Abstract
Xylella fastidiosa is a vector-borne, plant-pathogenic bacterium that causes disease in citrus (citrus variegated chlorosis [CVC]) and coffee (coffee leaf scorch [CLS]) plants in Brazil. CVC and CLS occur sympatrically and share leafhopper vectors; thus, determining whether X. fastidiosa isolates can be dispersed from one crop to another and cause disease is of epidemiological importance. We sought to clarify the genetic and biological relationships between CVC- and CLS-causing X. fastidiosa isolates. We used cross-inoculation bioassays and microsatellite and multilocus sequence typing (MLST) approaches to determine the host range and genetic structure of 26 CVC and 20 CLS isolates collected from different regions in Brazil. Our results show that citrus and coffee X. fastidiosa isolates are biologically distinct. Cross-inoculation tests showed that isolates causing CVC and CLS in the field were able to colonize citrus and coffee plants, respectively, but not the other host, indicating biological isolation between the strains. The microsatellite analysis separated most X. fastidiosa populations tested on the basis of the host plant from which they were isolated. However, recombination among isolates was detected and a lack of congruency among phylogenetic trees was observed for the loci used in the MLST scheme. Altogether, our study indicates that CVC and CLS are caused by two biologically distinct strains of X. fastidiosa that have diverged but are genetically homogenized by frequent recombination.
Emerging diseases have become increasingly important in recent decades (51). Molecular characterization of pathogens coupled with biological tests on host range and ecological parameters have proved to be essential in the development of quarantine, containment, and control practices for such organisms. Although much work on emerging diseases is focused on human and animal pathogens, plants have also been targeted by new and reemerging pathogens. Not surprisingly, biological invasions, climate change, and farming techniques have been identified as the main drivers of emerging plant diseases (4). In addition, for many plant disease systems the lack of basic knowledge about the genetic structure and biology of the pathogen(s) involved may limit options for management. Such knowledge is especially challenging to obtain for pathogens with wide host ranges.
Xylella fastidiosa is a xylem-limited, plant-pathogenic bacterium with a wide host range. This bacterium causes a growing number of diseases in many crops throughout the Americas, including grape, almond, citrus, peach, and coffee plants (21). It is disseminated in nature by sharpshooter leafhoppers (Hemiptera, Cicadellidae, Cicadellinae), which are xylem sap-feeding specialist insects with generally polyphagous behavior in relation to host plants (43). There is no evidence for any vector-pathogen specificity required for transmission, with different X. fastidiosa strains being transmitted by different sharpshooter species to different host plants (1). There are, nonetheless, differences in how efficiently transmission occurs, depending on the vector-host plant-pathogen strain combination (1). Thus, this is an ecologically unique pathosystem, with a bacterial pathogen that colonizes a wide range of hosts and is transmitted by polyphagous vectors.
Most X. fastidiosa-colonized plants are asymptomatic (e.g., reference 40). Strains are grouped primarily on the basis of the crop hosts in which they cause disease, and most strains do not cause disease in plants susceptible to other strains of X. fastidiosa. For example, almond plants are susceptible to isolates from grape plants but not vice versa (3). Oleander and grape X. fastidiosa clades are monophyletic (47), but neither causes disease in cross-inoculation experiments (41). Therefore, biological relationships must be tested and cannot be directly inferred from evolutionary history alone. Currently, X. fastidiosa strains are divided into four subspecies (45, 47): (i) X. fastidiosa subsp. piercei (grape), (ii) X. fastidiosa subsp. sandyi (oleander), (iii) X. fastidiosa subsp. multiplex (several hosts), and (iv) X. fastidiosa subsp. pauca (citrus). X. fastidiosa subsp. pauca is the only one from South America, where X. fastidiosa causes disease in citrus, coffee, and plum plants (39). Among such diseases, citrus variegated chlorosis (CVC) has been relatively well studied, but limited work has been done on coffee leaf scorch (CLS) and plum leaf scald.
