Abstract
Mercury-resistant bacteria may be important players in mercury biogeochemistry. To assess the potential for mercury reduction by two subsurface microbial communities, resistant subpopulations and their merA genes were characterized by a combined molecular and cultivation-dependent approach. The cultivation method simulated natural conditions by using polycarbonate membranes as a growth support and a nonsterile soil slurry as a culture medium. Resistant bacteria were pregrown to microcolony-forming units (mCFU) before being plated on standard medium. Compared to direct plating, culturability was increased up to 2,800 times and numbers of mCFU were similar to the total number of mercury-resistant bacteria in the soils. Denaturing gradient gel electrophoresis analysis of DNA extracted from membranes suggested stimulation of growth of hard-to-culture bacteria during the preincubation. A total of 25 different 16S rRNA gene sequences were observed, including Alpha-, Beta-, and Gammaproteobacteria; Actinobacteria; Firmicutes; and Bacteroidetes. The diversity of isolates obtained by direct plating included eight different 16S rRNA gene sequences (Alpha- and Betaproteobacteria and Actinobacteria). Partial sequencing of merA of selected isolates led to the discovery of new merA sequences. With phylum-specific merA primers, PCR products were obtained for Alpha- and Betaproteobacteria and Actinobacteria but not for Bacteroidetes and Firmicutes. The similarity to known sequences ranged between 89 and 95%. One of the sequences did not result in a match in the BLAST search. The results illustrate the power of integrating advanced cultivation methodology with molecular techniques for the characterization of the diversity of mercury-resistant populations and assessing the potential for mercury reduction in contaminated environments.
Mercury contamination is a widespread environmental problem due to the combined action of release processes such as volcanic eruptions and combustion of fossils fuels and atmospheric transport. Mercury bioaccumulates in food webs and is of concern at higher trophic levels because of its neurotoxicological effects even at low concentrations.
Mercury resistance has been described in several eubacterial phyla (Firmicutes, Actinobacteria, and Proteobacteria) and in several archaean genomes (3, 19). Resistance to mercury is conferred by the mer operon, where the merA-encoded mercuric reductase reduces Hg2+ (aq) to volatile and less toxic elemental Hg° (g) (3).
Exposure of microbial communities to mercury typically results in an initial decline in bacterial numbers, followed by a rapid growth of mercury-resistant subpopulations (2, 21). As a consequence of the outgrowth of only the resistant fraction of the community, the genetic diversity of mercury-acclimated communities is generally relatively low. Although the community's diversity may slowly recover (21), it may be lowered by a factor of as much as 1,000 (9).
By having evolved resistance mechanisms to detoxify several chemical forms of mercury, resistant microbes may play an important role in mercury biogeochemistry in mercury-contaminated environments (18). Thus, exploitation of microbial mercury resistance may potentially be used as a method of detoxifying mercury-contaminated sites. Several factors, however, influence microbial reduction processes. The bioavailability of mercury determines how much of the total Hg present is accessible to microorganisms. The bioavailability of mercury is dependent on the nature and concentration of the binding phase, which in turn are controlled by the redox status (14). Another important parameter is the ability of microbial communities to volatilize mercury, i.e., the abundance, diversity, and activity of the mercury-resistant members of the communities.
The aims of the present work were to characterize the diversity of mercury-resistant heterotrophic bacterial populations of subsurface mercury-contaminated soils and obtain an initial impression of the diversity of their merA genes. Since the characterization of microbial community structure by cultivation-independent approaches does not allow discrimination between mercury-resistant and mercury-sensitive bacteria and since traditional cultivation favors fast-growing and dominant species, we applied an integrated approach that combined an advanced cultivation methodology (7) with molecular techniques. Preincubation under simulated natural conditions in a soil substrate membrane system (SSMS) prior to plating on standard medium induced the growth of hard-to-culture mercury-resistant bacteria and enabled us to account for the majority of the mercury-resistant heterotrophic bacterial populations. Furthermore, the method permitted the discovery of new merA gene sequences.
MATERIALS AND METHODS
Soil characteristics.
Soil from two depths (35 to 55 cm and 80 to 100 cm) was collected at a mercury-contaminated site at the Lower East Fork Poplar Creek floodplain, Oak Ridge, TN. The soil was passed through a 2-mm-mesh sieve and maintained at 4°C until use.
The total concentration of Hg was measured with a Jerome 431-X Mercury Vapor Analyzer after shaking the soil samples in aqua regia (3 parts concentrated HCl to 1 part concentrated HNO3) as described by Kriger and Turner (15).
The bioavailable concentration of mercury was measured with a mer-lux whole-cell biosensor assay as described by Rasmussen et al. (22). Briefly, 1 g of soil was shaken with 10 ml of sterile water in a 300-ml Erlenmeyer flask (300 rpm, 15 min). Large soil particles were removed by centrifugation for 10 min at 12,000 × g (4°C). Appropriate dilutions of the soil slurry were added to 6-ml polystyrene tubes (Falcon; BD, Franklin Lakes, NJ) and mixed with 107 Escherichia coli HMS174/pRB28 biosensor cells. Light emission was recorded as the number of relative light units per 30 s with a BG-P portable luminometer (MGM Instruments, Hamden, CT) and correlated to a standard curve based on known concentrations of Hg.
Total numbers of bacteria in the soils were determined by acridine orange (AO) direct counting (AODC) by the procedure described below for the determination of numbers of microcolony-forming units (mCFU).
Cultivation of mercury-resistant bacteria.
Mercury-resistant soil bacteria were cultivated and isolated by two different approaches, (i) direct plating on 10% tryptic soy broth (TSB) agar (TSA) enriched with 4 μg Hg2+ ml−1 and (ii) preincubation for up to 28 days in an SSMS (7), prior to plating on 10% TSA medium enriched with 4 μg Hg2+ ml−1.
