1 glucose + 0.49 NH3 = 1.79 biomass + 1.40 ethanol + 0.11 formate + 1.30 CO2 | 0.36 | 1 | |
| Xylose | 1 xylose = 1.67 ethanol + 1.67 CO2 | 0.51 | 3 |
| 1 xylose = 1.67 ethanol + 1.67 CO2 + 0.67 ATP | 0.51 | 1 | |
| 1 xylose + 0.16 NH3 = 0.60 biomass + 1.47 ethanol + 0.04 H2 + 1.47 CO2 | 0.45 | 1 | |
| 1 xylose + 0.16 NH3 = 0.60 biomass + 1.47 ethanol + 0.04 formate + 1.43 CO2 | 0.45 | 1 |
The measure of biomass is 1 Cmol.
Hexose utilization.
The same set of deletion targets was identified for hexoses as the carbon and energy source and is applicable also for the coutilization of both sugar types (Tables 2 and 3 and Fig. 2C and D). It is interesting that the total number of anaerobic EMs is much higher for hexoses than for pentoses. However, after the deletion of two reactions encoded by zwf and ndh, the sets of biomass-producing EMs and EMs that produce both biomass and ethanol became the same for growth on pentoses and hexoses. In addition, growth on the PTS sugars (glucose and mannose) resulted in a lower minimal ethanol yield (0.36 g ethanol/g sugar) than growth on the non-PTS sugars (galactose, xylose, and arabinose) (Table 3). Since growth on the non-PTS sugars requires ATP to transport sugars prior to sugar phosphorylation, less energy is available for biomass synthesis. Therefore, the biomass yield decreases, and more available carbon can be used for ethanol production.
Coutilization of pentose and hexose.
The coutilization of xylose and glucose poses an interesting situation, since the metabolism of individual sugars exhibits opposite flux distributions in parts of the metabolic network. The EM analysis identified 92,593 EMs, which are considerably more than the sum of the total number of EMs that support utilization of each sugar alone (Table 2). The new EMs appear due to the coutilization of both glucose and xylose. Deletion of the same set of 7 reactions resulted in 18 remaining anaerobic EMs; 12 utilize either glucose or xylose individually as presented above, and the other 6 coutilize glucose and xylose. Deletion of this set of reactions effectively eliminated all lower yielding pathways and pushed the range of ethanol yields toward the upper limit (0.36 to 0.51 g ethanol/g sugars) (Table 2).
The set of EMs possible on a mixture of sugars consisting of glucose and pentoses can be further reduced to six by creating a strain unable to use glucose. Such a pentose-specific strain can be realized by removing, in addition to the previously described genes, the glucose phosphotransferase system (ptsG), glucose kinase (glk), and the mannose phosphotransferase system (manX), thus preventing glucose transport into the cell or glucose phosphorylation in the cell.
Strain comparisons.
Over the past 15 to 20 years, after the expression of the foreign ethanol-producing pathway from Zymomonas mobilis in E coli was successful (3), several ethanologenic E. coli strains have been developed to improve ethanol production through numerous rounds of modification. The modifications typically rely on intuitive understanding of cell metabolism and cell behavior (10, 11, 24, 30, 44). Unlike these approaches, our approach was rational, based on EM analysis to design cells with a minimal functionality dedicated to ethanol production. To demonstrate differences between the rationally designed strain TCS083 and other strains developed previously via intuition, we applied EM analysis to describe the effect of the gene mutations in these strains on the range of ethanol yields and on the reduction of inefficient ethanol-producing pathways (Table 4). The results show that the developed strains FBR3 (11), FBR5 (10), KO11 (LYO1) (30, 44), and LY168 (21) still contain a large portion of inefficient ethanol-producing EMs that support a large range of ethanol yields. The existence of inefficient ethanol-producing pathways in these strains can potentially reduce the ethanol yield. For instance, the characterization of KO11 in chemostat studies showed that during growth on xylose, KO11 loses its hyperethanologenicity at the expense of cell growth and acetate synthesis (12). Compared to each of these engineered strains, TCS083 contains a larger number and a different set of deleted reactions. It should be mentioned that unlike TCS083, all of these engineered strains with the exception of LY168 have pflB deleted. This gene belongs to the operon of pflABCD, which encodes pyruvate formate lyase that is active under anaerobic growth conditions (1, 46). According to EM analysis, deletion of this enzyme prevents growth under anaerobic conditions because the synthesis of acetyl-CoA, required for biomass synthesis, is blocked. For this reason, TCS083 does not contain a pflB deletion. However, the normal growth of FBR3, FBR5, and KO11 (LY01) likely still occurs, presumably due to the activity of other subunits of pyruvate formate lyase and the subsequent metabolic evolution of engineered strains (11, 30).