The relationships among coffee and citrus X. fastidiosa isolates in Brazil have been controversial. There are essentially three lines of evidence, each arguing either that these strains are the same or that they are different. CVC emerged in 1987 in northern Sao Paulo state (7, 18), and CLS was first reported in 1995 in the same region (35). However, several authors have argued that CLS symptoms were present in coffee plants long before 1987 and were misdiagnosed as nematode infections or plant nutritional imbalances (see reference 26 for discussion on that). In addition, X. fastidiosa was later found to infect coffee plants in other Brazilian regions, suggesting an old association with that host. Genetically, the relationships among coffee and citrus isolates are also poorly resolved. Results obtained by using different typing methods have consistently grouped isolates from these plants in a monophyletic clade, separately from North American X. fastidiosa isolates (32, 47). However, within South America, placement of citrus and coffee isolates has been variable and dependent on the number of tested isolates from each host plant and the methodology used (e.g., references 29, 30, and 42). In certain cases, for example, the fully sequenced citrus isolate 9a5c was found to be an outgroup to citrus X. fastidiosa (29). Lastly, to our knowledge only two biological cross-inoculation studies have been conducted with citrus and coffee X. fastidiosa isolates, again with inconclusive results. Li et al. (26) showed that CLS can be caused by both coffee and citrus X. fastidiosa isolates. On the other hand, another study showed that a CVC isolate of X. fastidiosa colonized coffee plants but died off over time, whereas no colonization of citrus plants was observed for a CLS isolate (37). Therefore, previous research has not resolved whether CVC and CLS are caused by the same strain or by different strains of X. fastidiosa.
In this study, we sought to clarify the relationships among citrus and coffee X. fastidiosa isolates collected from different regions in Brazil. We used microsatellite- and multilocus sequence typing (MLST)-based approaches to determine the genetic structure of this pathogen and conducted cross-inoculation assays to biologically characterize representative isolates. We tested the hypotheses that X. fastidiosa isolates from citrus and coffee plants are distinguished by host plant or geographical location, their phylogenetic placement in relation to North American X. fastidiosa, and colonization patterns in citrus and coffee hosts.
MATERIALS AND METHODS
Isolates.
Xylella fastidiosa isolates were collected from symptomatic citrus and coffee plants from four states in Brazil (Table 1). We cultured X. fastidiosa from samples, following standard protocols (2), and triply cloned each isolate prior to freezing cells at −80°C. We recovered frozen samples by plating suspensions on PWG medium (20) to extract DNA for molecular analyses (37) and to mechanically inoculate citrus and coffee plants for pathogenicity tests (see below). All samples were tested with the diagnostic primer set RST31-33 to confirm their identification as X. fastidiosa (31).
TABLE 1.
List of Xylella fastidiosa isolates used in this study, including cluster assignment based on MLST and microsatellite data
| Isolate | Host of origin | Genetic clustera according to:
|
City and stateb | Collector | ||
|---|---|---|---|---|---|---|
| MLST | Microsatellite analysis
|
|||||
| STRUCTURAMA | STRUCTURE | |||||
| 10 | Citrus | 1 | 1 | 1 | Pedregulho, SP | S. Lopes |
| 11 | Citrus | 1 | 1 | 1 | Pedregulho, SP | S. Lopes |
| 34 | Citrus | 1 | 1 | 1 | Araras, SP | J. Lopes |
| 35 | Citrus | 1 | 1 | 1 | Com. Gomes, MG | J. Lopes |
| 36 | Citrus | 1 | 1 | 1 | Matão, SP | S. Lopes |
| 37 | Citrus | 1 | 1 | 1 | Matão, SP | S. Lopes |
| 41 | Citrus | 1 | 1 | 1 | Taquaritinga, SP | J. Lopes |
| 42 | Citrus | 1 | 1 | 1 | Taquaritinga, SP | J. Lopes |
| 44 | Citrus | 1 | 1 | 1 | Ubirajara, SP | J. Lopes |