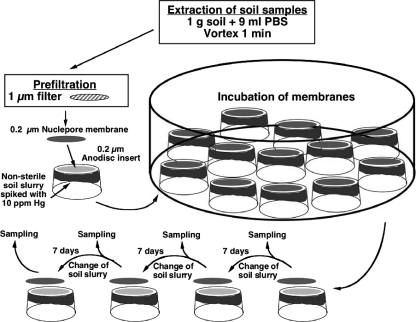
Samples were prepared by extracting 1 g soil with 9 ml salt buffer [KH2PO4 (0.25 g liter−1), MgSO4·7 H2O (0.125 g liter−1), NaCl (0.12 5 g liter−1), (NH4)2SO4 (0.2 g liter−1)] and vortexing for 1 min at maximum velocity on a Vibrofix VF1 Electronic vortexer (IKA Labortechnik, Stauffen, Germany). To remove large soil particles and clay material, extracts were prefiltered through a 1.0-μm syringe filter (Cameo; GE Osmonics). Subsamples of the prefiltered extracts were then either serially diluted and spread on 1/10-strength TSA or diluted with 10 ml salt buffer and filtered onto 0.2-μm polycarbonate membrane filters (25 mm in diameter; Nuclepore). The polycarbonate membranes with the bacterial cells facing upward were placed on the fixed 0.22-μm Anopore disks of inverted 25-mm Nunc tissue culture inserts (Nunc A/S, Roskilde, Denmark) containing a nonsterile soil substrate. The soil substrate was prepared by mixing 4 g air-dried soil from the same depths as the samples with 2 ml distilled MilliQ water spiked with HgCl2 (final concentration, 10 μg Hg2+ g−1). The wetted soil was placed inside the tissue culture inserts and gently vortexed until the soil texture was broken down to a muddy slurry. Replicate SSMSs were incubated in the dark at room temperature in closed containers with a moist filter paper to prevent desiccation of the soil substrate. At each sampling time (every 7 days), triplicate membranes were sacrificed for the determination of numbers of mCFU, extraction of DNA for denaturing gradient gel electrophoresis (DGGE), and isolation of culturable bacteria. The remaining nonsampled membranes were transferred to new tissue culture inserts containing fresh soil slurry. The experimental setup of the SSMS is outlined in Fig. 1. The rationale for pregrowing bacteria on the Nuclepore membranes was to induce growth by mimicking natural growth conditions. Since the bacteria were growing on nutrients diffusing up from the soil slurry, both the source and the concentration of nutrients were natural due to competition with the bacteria in the slurry. Furthermore, cross-feeding was allowed while, on the other hand, potential waste products were removed by diffusion into the soil slurry.
FIG. 1.
Experimental setup and sampling of the SSMS for inducing the growth of hard-to-culture mercury-resistant soil bacteria. Subsamples of 100 μl (20 μl for membranes to be incubated for 21 to 28 days) of the soil extract were diluted with 10 ml salt buffer before being filtered onto polycarbonate membranes. PBS, phosphate-buffered saline.
Determination of numbers of mCFU.
Microcolonies on the polycarbonate membranes were stained with AO by floating the membranes (microcolonies facing up) on a 0.025% AO solution for 3 min. Excess AO was removed by floating membranes on water for 2 min before mCFU were counted at a 630× magnification with a Zeiss Axioplan MC 100 epifluorescence microscope. Clusters of four or more cells were considered to be mCFU.
DGGE analysis.
The genetic diversity of the microcolonies on the polycarbonate membranes was assessed by DGGE. Microcolonies were dislodged in 1 ml sterile MilliQ water by vortexing for 1 min at maximum velocity. After removal of the filter, the cell suspension was centrifuged for 5 min at 12,000 × g. The supernatant was discarded, and the pellet was resuspended in 100 μl sterile MilliQ water. The entire volume was used for DNA extraction with the FastDNA SPIN Kit for Soil equipment as recommended by the manufacturer (Bio 101, Inc., Carlsbad, CA). Purified DNA was stored at −20°C.
PCR mixtures (50 μl) consisted of 1 μl template DNA, 1 U Taq polymerase (Roche, Mannheim, Germany), 2.5 pM each deoxynucleoside triphosphate (dNTP), and 2 μl of each primer solution (5 pmol μl−1). For a description of the primers used (P2 and P3), see reference 16. After PCR (5 min at 94°C, followed by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min; the last cycle was followed by 8 min at 72°C), products were verified by agarose gel electrophoresis.
DGGE was performed on a D-GENE System (Bio-Rad). PCR products (400 ng DNA in 25 μl) were loaded onto 7.5% (wt/vol) polyacrylamide gels made with denaturing gradients ranging from 40 to 70% (where 100% contained 7 M urea and 40% formamide). Gels were made with the Bio-Rad DGGE kit as described by the manufacturer. The electrophoresis was run at 60°C for 16 h at 70 V. After electrophoresis, gels were soaked in SYBR gold for 1 h (1:10,000 dilution; Molecular Probes, Eugene, OR) and digital images were obtained. Preliminary studies had shown very high reproducibility among the triplicate membranes. Hence, only one membrane was sacrificed for DGGE analysis at each sampling time in the experiment reported.
Extraction of mCFU for isolation of culturable bacteria on standard medium.
Bacteria on the polycarbonate membranes were dislodged in 1 ml salt buffer by vortexing for 1 min at maximum velocity. Appropriate dilutions were prepared in salt buffer, and 100 μl of each dilution was spread onto both 10% TSA and 10% TSA amended with 4 μg Hg2+ ml−1. To ensure that slow-growing strains were not overlooked, plates were incubated at ambient temperature and CFU were counted daily until the number of colonies became constant.
At each sampling, up to 100 colonies were picked randomly from the mercury-containing agar plates and streaked onto new plates. The isolates were restreaked at least three times to ensure purity. Isolates were stored frozen at −80°C in 30% (vol/vol) glycerol in liquid 10% TSB amended with Hg2+ (4 μg ml−1).
Cell material from all isolates was transferred into a 1.5-ml Eppendorf tube containing 100 μl distilled water. Tubes were vortexed and boiled in a heating block at 102°C for 10 min. After centrifugation (12,000 × g, 10 min, 4°C), 70 μl of the supernatant was transferred to a new 1.5-ml Eppendorf tube and stored at −20°C for later use as a DNA template for PCRs.