TABLE 4.
Elementary mode analysis of various ethanologenic E. coli strains using glucose as a carbon source under anaerobic conditionsa
| Strain | Total EMs | ETOH yield (g/g) | ETOH-producing EMs | ETOH yield of ETOH-producing EMs (g/g) | Reference(s) |
|---|---|---|---|---|---|
| KO11, LYO1b | 1,835 | 0.00-0.51 | 1,821 | <0.01-0.51 | 30, 44 |
| LY168c | 954 | 0.28-0.51 | 954 | 0.28-0.51 | 21 |
| FBR3d | 4,122 | 0.00-0.51 | 4,102 | <0.01-0.51 | 11 |
| FBR5e | 1,496 | 0.00-0.51 | 1,230 | 0.16-0.51 | 10 |
| TCS083f | 12 | 0.36-0.51 | 12 | 0.36-0.51 | This study |
For some strains, the number of anaerobic EMs is greater than that of ethanol-producing EMs because some elementary modes make by-products. ETOH, ethanol.
KO11 contains ΔfrdABCD ΔpflB::tet+. LY01 is a derivative of KO11 modified through metabolic evolution to improve ethanol tolerance.
LY168 is a derivative of KO11. LY168 contains ΔfrdABCD ΔldhA ΔackA::Tet+.
FBR3 contains ΔldhA ΔpflB.
FBR5 is a derivative of FBR3. FBR5 contains ΔfrdABCD ΔldhA ΔpflB.
TCS083 contains Δzwf Δndh ΔsfcA ΔmaeB ΔldhA ΔfrdA ΔpoxB Δpta::Kan.
Strain construction.
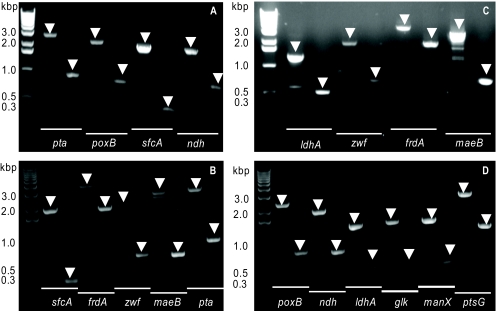
It should be noted that all design considerations up to this point were made on entirely theoretical grounds. This theoretical design strategy was validated by constructing strain TCS083. This strain was derived from the previously described strain TCS062, which already has six genes deleted (41), by adding two additional gene deletions involved in the acetate-producing pathway, poxB and pta. CT1101 is a derivative of TCS083 that possesses three additional deletions, which make it pentose specific. Both TCS083 and CT1101 had all targeted knockout genes removed from their chromosomes, as all DNA fragments were of the expected sizes after PCR amplification with the appropriate primer pairs (Fig. 3).
FIG. 3.
PCR of deleted genes for TCS083 and CT1101, with the wild type used as a positive control. Panels A and C show deleted genes tested for TCS083, including zwf, ndh, sfcA, maeB, ldhA, frdA, poxB, and pta. Panels B and D display deleted genes tested for CT1101, including zwf, ndh, sfcA, maeB, ldhA, frdA, poxB, pta, glk, manX, and ptsG. For each gene tested, the left lane shows the location of an amplified gene for the wild type and the right lane for the mutant. A shift to a smaller band size occurring in a lane of a mutant indicates that the tested gene is deleted. The arrow in each lane points to the location of the expected band size.
Strain characterization.
According to the strain design, TCS083/pLOI297 always coproduces biomass and ethanol in a growth-associated manner. Therefore, we first tested that cell growth and ethanol production are tightly coupled. Growth experiments in anaerobic shake tubes using either glucose or xylose as a carbon source confirmed that the control wild type could grow normally with or without the plasmid pLOI297, as expected. However, the mutant TCS083 could not grow without the plasmid pLOI297, as predicted. Growth was restored only when the plasmid was introduced into the mutant. Therefore, the result is consistent with the design of a cell with minimal metabolic functionality dedicated to ethanol production.