| 45 | Citrus | 1 | 1 | 1 | Gavião Peixoto, SP | R. Marques |
| 46 | Citrus | 1 | 1 | 1 | Gavião Peixoto, SP | R. Marques |
| 53 | Citrus | 1 | 1 | 1 | Com. Gomes, MG | R. Marques |
| 54 | Citrus | 1 | 1 | 1 | Frutal, SP | R. Marques |
| 55 | Citrus | 1 | 1 | 1 | Frutal, SP | R. Marques |
| 64 | Citrus | 1 | 1 | 1 | Rio Real, BA | F. Laranjeira |
| 65 | Citrus | 1 | 1 | 1 | Itapirucu, BA | F. Laranjeira |
| 6570 | Citrus | 1 | 1 | 1 | Bebedouro, SP | J. Lopes |
| 66 | Citrus | 1 | 1 | 1 | Rio Real, BA | F. Laranjeira |
| 67 | Citrus | 1 | 1 | 1 | Itapirucu, BA | F. Laranjeira |
| 84 | Citrus | 1 | 1 | 1 | Botucatu, SP | P. Paiva |
| 85 | Citrus | 1 | 1 | 1 | Itajú, SP | P. Paiva |
| 47 | Citrus | 1 | 1 | 2 | Gavião Peixoto, SP | R. Marques |
| 18 | Citrus | 2 | 2 | 2 | Neves Paulista, SP | S. Lopes |
| 86 | Citrus | 2 | 2 | 2 | São Carlos, SP | P. Paiva |
| 88 | Citrus | 2 | 2 | 2 | Cafelândia, SP | P. Paiva |
| 9a5c | Citrus | 2 | 2 | 2 | Macaubal, SP | Reference 7 |
| 29 | Coffee | 3 | 3 | 3 | Garça, SP | J. Lopes |
| 51 | Coffee | 3 | 3 | 3 | Lavras, MG | M. Bento |
| 1 | Coffee | 4 | 3 | 3 | Ribeirão Preto, SP | S. Lopes |
| 3124 | Coffee | 4 | 3 | 3 | Matão, SP | W. Li |
| 4 | Coffee | 4 | 3 | 3 | Cravinhos, SP | S. Lopes |
| 50 | Coffee | 4 | 3 | 3 | Lavras, MG | M. Bento |
| 56 | Coffee | 4 | 3 | 3 | Planaltina, DF | C. Oliveira |
| 57 | Coffee | 4 | 3 | 2 | Planaltina, DF | C. Oliveira |
| 58 | Coffee | 4 | 3 | 2 | Planaltina, DF | C. Oliveira |
| 59 | Coffee | 4 | 3 | 3 | Planaltina, DF | C. Oliveira |
| 61 | Coffee | 4 | 3 | 3 | Planaltina, DF | C. Oliveira |
| 62 | Coffee | 4 | 3 | 2 | Planaltina, DF | C. Oliveira |
| 6756 | Coffee | 4 | 3 | 3 | Matão, SP | R. Almeida |
| 73 | Coffee | 4 | 3 | 2 | São Gotardo, MG | L. Zambolim |
| 80 | Coffee | 4 | 3 | 2 | São Gotardo, MG | L. Zambolim |
| 32 | Coffee | 5 | 3 | 3 | Muritinga Sul, SP | J. Ottoboni |
| 68 | Coffee | 6 | 2 | 2 | São Gotardo, MG | L. Zambolim |
| 8 | Coffee | 7 | 2 | 3 | Pedregulho, SP | S. Lopes |
| 24 | Coffee | 7 | 3 | 2 | Garça, SP | J. Lopes |
| 33 | Coffee | 7 | 3 | 2 | Varginha, MG | S. Lopes |
See text for cluster assignment using different methods.
State abbreviations: BA, Bahia; DF, Distrito Federal; MG, Minas Gerais; SP, Sao Paulo.
Microsatellites.
We used the methods described by Coletta-Filho et al. (8) to conduct a study on the population genetic structure of citrus and coffee X. fastidiosa isolates. This method is based on fast-evolving microsatellites (also known as short sequence repeats [SSR] and variable number tandem repeats) that permit genetic studies at the pathogen population level. We analyzed six loci in this study, SSR20, -21, -26, -28, -30, and -40, which have been previously found to be informative for CVC population genetics studies (8). Analysis of gels was performed using Quantity One software (Bio-Rad, Hercules, CA) to assign the fragment size for each sample locus, followed by manual checking of assignments.
Microsatellite AMOVA.
We tested the hypotheses that citrus and coffee X. fastidiosa isolates were genetically clustered in different groups based on host plant or geographical location. Populations were defined as isolates from the same host plant collected in one state (for example, coffee isolates collected in Minas Gerais state). We used analyses of molecular variance (AMOVA), as implemented in ARLEQUIN 3.11 (12), to determine the covariance among groups, among populations within groups, and among individuals within populations. Two analyses were performed, one by grouping populations by host plant and the other by grouping them by the state where they were collected. The genetic separation among populations (a total of six populations, three from citrus and three from coffee plants) was determined by comparing population pairwise FST values, using ARLEQUIN 3.11. One thousand permutations were used to test the significance of AMOVA and population pairwise FST comparisons.
Microsatellite clustering analyses.