Genetic fingerprinting of isolates by RAPD.
All isolates were characterized by their random amplification of polymorphic DNA (RAPD) band patterns according to Hansen et al. (10). RAPD-PCR mixtures (50 μl) consisted of 1 U Taq polymerase (Roche, Mannheim, Germany), 2.5 pM each dNTP, 2 μl template DNA, and 1.2 μM OPA-09 primer (5′-GGGTAACGCC-3′; Qiagen, Cologne, Germany). PCR was performed on a PCR Express (Hybaid), i.e., 30 s at 4°C, followed by 45 cycles of 94°C for 5 s, 32°C for 1 min, and 72°C for 2 min. PCR products were visualized on a 1.5% agarose gel (SeaKem GTG) in 1× Tris-borate-EDTA buffer. Gels were stained with ethidium bromide, and isolates having identical band patterns were identified and grouped.
Sequencing of the 16S rRNA genes of bacterial isolates.
DNA coding for the 16S rRNA gene was amplified by PCR (5 min at 94°C, followed by 35 cycles of 94°C for 1 min, 46°C for 1 min, and 72°C for 3 min; the last cycle was followed by 10 min at 72°C) with primers GM3F and GM4R, which amplify the fragment encompassing positions 8 to 1507 (17). The PCR mixture (50 μl) consisted of 1 μl template DNA, 1 U Taq polymerase (Roche, Mannheim, Germany), 2.5 pM each dNTP, and 2 μl of each primer solution (5 pmol μl−1). Purification of the PCR product and sequencing were performed by Macrogen Inc. (Seoul, South Korea) in one direction with the GM3F primer.
16S rRNA gene sequences were trimmed at conserved regions, resulting in fragments of approximately 860-bp lengths. Sequences were aligned against the nearest neighbors found in GenBank by a BLAST search (1). A bootstrap analysis with 1,000 sample replications was performed with ClustalW (1.83) (27).
Sequencing of merA genes.
Selected isolates representing different phylogenetic groups were grown in liquid 10% TSB amended with 2 μg Hg2+ ml−1 (25°C, 160 rpm) until dense growth was observed. Genomic DNA was extracted with the High Pure PCR Template Kit (Roche Diagnostics GmbH, Mannheim, Germany) (isolates SEQ 1, 11, 21, 23, 24, and 25) or by boiling (isolates SEQ 16 and 22). Amplification of merA was performed with Phusion DNA polymerase (Finnzymes, Espoo, Finland). The Phusion PCR mixture contained 2 μl extracted DNA, appropriate phylum-specific merA primers (19) (final concentration, 0.5 μM), nucleotides (200 μM), HF buffer (1×), polymerase (0.02 U μl−1), and distilled H2O to a final volume of 30 μl. For high-GC DNA (all except Firmicutes and Bacteroidetes), 3% dimethyl sulfoxide was used in the PCR according to the manufacturer's protocol (Finnzymes, Espoo, Finland). The PCR cycling conditions varied with the primers used. All PCRs included an initial denaturing step at 98°C for 3 min, followed by 38 cycles of denaturing, annealing, and elongation in two or three steps, after which the PCR product was stored at −20°C until further use. The denaturing step in the 38 cycles was always conducted at 98°C for 10 s. For Alphaproteobacteria, annealing and elongation occurred at 72°C for 22 s (primers Al-Fw and Al-Rv; amplicon of approximately 812 bp). For Beta- and Gammaproteobacteria, annealing was at 62°C for 10 s and elongation was at 72°C for 42 s (primers BG-Fw349 and #54; amplicon of approximately 1,225 bp). For Firmicutes, annealing was at 63°C for 10 s and elongation was at 72°C for 18 s (primers Fir-Fw and Fir-Rv1892; amplicon of approximately 455 bp). For Actinobacteria, annealing was at 66°C for 10 s and elongation was at 72°C for 18 s (primers Act-Fw and Act-Rv; amplicon of approximately 391 bp). Primers targeting merA of Bacteroidetes were developed on the basis of the only known putative merA gene of the Bacteroidetes phylum found in the sequenced genome of marine isolate Leeuwenhoekiella blandensis (20). Primers Bact-Fw436 (5′-GGT GGC ACT TGT GTT AAT G-3′) and Bact-Rv (5′-CTT GCA CAA CAA CTC AAT TTA G-3′) were tested on genomic DNA extracted from L. blandensis. Annealing and elongation occurred at 59°C for 10 s and 72°C for 40 s, respectively (expected amplicon of approximately 1,295 bp).
Entire PCR mixtures were gel electrophoresed (1.0% agarose), and following ethidium bromide staining, merA amplicons were cut out and purified with Gel-Out (AA Biotechnology, Gdynia, Poland). Sequencing was done by Macrogen Inc. (Seoul, South Korea). The chromatograms were edited with FinchTV (www.geospiza.com), and similarity to merA genes or proteins was examined by BLASTN and BLASTP searches (1).
Nucleotide sequence accession numbers.
The 33 different 16S rRNA gene sequences found in this study have been deposited in GenBank and given accession numbers DQ401833 to DQ401840 (CFU SEQ 1 to 8) and DQ401841 to DQ401865 (SEQ 1 to 25). The merA sequences have been deposited as EU332700 (SEQ 11), EU332701 (SEQ 16), EU332702 (SEQ 21), EU332703 (SEQ 23), EU332704 (SEQ 24), and EU332705 (SEQ 25).
RESULTS
Soil characteristics.
The total mercury concentration at 35- to 55-cm depth was almost eight times higher than at 80 to 100 cm (Table 1). Similarly, the concentration of bioavailable mercury was highest in the upper soil (Table 1). In both soils, however, the concentration of bioavailable mercury was less than 0.02% of the total concentration.
TABLE 1.