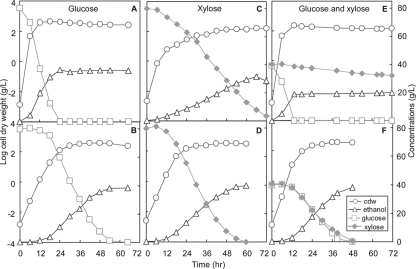
We tested the performance of strain TCS083/pLOI297 together with MG1655/pLOI297 as a control, first on individual xylose and glucose sugars and then on sugar mixtures in controlled batch bioreactors. Figure 4A and B shows the batch reactor time profiles of the wild-type MG1655/pLOI297 and TCS083/pLOI297 for growth on 80 g/liter glucose. MG1655/pLOI297 achieved an ethanol yield of 0.46 ± 0.01 (g ethanol/g glucose) and an ethanol titer of 36.53 ± 0.31 g/liter. Under identical growth conditions, TCS083/pLOI297 reached a cell dry weight of 3.11 ± 0.47 g/liter. Because the stoichiometries of the two biomass-producing pathways and the four remaining pathways that produce only ethanol are known (Table 3), we could compute on the basis of biomass formed that 16% ± 2% of the total glucose was consumed in the pathway supporting growth and 84% ± 2% in the pathways producing only ethanol. Furthermore, with these weighting factors together with the pathway stoichiometries, we could predict that 80 g/liter of consumed glucose should result in a final ethanol titer of 39.20 ± 0.34 g/liter. This value is in excellent agreement with the measured ethanol titer of 38.77 ± 0.63 (g/liter) (Table 5).
FIG. 4.
Time profiles for glucose, xylose, cell dry weight (cdw), and ethanol for the wild type (upper three panels) and mutant TCS083/pLOI297 (lower three panels). The strains were anaerobically cultivated in controlled 10-liter bioreactors where they were sparged with nitrogen. The initial sugar concentration was 80 g/liter. In an experiment with mixed sugars, 40 g/liter of each sugar was provided. Note that the figure shown is representative of a single-batch bioreactor run of duplicate experiments and that each data point represents the mean ± standard deviation of three independent measurements.
TABLE 5.
Prediction of ethanol titers of TCS083/pLOI297 and CT1101/pLOI297a
| Strain | Substrate(s) | Titer of biomass (g/liter) | w1c (%) | w2c (%) | Predicted amt of ethanolb (g/liter) | Amt of ethanol measured (g/liter) |
|---|---|---|---|---|---|---|
| TCS083/pLOI297 | Glucose | 3.11 ± 0.47 | 16 ± 2 | 84 ± 2 | 39.20 ± 0.34 | 38.77 ± 0.63 |
| TCS083/pLOI297 | Xylose | 3.25 ± 0.12 | 42 ± 1 | 58 ± 1 | 39.22 ± 0.09 | 39.07 ± 0.59 |
| TCS083/pLOI297 | Glucose and xylose | 3.54 ± 0.16 | 32 ± 1 | 68 ± 1 | 39.46 ± 0.57 | 38.81 ± 0.91 |
| CT1101/pLOI297 | Glucose and xylose | 1.48 ± 0.02 | 39 ± 1 | 61 ± 1 | 19.32 ± 0.73 | 20.03 ± 0.18 |
Predictions are based on the overall stoichiometric equations of the efficient ethanol-producing pathways (Table 1). The values are reported as means ± standard deviations of six independent measurements from duplicate runs of batch bioreactors.
The predicted amount of ethanol can be calculated as follows: 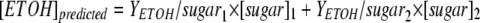 , where [ETOH]predicted (g/liter) is the ethanol titer predicted, [sugar] (g/g) is the amount of sugar consumed, and YETOH/sugar (g ethanol/g sugar) is the theoretical yield of ethanol on sugar calculated by using stoichiometric equations. Subscripts “1” and “2” refer to the growth-associated phase and non-growth-associated phase, respectively.
, where [ETOH]predicted (g/liter) is the ethanol titer predicted, [sugar] (g/g) is the amount of sugar consumed, and YETOH/sugar (g ethanol/g sugar) is the theoretical yield of ethanol on sugar calculated by using stoichiometric equations. Subscripts “1” and “2” refer to the growth-associated phase and non-growth-associated phase, respectively.
w1 and w2 are the weighting factors of glucose (xylose) channeled into either the growth-associated ethanol-producing pathway or the non-growth-associated ethanol-producing pathway, respectively. w1 can be calculated as follows: w1 = 100% × [sugar]1/[sugar], and w2 = 100% − w1. [sugar]1 can be determined by the following formula: 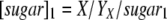 , where X (g/liter) is the experimentally determined biomass and
, where X (g/liter) is the experimentally determined biomass and 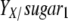 (g biomass/g sugar) is the yield of biomass on sugar calculated by using stoichiometric equations.