Linkage equilibrium, the statistical independence of the loci tested, suggests high rates of recombination in the case of bacteria. Because the clustering methods that we used assume linkage equilibrium, we tested for its presence in our dataset by using LIAN 3.0 (19). LIAN 3.0 was also used to determine the diversity in samples. To determine the number of genetic clusters in the microsatellite dataset, we used the software package STRUCTURE 2.2 (38), despite the fact that it makes assumptions violated by our dataset (i.e., linkage equilibrium). We tested the posterior likelihood of the samples being divided into each number of genetic clusters (k) between one and five by resampling the dataset 10 times (burn-in, 10,000 steps; run, 100,000 steps). In addition to determining the k value with the highest likelihood, we also determined ΔK, which provides an alternative on how to infer k rather than examining only the likelihood values obtained from runs with different k values (11). In addition, STRUCTURAMA (http://www.structurama.org/) was also used to infer the population structure of the samples; however, it also assumes linkage equilibrium. In STRUCTURAMA, samples are initially not assigned to any specific population. We ran it for 100,000 generations, varying the number of fixed populations from one to six. We also used BAPS 2.2 (Bayesian Analysis of Population Structure) (9) to infer the number of genetic clusters in the dataset. BAPS treats the number of populations and locus frequencies as random variables. We used the enumerative calculation to estimate population structure, varying the number of putative populations (assigned as described above) from one to six. In BAPS, populations were compared rather than individuals within populations.
MLST.
We sequenced 5 of 10 loci that were part of an MLST scheme previously used for X. fastidiosa (44). The loci sequenced were lacF, rfbD, petC, cysG, and leuA. Primers for the MLST scheme were originally designed based on North American X. fastidiosa isolates, and not all functioned well with our samples. We designed new reverse primers for rfbD (5′-TCCATAAACGGCGCCTTC-3′) and cysG (5′-GCATATGTCTGTGCGGTGTGC-3′) due to poor amplification with the published primers. We were not able to amplify holC for most isolates, and that locus was eliminated from the analysis. We did not test the other four loci that were part of that MLST scheme. DNA was amplified using HotStart HiFidelity polymerase (Qiagen, Valencia, CA), the sizes of the amplicons were checked in agarose gels, and the DNA was later purified for sequencing with a Qiagen PCR purification kit. Sequencing was performed at the DNA sequencing facility of the University of California, Berkeley, CA. Alignments were prepared with BIOEDIT 7.0.4 (15) for analyses.
We used START2 (23) to obtain a summary of the MLST data and the allelic profile for all samples. LIAN 3.0 was used to determine genetic diversity and linkage equilibrium in the dataset. The different allelic profiles were identified as seven sequence types (STs; ST1 to ST7) and these assigned STs used for the analyses described below. We also ran an AMOVA using ARLEQUIN 3.11 to determine the genetic structure of populations sampled, using the DNA sequences of the five loci (individually, not concatenated) as sources of data. As for the microsatellites, we assigned X. fastidiosa to different populations based on the host plants and states where they were collected. We also conducted FST pairwise comparisons among the six populations.
Phylogenetic analyses.
To test for phylogenetic congruency among the loci sequenced, we conducted analyses using the citrus and coffee X. fastidiosa STs and an outgroup (the genome-sequenced grape X. fastidiosa isolate Temecula; GenBank accession no. AE009442) (50). We conducted phylogenetic analyses by using PAUP* 4.0b.1 (D. L. Swofford, Sinauer, Sunderland, MA) and performing maximum parsimony and maximum likelihood searches on individual loci and concatenated sequences. We used Modeltest (36) to determine the best evolutionary model for the dataset by using the Akaike information criterion for each likelihood search (concatenated sequences, GTR+I; lacF, TrN; petC, TVM; rfbD, TrN+I; cysG, HKY; and leuA, HKY+I). Support for analyses was obtained with 1,000 (maximum parsimony) and 100 (maximum likelihood) bootstrap replicates. To determine significant incongruence among tree topologies, we used the Shimodaira-Hasegawa likelihood ratio test (48), as implemented in PAUP*.