Characteristics of the soils used in this study
| Characteristic | Value at a soil depth (cm) of:
|
|
|---|---|---|
| 35-55 | 80-100 | |
| Total Hg (mean no. of μg/g soil ± SEM) | 7.6 ± 1.0 | 1.0 ± 0.2 |
| Bioavailable Hg (mean no. of ng/g soil ± SEM) | 0.8 ± 0.4 | BDa |
| Culturability, unfiltered (%)b | 0.03 | 0.006 |
| Culturability, 1.0-μm filtrate (%)b | 0.08 | 0.02 |
| Mercury resistance (%)c | 2.9 | 2.3 |
BD, below the detection limit of 0.2 ng Hg/g soil.
Calculated as CFU/AODC.
Calculated as CFU-Hgr/CFU.
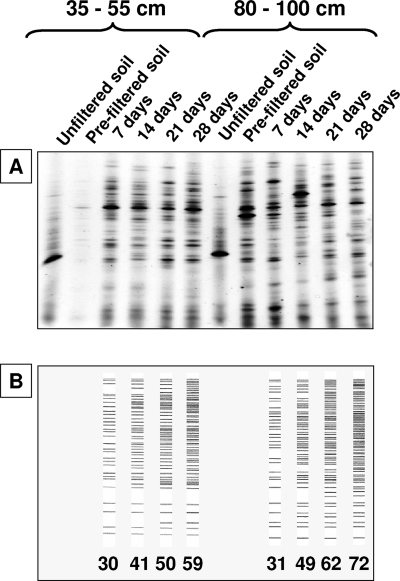
The total bacterial density as determined by AODC in the soils ranged between 1.0 × 109 cells g−1 (80 to 100 cm) and 1.3 × 109 cells g−1 (35 to 55 cm). Approximately 99.4% of the bacteria were removed by the 1.0-μm prefiltration step. The bacteria removed probably belonged to dominant bacterial populations, as evidenced by a comparison of DGGE band patterns obtained before and after filtration (see Fig. 3A). The fraction of directly culturable bacteria (CFU/AODC) varied between 0.006% and 0.03%, with the highest culturability at a 35- to 55-cm depth (Table 1). In the 1.0-μm-filtered fraction, culturability was higher than in the unfiltered sample, ranging from 0.02% (80 to 100 cm) to 0.08% (35 to 55 cm).
FIG. 3.
Genetic diversity as assessed by DGGE profiling. (A) DGGE profiles of microcolonies on polycarbonate membranes and unfiltered and prefiltered soil samples. (B) Cumulative numbers of DGGE bands observed during the incubation of the membranes.
The level of mercury resistance was found to be relatively low. At both soil depths, the percentage of the total number of CFU that were Hg resistant (CFU-Hgr/CFU) was less than 3% (Table 1).
Number of mCFU on membranes.
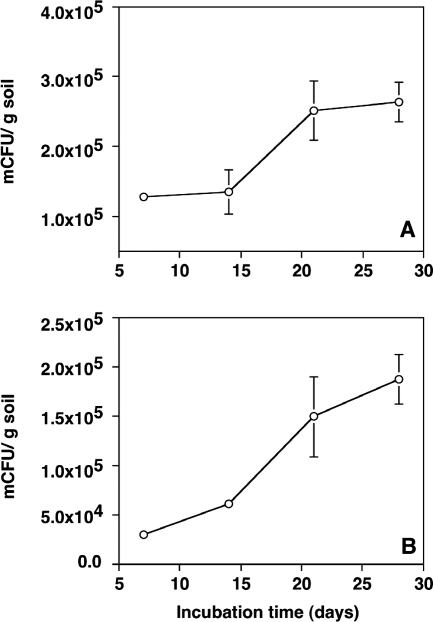
The bacterial cells on the Nuclepore membranes formed distinct microcolonies with diverse morphologies. At the 35- to 55-cm depth, the number of mCFU doubled (t test; P < 0.01) from 1.3 × 105 ± 0.1 × 105 to 2.6 × 105 ± 0.3 × 105 mCFU g−1 between days 7 and 28 (Fig. 2A). All microcolonies were mercury resistant, as evidenced by plating of dislodged cells on 10% TSA with and without 4 μg Hg2+ ml−1 (data not shown). On day 28, the numbers of mercury-resistant mCFU accounted for almost 3.5% of the total bacterial counts in the 1.0-μm soil filtrate. For comparison, the number of mercury-resistant CFU obtained by traditional direct plating only accounted for approximately 0.002%. Thus, the numbers of mercury-resistant mCFU were up to 1,580 times higher than the numbers of resistant CFU.
FIG. 2.
Density of mCFU formed on polycarbonate membranes during incubation in the SSMS. (A) Sample from a 35- to 55-cm depth. (B) Sample from an 80- to 100-cm depth. Error bars indicate the standard error of the mean.
In the 80- to 100-cm soil, the number of mercury-resistant mCFU increased by a factor of 6 (t test; P < 0.01), from 3.1 × 104 ± 0.7 × 104 to 1.9 × 105 ± 0.3 × 105 mCFU g−1 (Fig. 2B), and at day 28 was equivalent to 3% of the total number of bacteria. The number of mercury-resistant CFU obtained by direct plating, on the other hand, only accounted for approximately 0.001%. Thus, the number of mercury-resistant mCFU was more than 2,800-fold higher than the number of resistant CFU.
Genetic diversity of microcolonies on membranes.
The diversity of the mCFU on the polycarbonate membranes changed during incubation (Fig. 3). At first glance, the DGGE band patterns appeared rather similar throughout the 28-day incubation period (Fig. 3A). To enable a more detailed assessment of the potential succession in the genetic diversity of the microcolonies, the cumulative number of DGGE bands was determined (Fig. 3B). As shown in Fig. 3B, the number of bands identified increased with incubation time. In the 35- to 55-cm soil, the total number of bands recorded after 28 days was twice as high as the number observed on day 7. In the 80- to 100-cm soil, the number of bands increased by a factor of 2.3.
Genetic fingerprinting of isolated bacteria.