(g biomass/g sugar) is the yield of biomass on sugar calculated by using stoichiometric equations.
MG1655/pLOI297 exhibited a completely different phenotype for growth on 80 g/liter xylose. The xylose consumption rate was significantly slower than the glucose consumption rate. As shown in Fig. 4C, MG1655/pLOI297 could not completely consume all xylose, with about 4% of the xylose remaining unused after 72 h. MG1655/pLOI297 achieved an ethanol yield of 0.46 ± 0.01 (g ethanol/g xylose) and an ethanol titer of 33.08 ± 2.15 (g/liter). In contrast, the TCS083/pLOI297 phenotype for growth on 80 g/liter xylose was similar to that for growth on glucose. It could consume all xylose after 54 h and reached a cell dry weight of 3.25 ± 0.12 g/liter (Fig. 4D). From the pathway stoichiometries, we deduced again that 42% ± 1% of the xylose consumed was used in the reactions leading to biomass formation. The stoichiometry and the weighted average of the two pathway types resulted in a final ethanol titer of 39.22 ± 0.09 g/liter, which is again very close to the experimentally determined ethanol titer of 39.07 ± 0.59 (g/liter) (Table 5).
Determination of the ethanol production kinetics on a mixture of 40 g/liter glucose and 40 g/liter xylose showed that MG1655/pLOI297 consumed first glucose and then xylose in a sequential manner. As shown in Fig. 4E, it took MG1655/pLOI297 only about 12 h to completely consume glucose but 72 h to consume just 20% of 40 g/liter xylose. The slow xylose consumption rate of the wild type after the glucose was consumed was probably due to a combination of cellular inhibitory effects resulting from the formation of by-products such as succinic acid, lactic acid, acetic acid, ethanol, and formic acid. MG1655/pLOI297 achieved an ethanol yield of 0.40 ± 0.02 (g ethanol/g sugar) and an ethanol titer of 18.66 ± 1.01 (g/liter). In contrast, TCS083/pLOI297 did not show pronounced diauxic growth behavior. It could simultaneously consume both glucose and xylose and completely utilize all sugars within 48 h. TCS083/pLOI297 reached a biomass concentration of 3.54 ± 0.16 g/liter (Fig. 4F). Assuming that both sugars equally contributed to the biomass formation reaction, this translates to the use of 32% ± 1% of the sugars for growth and the conversion of the rest into ethanol at theoretical values to reach a final titer of 39.46 ± 0.57 g/liter. The experimentally determined final ethanol titer was 38.81 ± 0.91 (g/liter) (Table 5).
Catabolite repression.
Fig. 5C shows the consumption of only pentoses by CT1101/pLOI297 in a mixture of xylose and glucose. This strain evidently has all catabolite repression removed and can grow on pentoses even in the presence of glucose. The strain reached a biomass concentration of 1.48 ± 0.02 g/liter. According to pathway stoichiometries, 39 ± 1% of the total xylose consumed was used for growth. The computed final ethanol titer was 19.32 ± 0.73 g/liter and compared well with the experimental value of 20.03 ± 0.18 (g/liter) (Table 5).
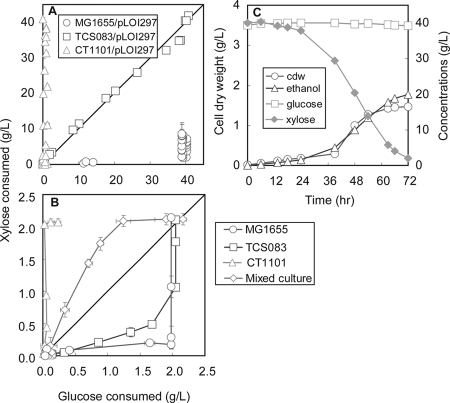
FIG. 5.
Time profiles for glucose, xylose, cell dry weight (cdw), and ethanol for mutant CT1101/pLOI297 growing on a sugar mixture (lower panel) (see the legend to Fig. 4 for growth conditions). The relationship between consumed xylose and consumed glucose is shown for anaerobic growth (upper left panel) and aerobic growth (upper right panel) for the wild-type MG1655 and mutant strains TCS083 and CT1101 and a 1:4 initial mixture of strains TCS083 and CT1101.
The diauxic growth behavior of the wild type and the absence of catabolite repression is clearly shown in the phase plot that relates the consumed xylose to the consumed glucose (Fig. 5A). In the wild type, the glucose was completely exhausted before consumption of xylose could be observed, while the constructed mutant TCS083 consumed both sugars simultaneously and strain CT1101 used only xylose even when glucose was present.