To determine the relationship between X. fastidiosa isolates in South and North America, we randomly selected two isolates from each one of the six clonal clusters identified in North America by Schuenzel et al. (47), in addition to the STs identified in this study. We used the five-locus concatenated sequence for the analyses. Maximum parsimony and maximum likelihood searches were performed as described above (model HKY+I was selected for the maximum likelihood search). Because of the evidence for recombination in X. fastidiosa presented by Scally et al. (44) and this work, we also used SplitsTree 4 (22) to perform a phylogenetic network analysis with the neighbor-net method. Phylogenetic networks better represent the evolutionary relationships among recombining bacteria, as conflicting signals can be represented as a network instead of a bifurcating tree. We used ClonalFrame 1.1 (10) to infer the relationships among these taxa. ClonalFrame incorporates recombination into the phylogenetic analysis, accounting for both mutation and recombination rates when estimating the posterior probabilities of trees. It assumes that mutation and recombination rates are constant throughout the tree. We ran two chains on ClonalFrame, with 100,000 burn-in steps followed by 500,000 iterations, recording samples every 100 iterations. To assess for convergence of parameters among runs, we used the Gelman and Rubin convergence test (14), as implemented in ClonalFrame. PHItest (6), as implemented in SplitsTree 4 (22), was used to determine the presence of recombination in the dataset. We also used TREE-PUZZLE 5.2 (46) to perform a maximum likelihood mapping analysis (49) under the HKY model for the same dataset (although here we used both concatenated and single-locus sequences) by subdividing the taxa into four subspecies as previously proposed (45, 47). This method determines the genetic relationships among the groups we arbitrarily assigned instead of individual taxa (ST) and can be used to test the monophyly of tested groups.
Host colonization assays.
We conducted two tests to determine if CVC- and CLS-causing isolates collected from symptomatic plants in commercial coffee plantations and citrus orchards were able to colonize citrus and coffee plants under greenhouse conditions. Although citrus plants infected with X. fastidiosa readily show diagnostic CVC symptoms under greenhouse conditions (2), typical symptoms of CLS (e.g., shortening of internodes, leaf size reduction, leaf scorch, and yellowing) are not evident in less than 1 to 2 years if the pinprick inoculation method is used (37). Therefore, we focused our assays on determining X. fastidiosa colonization of host plants and not symptom development (i.e., disease); however, we recorded the presence of symptoms on citrus plants. Because all isolates were collected from symptomatic coffee or citrus plants, we assumed they were pathogenic. In the first experiment, we tested if isolates maintained successful infections in plants over time by quantifying their populations in an apical leaf 11 months after inoculation. Healthy seedlings of Citrus sinensis (L.) Osbeck cv. Caipira and Coffea arabica L. cv. Catuaí vermelho (clone 99) were pin inoculated in a single point of the stem with 5 μl of 109- to 1010-CFU/ml X. fastidiosa suspensions, following methods previously described (2, 37); plants were maintained in 1-liter pots containing a mixture of soil, manure, and sand (3:2:1), inside an insect-free greenhouse at ESALQ, University of Sao Paulo, Piracicaba, Brazil. Eleven months after inoculation, we tested all plants for the presence of X. fastidiosa by the culturing method, which also allows for the quantification of live cells in the sampled tissue (2). We sampled an apical but mature leaf from plants because detection in these samples would imply cell multiplication and movement (i.e., colonization) of host plants. Each sample consisted of 0.1 to 0.15 g of the leaf petiole and main vein; primary isolation and quantification of the number of CFU per gram of leaf tissue were carried out by dilution plating on solid PWG medium as described before (2). Samples of recovered bacterial colonies were confirmed as X. fastidiosa by PCR using the diagnostic primer set RST31-33 (31). Table 7 lists the isolates used and the numbers of plants tested. Twenty coffee and seven negative-control citrus plants were buffer inoculated and tested negative for X. fastidiosa.
TABLE 7.