A total of 403 bacteria survived subculturing and were isolated to purity on 10% TSA amended with 4 ppm Hg2+ following preincubation in the SSMS. In addition, 165 isolates were obtained by the direct-plating approach.
The bacterial isolates obtained by the SSMS approach resulted in 133 different RAPD fingerprint groups, each containing between 1 and 82 isolates. Of the different groups, 77 were unique to the 35- to 55-cm soil depth while 46 were unique to the 80- to 100-cm soil depth. Only 10 RAPD fingerprints were found at both soil depths. The isolates from the direct cultivation gave rise to 30 different RAPD fingerprints.
Partial 16S rRNA gene sequencing showed that several RAPD fingerprint groups could be attributed to the same species. Thus, the 133 groups from the SSMS approach resulted in the identification of 25 different partial 16S rRNA gene sequences while the 30 different groups from the direct-plating approach revealed 8 different 16S rRNA gene sequences. Sequencing of multiple members within a RAPD fingerprinting group always resulted in the same 16S rRNA gene sequence.
During incubation in the SSMS, new isolates (16S rRNA gene sequences) continued to emerge throughout the incubation period (Table 2). Thus, similar to the increasing number of observed DGGE bands, plating of dislodged cells on standard medium suggested a succession in the diversity of the microcolonies on the membranes. Some isolates were found throughout the incubation period (e.g., SEQ 5), whereas others were only found by the end (e.g., SEQ 16 and 22). Of the 25 different sequences, 9 were exclusively found in the 35- to 55-cm soil, 8 were only found in the 80- to 100-cm soil, while 8 were found at both soil depths. Eleven of the sequences were unique for a single sampling time and soil depth (Table 2).
TABLE 2.
Distribution over time of the 25 different 16S rRNA gene sequences obtained from cultivable bacteria following SSMS preincubation
| Isolate or parameter | Bacterial phyluma | Sampleb from 35-55 cm on day:
|
Sampleb from 80-100 cm on day:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 7 | 14 | 21 | 28 | 7 | 14 | 21 | 28 | ||
| SEQ 1 | CFB | + | + | + | |||||
| SEQ 2 | CFB | + | + | ||||||
| SEQ 3 | CFB | − | |||||||
| SEQ 4 | Betaproteobacteria | + | + | + | + | + | |||
| SEQ 5 | Betaproteobacteria | + | + | + | + | + | + | + | |
| SEQ 6 | Betaproteobacteria | + | + | ||||||
| SEQ 7 | Betaproteobacteria | − | |||||||
| SEQ 8 | Betaproteobacteria | − | |||||||
| SEQ 9 | Betaproteobacteria | + | + | ||||||
| SEQ 10 | Betaproteobacteria | − | |||||||
| SEQ 11 | Betaproteobacteria | − | |||||||
| SEQ 12 | Betaproteobacteria | + | + | + | |||||
| SEQ 13 | Betaproteobacteria | + | + | + | |||||
| SEQ 14 | Betaproteobacteria | + | + | + | + | + | + | ||
| SEQ 15 | Betaproteobacteria | − | |||||||
| SEQ 16 | Betaproteobacteria | − | |||||||
| SEQ 17 | Betaproteobacteria | + | + | + | + | ||||
| SEQ 18 | Gammaproteobacteria | − | |||||||
| SEQ 19 | Gammaproteobacteria | + | + | ||||||
| SEQ 20 | Gammaproteobacteria | − | |||||||
| SEQ 21 | Alphaproteobacteria | − | |||||||
| SEQ 22 | Firmicutes | − | |||||||
| SEQ 23 | Actinobacteria | + | + | + | + | + | |||
| SEQ 24 | Actinobacteria | + | + | ||||||
| SEQ 25 | Actinobacteria | + | + | ||||||
| No. of sequences | 8 | 3 | 8 | 12 | 3 | 7 | 11 | 7 | |
| No. of 1st appearances | 8 | 2 | 2 | 5 | 3 | 6 | 6 | 1 | |
CFB, Cytophaga-Flavobacterium-Bacteroidetes. Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria are subphyla of Proteobacteria.
+, presence of a specific sequence; −, unique sequence not seen at another sampling time.
Phylogenetic comparison of culturable bacteria acquired by SSMS and direct plating.
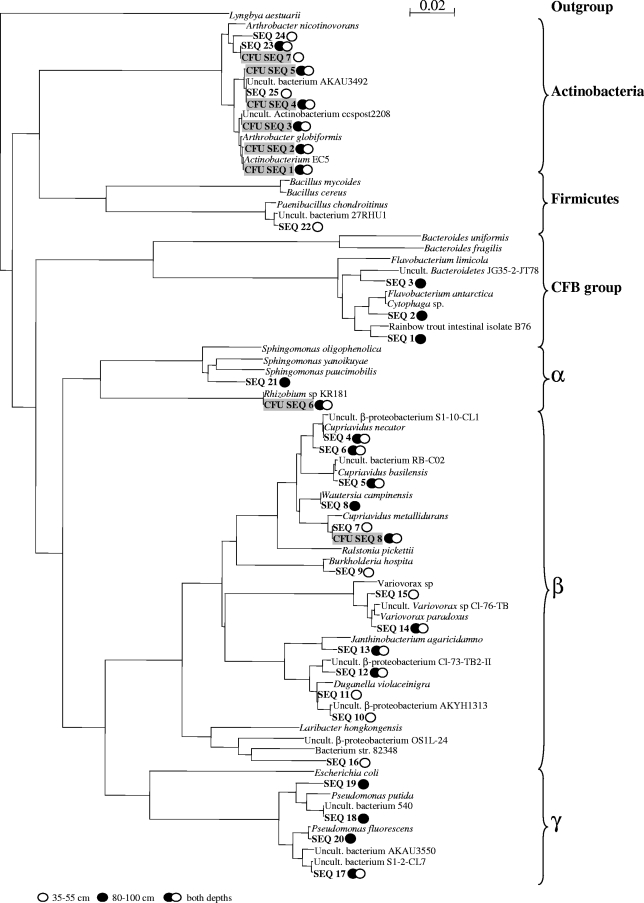
The diversity of the isolates obtained by the SSMS approach was higher than that of the isolates obtained by direct cultivation. This suggests that preincubation in the SSMS induced the growth of bacteria that were not readily cultivable on standard medium (Table 3 and Fig. 4). Three different phylogenetic groups (Actinobacteria and Alpha- and Betaproteobacteria) were identified on the basis of direct cultivation. By contrast, six different phylogenetic groups (Actinobacteria, Firmicutes, Bacteroidetes, and Alpha-, Beta-, and Gammaproteobacteria) were represented if the soil bacteria were preincubated in the SSMS prior to plating. Not only were the species compositions of the isolates from the two cultivation approaches different, the representations of the individual phylogenetic groups also differed (Table 3). Actinobacteria were the dominant group among the readily culturable bacteria, whereas Betaproteobacteria were most common among the SSMS isolates. In only three cases were identical isolates retrieved by both the SSMS and direct-cultivation approaches (Fig. 4).