DISCUSSION
The presented minimal strains have to be considered minimal in terms of metabolic functionality that has been optimized for ethanol formation. Their genomes still contain most of the genes present in wild-type cells. However, the combination of expressed gene products supports only a minimum number of possible pathways. It has recently been shown that removal of unneeded genome sequences has little effect on the phenotype of the remaining cells. For instance, removal of 10% of the genome does not result in any noticeable change in the specific growth rate of the cells (32). This finding indicates that the cellular synthetic capacity can easily handle the synthetic burden of additional DNA. Therefore, a reduction of the genome to the absolute minimum is not expected to increase the specific growth rates of cells. The experimental performance of the constructed strains validates the theoretical EM-based design of the strains with minimized functionality. The results agree with those of a previous study involving a minimal strain, optimized for cell growth, that was constructed with six knockout mutations (41). While the wild-type strain has over 1,000 pathways available to convert the sugars into ethanol, evidently the derived minimal strains can do so only on the basis of six well-defined pathways.
The theoretical framework for designing a minimal cell is based on EM analysis. It can dissect a metabolic network into unique, nondecomposable pathways that present all possible physiological states of cells at steady-state conditions (35, 36). A different approach based on flux balance analysis using optimization frameworks such as OptKnock and MOMA has also been applied by other research groups to identify gene knockouts to optimize both growth and production of a metabolite (2, 7, 13, 37). This approach does not identify the complete set of alternative optimal solutions nor other subsets of suboptimal solutions. Therefore, even though the set of deleted genes identified by flux balance analysis can identify an optimal pathway, there is no guarantee that the engineered cells can function according to this optimal pathway.
A remarkable phenotype of TCS083/pLOI297 is the ability to utilize both xylose and glucose simultaneously (Fig. 4F and 5A). This phenotype clearly does not exist in the wild type (Fig. 4E, 5A), which shows pronounced diauxic growth behavior presumably mediated through glucose catabolite repression (25). It was previously shown that in E. coli, deletion of the ptsG gene can remove catabolite repression at the expense of a lower glucose uptake rate (29). In our minimal strains, ptsG is still present, but the specific growth rate on glucose is reduced, indicating that catabolite repression is closely linked to the growth dynamics, and likely to the glycolytic flux, that determine the concentrations and the effects of specific regulation factors in the strains. This finding is consistent with experiments that have shown that blockage of the glycolytic flux triggers the degradation of ptsG mRNA (23). Moreover, when it grows at a high specific growth rate, our minimal strain does show pronounced catabolite repression and preferential glucose consumption under aerobic growth conditions comparable to those of the wild-type strain (Fig. 5B).
TCS083/pLOI297 was rationally designed for efficient ethanol production based on EM analysis. The performance of TCS083/pLOI297 was tested experimentally. The mutant outperformed the wild type in the efficient conversion of sugars into ethanol and closely matched the theoretical prediction. Based on published data, we compared the mutant TCS083/pLOI297 with other engineered ethanologenic E. coli strains such as KO11 (44) and FBR5/pLOI297 (29) under similar growth conditions. For a similar period of xylose fermentation, both KO11 and FBR5/pLOI297 achieved a similar ethanol yield of 0.46 ± 0.01 (g/g), for a range of 75 to 90 g/liter xylose consumed, while TCS083/pLOI297 reached a higher ethanol yield of 0.49 ± 0.01 (g/g). In addition, FBR5/pLOI297 obtained an ethanol yield of 0.46 ± 0.03 (g/g) for a similar period of glucose fermentation, while TCS083/pLOI297 achieved a higher ethanol yield of 0.49 ± 0.01 (g/g). Furthermore, TCS083/pLOI297 can simultaneously consume pentoses and hexoses, while neither KO11 nor FBR5 can. Overall, TCS083/pLOI297 appears to perform more favorably than KO11 and FBR5/pLOI297 under similar growth conditions. The performance of TCS083/pLOI297 will be further characterized and compared with those of KO11 and FBR5/pLOI297 under different growth conditions.
TCS083 is expected to be useful for other biotechnological applications. Under completely aerobic growth conditions, the biomass yields of TCS083 are maximized, similar to those of TCS062, since CO2 is the only by-product and its formation is minimized. In large-scale reactor operations, oxygen-limiting growth conditions are typically created due to mixing inhomogeneities, resulting in the secretion of acetic acid. In such cases, TCS083 should perform better than TCS062, since acetate-producing pathways are disrupted. Indeed, shake flask experiments, in which completely aerobic conditions cannot be achieved, have already confirmed that TCS083 does indeed outperform not only TCS062 and the wild-type strain but also the industrial strain BL21 (38) and the reduced-genome strain MDS42 (32) typically used for protein production (Fig. 6).
FIG. 6.
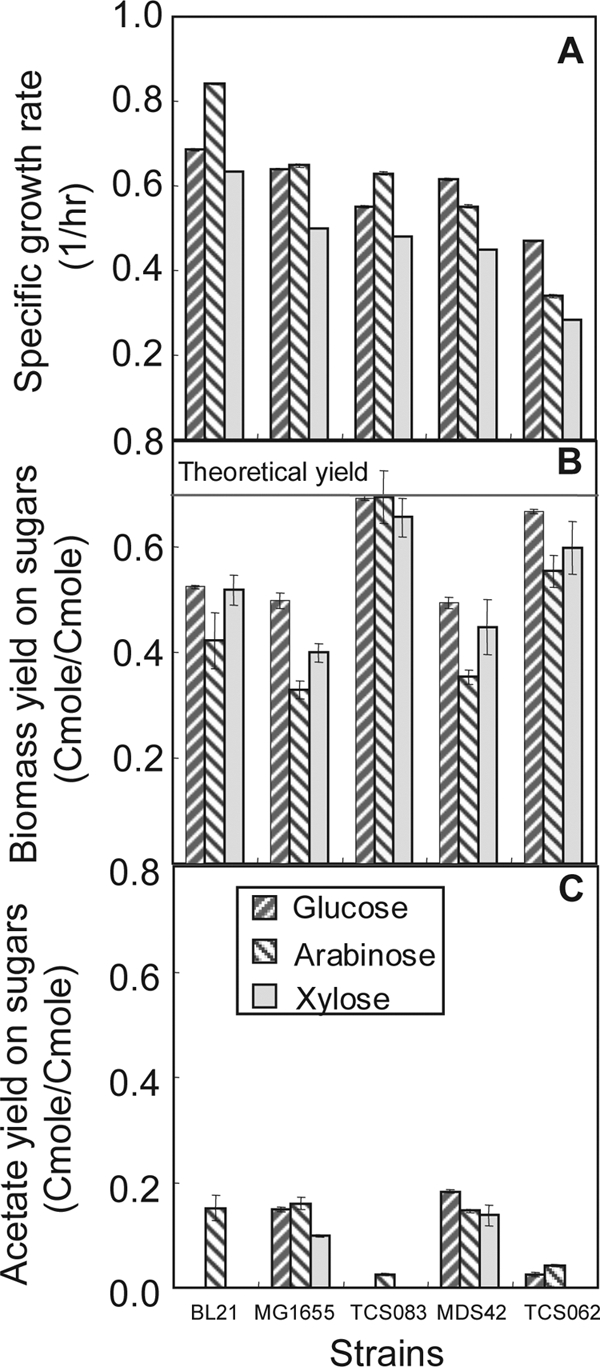
Growth characteristics of different E. coli strains, including BL21 MDS42, MG1655, TCS062, and TCS083. (A) Specific growth rates. (B) Biomass yields on glucose. (C) Acetate yields on glucose. The experiments were conducted in baffled shake flasks containing defined medium supplied with various pentoses and hexoses under aerobic conditions. Each value represents the mean ± standard deviation of the results of triplicate experiments.
Lignocellulosic hydrolysates considered for ethanol production contain a mixture of pentoses and hexoses. Due to the effect of carbon catabolite repression when microorganisms grow in mixtures of sugars containing glucose (34) and the sequential utilization of sugar mixtures, ethanol productivity is severely affected. In addition, different compositions of pentoses and hexoses present in different sources of biomasses, for example, also pose a challenge to the control of efficient ethanol productivities. To address this problem, the efficient pentose-specific ethanologenic E. coli strain CT1101 can be combined with a hexose-utilizing organism to arbitrarily adjust the individual sugar consumption rates in a mixed culture, as the relative consumption rates depend on the ratio of organisms used.
The design of a strain with minimal metabolic functionality must always be coordinated with the purpose of the functionality. Here, the purpose was the most efficient production of ethanol. However, the methodology applied is general and should prove useful for the design and construction of many different minimal strains tailored for applications in biotechnology and for fundamental studies. Such strains are attractive due to the efficiency and simplicity of their metabolic pathways.
Supplementary Material
Acknowledgments
We acknowledge the National Institutes of Health (grant GM077529) and the Digital Technology Center of the University of Minnesota for financial support.