TABLE 3.
Diversity of soil isolates obtained by direct cultivation on 10% TSA and by preincubation in an SSMS prior to plating on 10% TSA
| Bacterial phyluma | % of total community after:
|
|
|---|---|---|
| Direct cultivation | SSMS preincubation | |
| Actinobacteria | 93 | 11 |
| Alphaproteobacteria | 2 | 0.5 |
| Betaproteobacteria | 5 | 73 |
| Gammaproteobacteria | 11 | |
| CFB | 0.5 | |
| Firmicutes | 4 | |
CFB, Cytophaga-Flavobacterium-Bacteroidetes. Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria are subphyla of Proteobacteria.
FIG. 4.
Neighbor-joining tree showing the phylogeny of the 16S rRNA gene sequences of the bacteria isolated. The tree has arbitrarily been rooted on the cyanobacterium Lyngbya aestuarii. Gray shading indicates isolates found by direct plating on 10% TSA. CFB, Cytophaga-Flavobacterium-Bacteroidetes. Uncult., environmental clones that have not been cultivated.
merA gene sequences.
Partial sequencing of merA from selected isolates led to the discovery of new merA sequences, expanding the known diversity of the locus. PCR products were obtained for Alpha- and Betaproteobacteria and Actinobacteria but not for Bacteroidetes (SEQ 1) and Firmicutes (SEQ 22). The similarity to sequences found in the databases ranged between 89 and 95% at the DNA level and from 58 to 94% at the amino acid level (Table 4). One of the sequences obtained did not result in a match in the BLAST search (SEQ 21; Table 4).
TABLE 4.
Similarities of partial merA gene fragments to sequences reported in the literature
| Isolate | Phyluma | Size of merA sequence (bp) | BLAST result
|
|||
|---|---|---|---|---|---|---|
| Similarity (%)
|
Protein accession no.b | Closest relative | ||||
| DNA | Amino acids | |||||
| SEQ 1 | CFB | |||||
| SEQ 11 | Betaproteobacteria | 1,138 | 89 | 94 | YP_743748 | Nitrosomonas eutropha |
| SEQ 16 | Betaproteobacteria | 1,153 | 100 | 100 | AAR31066 | Ralstonia eutropha |
| SEQ 21 | Alphaproteobacteria | 706 | 58 | ABO45918 | Sphingomonas sp. | |
| SEQ 22 | Firmicutes | |||||
| SEQ 23 | Actinobacteria | 359 | 93 | 93 | ABO77624 | Unknown |
| SEQ 24 | Actinobacteria | 359 | 92 | 93 | ABO77624 | Unknown |
| SEQ 25 | Actinobacteria | 359 | 92 | 93 | ABO77624 | Unknown |
CFB, Cytophaga-Flavobacterium-Bacteroidetes. Betaproteobacteria and Alphaproteobacteria are subphyla of Proteobacteria.
Accession number of closest related merA protein.
TheActinobacteria merA genes were identical in isolates SEQ 24 and SEQ 25, whereas SEQ 23 had four unique nucleotides within the 359 sequence. The partial protein sequences of all three isolates, however, did not differ, as determined by virtual translation (http://www.expasy.ch/tools/dna.html). The Actinobacteria merA sequences were most closely related to cloned merA sequences obtained from topsoil from the same location as the subsoils used in this study (19). Blasting of the merA gene of alphaproteobacterial isolate SEQ 21 did not result in a match, but the partial protein sequence showed the highest similarity to the merA gene of another Sphingomonas isolate from Lower East Fork Poplar Creek floodplain soil (19). The merA gene of one of the two betaproteobacterial isolates (SEQ 16) was identical to the omnipresent Tn501 merA gene found on, e.g., pJP4 (5), whereas the other showed the highest similarity to a merA gene found in Nitrosomonas eutropha (YP_743748).
DISCUSSION
The results presented here demonstrated that preincubation under natural conditions in the SSMS was suitable as a cultivation strategy to characterize subsurface mercury-resistant bacterial communities. A diverse collection of mercury-resistant heterotrophic bacteria was obtained, and new merA sequences, expanding the known diversity of the merA locus, were discovered.
Molecular methods are ideal to acquire an overview of the total species diversity of a microbial community and to assess the diversity of functional genes (e.g., merA) among the members of the community. However, it is not possible to link the functional genes to specific members of the bacterial community by molecular approaches. Discrimination between mercury-resistant and mercury-sensitive subpopulations requires a cultivation-based approach. Obvious drawbacks of traditional cultivation-based approaches are the well-known facts that less than 1% of the soil bacteria can readily be cultivated on standard medium and that the culturable proportion tends to be unrepresentative and less diverse than the total phylogenetic diversity, as determined by DNA-based molecular methods (13, 23).
Recently, methods that take advantage of growing bacteria under conditions that simulate the natural environment, or in an environment with limited nutrients, have enabled the growth of bacteria that have been recalcitrant to cultivation on standard media (6, 11, 25). To bypass the limitations of traditional cultivation and, hence, ensure the inclusion of hard-to-culture soil bacteria, we applied an SSMS in which the bacteria were preincubated under simulated natural conditions prior to plating on standard medium. The SSMS has previously been used to cultivate methanotrophs (26) and hitherto uncultured bacteria of candidate division TM7 (7). We used an approach in which polycarbonate membranes were transferred to new sediment slurry every 7 days. This change-of-slurry approach was selected because a preliminary experiment, in which the membranes were incubated on the same soil slurry for 28 days, had indicated that the diversity (DGGE profiles) of the bacteria growing on the membranes was lower if the soil slurry was not changed on a weekly basis (data not shown). Possibly, extended incubation on the same medium resulted in a depletion of nutrients at the slurry-membrane interface, limiting the growth of the bacteria on the membranes. The advantage of applying a continuous-cultivation approach has also been suggested by Bollmann et al. (4), who demonstrated that continuous cultivation in diffusion chambers may adapt some microorganisms for growth on standard nutrient-rich media.
The percentage of the total number of readily culturable bacteria that were mercury resistant (CFU-HgR/CFU) was 2.3 to 2.9% (Table 1). If this frequency of resistance among the readily culturable component of the soil bacteria is representative of the total community, the density of all mercury-resistant bacteria can be estimated as AODC × (CFU-Hgr/CFU), i.e., 2.2 × 105 and 1.4 ×105 cells g−1 for the 35- to 55-cm and 80- to 100-cm depths, respectively. These values are equivalent to the numbers of mCFU (Fig. 2), suggesting 100% recovery of the mercury-resistant bacteria as microcolonies on the polycarbonate membranes. This high recovery rate stands in contrast to the findings of Kaeberlein et al. (13). They used diffusion chambers simulating a marine sediment and found recovery rates ranging from 2 to 40% of the inoculated cells. Also, with respect to an increase in culturability relative to traditional plating, the SSMS method was very efficient, as we found an up to 2,800-fold increase in culturability. For comparison, Connon and Giovannoni (6), by using an extinction culturing technique to cultivate marine bacteria in liquid culture in microtiter wells at nearly in situ substrate concentrations, found a 14- to 1,400-fold increase in culturability relative to traditional cultivation techniques. Thus, the SSMS method proved highly suitable for growing hard-to-culture mercury-resistant bacteria.
The diversity (species richness) of the cultivable mercury-resistant bacteria obtained after preincubation in the SSMS included six different phyla (Table 3). In comparison, a clone library generated from the same soils revealed 15 different phyla (unpublished data). Thus, despite the fact that the mercury-resistant bacteria were obtained from 1.0-μm-prefiltered soil samples, the diversity of the mercury-resistant bacteria constituted a relatively large fraction (∼40%) of the total community diversity. Since the diversity of bacterial communities generally is negatively affected by mercury stress (9, 21), this high diversity of the mercury-resistant subpopulations was surprising. Mercury has been shown to negatively affect the conjugal transfer of plasmids (12). However, considering the long history of mercury contamination, the low concentration of bioavailable mercury, and the likely presence of nonexposed microniches, a plausible explanation for the high diversity of the mercury-resistant subpopulations is horizontal transfer of mobile genetic elements carrying the mer operon (19, 24).
Some of the isolates obtained following preincubation in the SSMS had their merA genes partially sequenced. Most of the gene sequences obtained were distinct from sequences in the NCBI database, with similarities lower than 93% (Table 4). By a cultivation-independent approach, Oregaard and Sørensen (19) examined the diversity of merA genes from the same soils as in this study. They found that only 1 out of 62 cloned Proteobacteria-related merA sequences was more than 95% similar to a GenBank sequence and concluded that sequences obtained from cultivable soil bacteria are not representative of what is found in nature. The fact that we, in agreement with these previous data, found merA gene sequences with very low similarity to known sequences emphasizes that the SSMS approach for cultivating bacteria does indeed overcome many of the limitations of traditional cultivation by including hitherto uncultured bacteria.
Mercury resistance is not well described in the Bacteroidetes phylum. To the best of our knowledge, only one report of a mercury-resistant Flavobacterium exists (8). Recently, however, a putative merA gene was identified in a Flavobacterium isolate from the Mediterranean Sea (20) and the strain was confirmed to be mercury resistant by growth on marine broth agar supplemented with 30 μM HgCl2 (19). We used primers targeting the merA locus of this isolate to amplify the mercuric reductase gene of one of the three Bacteroidetes isolates (SEQ 1) but, despite several attempts, failed to obtain a PCR fragment. Similar to the failure to amplify the Bacteroidetes-type merA locus, no PCR fragment from the Firmicutes isolate (SEQ 22) was observed. The failure to amplify merA from SEQ 1 and SEQ 22 could be due to problems with the extraction of DNA from the cells. However, as DNA was easily extracted for 16S rRNA gene sequencing, we consider this unlikely. The PCR primer sequences used were based on known merA sequences present in available databases. Probably, the sequence divergence between our isolates and the known merA sequences was too large for annealing of the primers. In accordance with our results, Oregaard and Sørensen (19) were unable to detect Firmicutes-type merA in the same soils with the same primer set as in this study but by a culture-independent approach.
The three Actinobacteria isolates all tested positive for merA; however, the similarity to database sequences was low. The partial merA sequences were most similar to cloned merA sequences found in topsoil (0- to 5-cm depth) at the same site as our soil samples (19) Interestingly, Oregaard and Sørensen (19), by using a cultivation-independent approach, were, contrary to this study, unable to detect Actinobacteria-type merA in subsurface soil.
Disregarding the isolates that did not yield a PCR product, the partial merA sequence of the Alphaproteobacteria isolate (SEQ 21) had the lowest similarity (Table 4) to the existing sequences. The partial merA sequence of another Sphingomonas sp. isolated from the same site as in our study by Oregaard and Sorensen (unpublished) showed only very limited similarity. The primers used to amplify the partial merA gene of the Alphaproteobacteria were recently designed on the basis of putative merA genes in the sequenced genomes of Alphaproteobacteria (19). Therefore, caution should be taken when identifying the sequences as merA genes, since none of these putative merA genes have been cloned and expressed heterologously to confirm phenotypic mercury resistance. Phenotypic mercury resistance has, however, been described in one marine Alphaproteobacteria isolate (28), but a merA gene was not recognized.