We thank the Minnesota Supercomputing Institute for use of their supercomputing facilities.
Footnotes
Published ahead of print on 18 April 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alexeeva, S., B. de Kort, G. Sawers, K. J. Hellingwerf, and M. J. de Mattos. 2000. Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. J. Bacteriol. 182:4934-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alper, H., Y. Jin, J. F. Moxley, and G. Stephanopoulos. 2005. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 7:155-164. [DOI] [PubMed] [Google Scholar]
- 3.Alterthum, F., and L. O. Ingram. 1989. Efficient ethanol production from glucose, lactose, and xylose by recombinant Escherichia coli. Appl. Environ. Microbiol. 55:1943-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:msb4100050-E1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 6.Bothast, R. J., N. N. Nichols, and B. S. Dien. 1999. Fermentations with new recombinant organisms. Biotechnol. Prog. 15:867-875. [DOI] [PubMed] [Google Scholar]
- 7.Burgard, A. P., P. Pharkya, and C. D. Maranas. 2003. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 84:647-657. [DOI] [PubMed] [Google Scholar]
- 8.Carlson, R., and F. Srienc. 2004. Fundamental Escherichia coli biochemical pathways for biomass and energy production: identification of reactions. Biotechnol. Bioeng. 85:1-18. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dien, B. S., N. N. Nichols, P. J. O'Bryan, and R. J. Bothast. 2000. Development of new ethanologenic Escherichia coli strains for fermentation of lignocellulosic biomass. Appl. Biochem. Biotechnol. 84-86:181-196. [PubMed] [Google Scholar]
- 11.Dien, B. S., R. B. Hespell, H. A. Wyckoff, and R. J. Bothast. 1998. Fermentation of hexose and pentose sugars using a novel ethanologenic Escherichia coli strain. Enzyme Microb. Technol. 23:366-371. [Google Scholar]
- 12.Dumsday, G. J., B. Zhou, W. Yaqin, G. A. Stanley, and N. B. Pamment. 1999. Comparative stability of ethanol production by Escherichia coli KO11 in batch and chemostat culture. J. Ind. Microbiol. Biotechnol. 23:701-708. [DOI] [PubMed] [Google Scholar]
- 13.Fong, S. S., A. P. Burgard, C. D. Herring, E. M. Knight, F. R. Blattner, C. D. Maranas, and B. O. Palsson. 2005. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol. Bioeng. 91:643-648. [DOI] [PubMed] [Google Scholar]
- 14.Forster, A. C., and G. M. Church. 2006. Towards synthesis of a minimal cell. Mol. Syst. Biol. 2:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 16.Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, N. W., Z. Chen, and A. P. Brainard. 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64:1852-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 19.Ingram, L. O., and T. Conway. 1988. Expression of different levels of ethanologenic enzymes from Zymomonas mobilis in recombinant strains of Escherichia coli. Appl. Environ. Microbiol. 54:397-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingram, L. O., H. C. Aldrich, A. C. Borges, T. B. Causey, A. Martinez, F. Morales, A. Saleh, S. A. Underwood, L. P. Yomano, S. W. York, J. Zaldivar, and S. Zhou. 1999. Enteric bacterial catalysts for fuel ethanol production. Biotechnol. Prog. 15:855-866. [DOI] [PubMed] [Google Scholar]
- 21.Jarboe, L. R., T. B. Grabar, L. P. Yomano, K. T. Shanmugan, and L. O. Ingram. 2007. Development of ethanologenic bacteria. Adv. Biochem. Eng. Biotechnol. 108:237-261. [DOI] [PubMed] [Google Scholar]
- 22.Keseler, I. M., J. Collado-Vides, S. Gama-Castro, J. Ingraham, S. Paley, I. T. Paulsen, M. Peralta-Gil, and P. D. Karp. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33:D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimata, K., Y. Tanaka, T. Inada, and H. Aiba. 2001. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J. 20:3587-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindsay, S. E., R. J. Bothast, and L. O. Ingram. 1995. Improved strains of recombinant Escherichia coli for ethanol production from sugar mixtures. Appl. Microbiol. Biotechnol. 43:70-75. [DOI] [PubMed] [Google Scholar]
- 25.Magasanik, B., and F. C. Neidhardt. 1956. Inhibitory effect of glucose on enzyme formation. Nature 178:801-802. [DOI] [PubMed] [Google Scholar]
- 26.Mayer, C., and W. Boos. March 2005, posting date. Chapter 3.4.1. Hexose/pentose and hexitol/pentitol metabolism. In R. Curtiss III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 27.Mushegian, A. R., and E. V. Koonin. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 93:10268-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neidhardt, F. C., J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.). 1987. Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, DC.
- 29.Nichols, N. N., B. S. Dien, and R. J. Bothast. 2001. Use of catabolite repression mutants for fermentation of sugar mixtures to ethanol. Appl. Microbiol. Biotechnol. 56:120-125. [DOI] [PubMed] [Google Scholar]
- 30.Ohta, K., D. S. Beall, J. P. Mejia, K. T. Shanmugam, and L. O. Ingram. 1991. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl. Environ. Microbiol. 57:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pósfai, G., M. D. Koob, H. A. Kirkpatrick, and F. R. Blattner. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pósfai, G., G. Plunkett III, T. Fehér, D. Frisch, G. M. Keil, K. Umenhoffer, V. Kolisnychenko, B. Stahl, S. S. Sharma, M. de Arruda, V. Burland, S. W. Harcum, and F. R. Blattner. 2006. Emergent properties of reduced-genome Escherichia coli. Science 312:1044-1046. [DOI] [PubMed] [Google Scholar]
- 33.Ragauskas, A. J., C. K. Williams, B. H. Davison, G. Britovsek, J. Cairney, C. A. Eckert, W. J. Frederick, Jr., J. P. Hallett, D. J. Leak, C. L. Liotta, J. R. Mielenz, R. Murphy, R. Templer, and T. Tschaplinski. 2006. The path forward for biofuels and biomaterials. Science 311:484-489. [DOI] [PubMed] [Google Scholar]
- 34.Saier, M. H., Jr., T. M. Ramseier, and J. Reizer. 1996. Regulation of carbon utilization, p. 1325-1343. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 35.Schuster, S., C. Hilgetag, J. H. Woods, and D. A. Fell. 2002. Reaction routes in biochemical reaction systems: algebraic properties, validated calculation procedure and example from nucleotide metabolism. J. Math. Biol. 45:153-181. [DOI] [PubMed] [Google Scholar]
- 36.Schuster, S., D. A. Fell, and T. Dandekar. 2000. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat. Biotechnol. 18:326-332. [DOI] [PubMed] [Google Scholar]
- 37.Segrè, D., D. Vitkup, and G. M. Church. 2002. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl. Acad. Sci. USA 99:15112-15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiloach, J., J. Kaufman, A. S. Guillard, and R. Fass. 1996. Effect of glucose supply strategy on acetate accumulation, growth, and recombinant protein production by Escherichia coli BL21 (λDE3) and Escherichia coli JM109. Biotechnol. Bioeng. 49:421-428. [DOI] [PubMed] [Google Scholar]
- 39.Stelling, J., U. Sauer, Z. Szallasi, F. J. Doyle III, and J. Doyle. 2004. Robustness of cellular functions. Cell 118:675-685. [DOI] [PubMed] [Google Scholar]
- 40.Stelling, J., S. Klamt, K. Bettenbrock, S. Schuster, and E. D. Gilles. 2002. Metabolic network structure determines key aspects of functionality and regulation. Nature 420:190-193. [DOI] [PubMed] [Google Scholar]
- 41.Trinh, C. T., R. Carlson, A. Wlaschin, and F. Srienc. 2006. Design, construction and performance of the most efficient biomass producing E. coli bacterium. Metab. Eng. 8:628-638. [DOI] [PubMed] [Google Scholar]
- 42.von Kamp, A., and S. Schuster. 2006. Metatool 5.0: fast and flexible elementary modes analysis. Bioinformatics 22:1930-1931. [DOI] [PubMed] [Google Scholar]
- 43.Wlaschin, A. P., C. T. Trinh, R. Carlson, and F. Srienc. 2006. The fractional contributions of elementary modes to the metabolism of Escherichia coli and their estimation from reaction entropies. Metab. Eng. 8:338-352. [DOI] [PubMed] [Google Scholar]
- 44.Yomano, L. P., S. W. York, and L. O. Ingram. 1998. Isolation and characterization of ethanol-tolerant mutants of Escherichia coli KO11 for fuel ethanol production. J. Ind. Microbiol. Biotechnol. 20:132-138. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, M., C. Eddy, K. Deanda, M. Finkelstein, and S. Picataggio. 1995. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267:240-243. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, J., and K. Shimizu. 2004. The effect of pfl gene knockout on the metabolism for optically pure d-lactate production by Escherichia coli. Appl. Microbiol. Biotechnol. 64:367-375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.