Conclusion.
Use of the SSMS microcultivation approach significantly enhanced the culturability of hard-to-culture mercury-resistant subsurface soil bacteria and not only permitted a successful characterization of the diversity of the resistant populations but also led to the discovery of merA gene sequences exceeding the diversity of known merA genes. Because mercury-resistant isolates were obtained, a further delineation of the merA genes is possible. Future work should focus on cloning the mer operons of Alphaproteobacteria, Bacteroidetes, and Firmicutes isolates and test if they confer mercury resistance when expressed in mercury-sensitive strains. Also, the activity of isolates in the laboratory and the environment should be determined to allow assessment of the potential for mercury reduction in subsurface soils.
Acknowledgments
We thank Tina Thane for excellent technical assistance.
This work was supported by the U.S. Department of Energy, Environmental Remediation Sciences Program (grant 1022498).
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Altschul, S. F., T. K. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkay, T., N. Kroer, L. D. Rasmussen, and S. J. Sørensen. 1995. Conjugal transfer at natural population densities in a microcosm simulating an estuarine environment. FEMS Microb. Ecol. 16:43-54. [Google Scholar]
- 3.Barkay, T., S. M. Miller, and A. O. Summers. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microb. Ecol. 27:355-384. [DOI] [PubMed] [Google Scholar]
- 4.Bollmann, A., K. Lewis, and S. S. Epstein. 2007. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl. Environ. Microbiol. 73:6386-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clément, P., D. H. Pieper, and B. González. 2001. Molecular characterization of a deletion/duplication rearrangement in tfd genes from Ralstonia eutropha JMP134(pJP4) that improves growth on 3-chlorobenzoic acid but abolishes growth on 2,4-dichlorophenoxyacetic acid. Microbiology 147:2141-2148. [DOI] [PubMed] [Google Scholar]
- 6.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari, B. C., S. J. Binnerup, and M. Gillings. 2005. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl. Environ. Microbiol. 71:8714-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gachhui, R., J. Chaudhuri, S. Ray, K. Pahan, and A. Mandal. 1997. Studies on mercury-detoxicating enzymes from a broad-spectrum mercury-resistant strain of Flavobacterium rigense. Folia Microbiol. 42:337-343. [DOI] [PubMed] [Google Scholar]
- 9.Gans, J., M. Wolinsky, and J. Dunbar. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387-1390. [DOI] [PubMed] [Google Scholar]
- 10.Hansen, B. M., P. H. Damgaard, J. Eilenberg, and J. C. Pedersen. 1998. Molecular and phenotypic characterization of Bacillus thuringiensis isolated from leaves and insects. J. Invertebr. Pathol. 71:106-114. [DOI] [PubMed] [Google Scholar]
- 11.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnsen, A. R., and N. Kroer. 2007. Effects of stress and other environmental factors on horizontal plasmid transfer assessed by direct quantification of discrete transfer events. FEMS Microb. Ecol. 59:718-728. [DOI] [PubMed] [Google Scholar]
- 13.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 14.Kim, E.-H., R. P. Mason, E. T. Porter, and H. L. Soulen. 2006. The impact of resuspension on sediment mercury dynamics, and methylmercury production and fate: a mesocosm study. Mar. Chem. 102:300-315. [Google Scholar]
- 15.Kriger, A. A., and R. R. Turner. 1995. Field analysis of mercury in water, sediment and soil using static headspace analysis. Water Air Soil Pollut. 80:1295-1304. [Google Scholar]
- 16.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 18.Ní Chadhain, S. M., J. K. Schaefer, S. Crane, G. J. Zylstra, and T. Barkay. 2006. Analysis of mercuric reductase (merA) gene diversity in an anaerobic mercury-contaminated sediment enrichment. Environ. Microbiol. 8:1746-1752. [DOI] [PubMed] [Google Scholar]
- 19.Oregaard, G., and S. J. Sørensen. 2007. High diversity of bacterial mercuric reductase genes from surface and sub-surface floodplain soil (Oak Ridge, USA). ISME J. 1:453-467. [DOI] [PubMed] [Google Scholar]
- 20.Pinhassi, J., J. P. Bowman, O. I. Nedashkovskaya, I. Lekunberri, L. Gomez-Consarnau, and C. Pedros-Alio. 2006. Leeuwenhoekiella blandensis sp. nov., a genome-sequenced marine member of the family Flavobacteriaceae. Int. J. Syst. Evol. Microbiol. 56:1489-1493. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen, L. D., and S. J. Sørensen. 2001. Effects of mercury contamination on the culturable heterotrophic, functional and genetic diversity of the bacterial community in soil. FEMS Microbiol. Lett. 36:1-9. [DOI] [PubMed] [Google Scholar]
- 22.Rasmussen, L. D., S. J. Sørensen, R. R. Turner, and T. Barkay. 2000. Application of a mer-lux biosensor for estimating bioavailable mercury in soil. Soil Biol. Biochem. 32:639-646. [Google Scholar]
- 23.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 24.Sørensen, S. J., M. Bailey, L. H. Hansen, N. Kroer, and S. Wuertz. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3:700-710. [DOI] [PubMed] [Google Scholar]
- 25.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svenning, M. M., I. Wartiainen, A. G. Hestnes, and S. J. Binnerup. 2003. Isolation of methane oxidizing bacteria from soil by use of a soil substrate membrane system. FEMS Microb. Ecol. 44:347-354. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vetriani, C., Y. S. Chew, S. M. Miller, J. Yagi, J. Coombs, R. A. Lutz, and T. Barkay. 2005. Mercury adaptation among bacteria from a deep-sea hydrothermal vent. Appl. Environ. Microbiol. 71:220-226. [DOI] [PMC free article] [PubMed] [Google Scholar]