Long-term patterns of citrus and coffee colonization by Xylella fastidiosa isolatesa
| Host and MLSTb | Isolate | Rate for:
|
|||
|---|---|---|---|---|---|
| Citrus host
|
Coffee host
|
||||
| Infectionc | CVC symptomsd | Infectionc | CLS symptomsd,e | ||
| Citrus | |||||
| 1 | 10 | 1/2 (6.1) | 1/2 | 0/20 | 0/20 |
| 1 | 11 | 4/13 (5.7 ± 0.5) | 0/13 | 0/20 | 0/20 |
| 1 | 35 | 9/11 (6.3 ± 0.1) | 1/11 | 0/20 | 0/20 |
| 1 | 36 | 9/19 (4.9 ± 0.3) | 1/19 | 0/20 | 0/20 |
| 1 | 37 | 8/15 (5.4 ± 0.3) | 3/15 | 0/20 | 0/20 |
| 1 | 6570 | 8/11 (5.4 ± 0.3) | 1/11 | 0/20 | 0/20 |
| 2 | 18 | 3/6 (6.1 ± 0.4) | 3/6 | 0/20 | 0/20 |
| Coffee | |||||
| 3 | 29 | 0/14 | 0/14 | 7/20 (4.9 ± 0.2) | 0/20 |
| 4 | 1 | 0/13 | 0/13 | 18/20 (5.2 ± 0.1) | 0/20 |
| 4 | 4 | 0/16 | 0/16 | 12/20 (5.1 ± 0.1) | 0/20 |
| 4 | 3124 | 0/16 | 0/16 | 6/20 (4.8 ± 0.2) | 0/20 |
| 5 | 32 | 0/13 | 0/13 | 11/20 (5.4 ± 0.2) | 0/20 |
| 7 | 8 | 0/15 | 0/15 | 16/20 (4.9 ± 0.1) | 0/20 |
| 7 | 24 | 0/8 | 0/8 | 14/20 (4.7 ± 0.2) | 0/20 |
| 7 | 33 | 0/7 | 0/7 | 11/20 (5.4 ± 0.1) | 0/20 |
Plants were sampled at one location (the leaf petiole on the apical portion of the plant) for X. fastidiosa culturing at 11 months after inoculation.
See Table 1 for additional information on isolates.
Number of infected plants/number of inoculated plants. In parenthesis is the mean number of CFU per gram of plant tissue (log10 value ± standard error).
Number of plants expressing symptoms/number of inoculated plants.
Typical CLS symptoms were not recorded for coffee plants within 11 months after inoculation.
Because a recent report showed that one citrus isolate multiplied in coffee plants and maintained low populations when inoculated at high titers (37), we conducted a second bioassay that tested plants for bacterial colonization within a shorter period (4 months) after inoculation at different distances from the inoculation site. Our goal was to determine if early multiplication and movement of isolates occur in nonreciprocal crosses. For this purpose, we mechanically inoculated 10 healthy citrus and coffee seedlings in the stem at the base of the fourth mature leaf (as in reference 2), with 7-μl drops of 107-CFU/ml suspensions of representative X. fastidiosa isolates (see Table 8). We also inoculated 10 citrus and coffee plants with buffer as negative controls. The citrus and coffee cultivars used, as well as the potting and greenhouse conditions, were the same as those in the first bioassay. The plants were evaluated by culturing 4 months after inoculation at two different sites, the first leaf above the inoculation point and a leaf 9 cm above that point.
TABLE 8.
Short-term patterns of citrus and coffee colonization by Xylella fastidiosa isolatesa
| Host and MLSTb | Isolate | Ratec for infection of:
|
|||
|---|---|---|---|---|---|
| Citrus host
|
Coffee host
|
||||
| First leaf | 9 cm above IP | First leaf | 9 cm above IP | ||
| Citrus | |||||
| 1 | 11 | 3/10 (4.8 ± 0.4) | 0/10 | 0/10 | 0/10 |
| 1 | 35 | 2/10 (6.0 ± 0.6) | 0/10 | 2/10 (4.6 ± 0.2) | 0/10 |
| 1 | 85 | 3/10 (4.9 ± 0.2) | 0/10 | 0/10 | 0/10 |
| 1 | 6570 | 6/10 (5.2 ± 0.1) | 4/10 (5.3 ± 0.4) | 0/10 | 0/10 |
| 2 | 9a5c | 4/10 (5.4 ± 0.4) | 1/10 (6.4) | 2/10 (4.6 ± 0.8) | 0/10 |
| Coffee | |||||
| 4 | 3124 | 0/10 | 0/10 | 0/10 | 3/10 (3.8 ± 0.4) |
| 4 | 61 | 0/10 | 0/10 | 1/10 (4.9) | 4/10 (6.4 ± 0.1) |
| 4 | 73 | 0/10 | 0/10 | 1/10 (4.5) | 0/10 |
| 7 | 24 | 0/10 | 0/10 | 0/10 | 0/10 |
| 7 | 33 | 0/10 | 0/10 | 0/10 | 0/10 |
Plants were sampled at two locations above the inoculation point (IP) for X. fastidiosa culturing at 4 months after inoculation.
See Table 1 for additional information on isolates.
Number of infected plants/number of inoculated plants. In parenthesis is the mean number of CFU per gram of plant tissue (log10 value ± standard error).
Multiplex PCR detection of coffee and citrus X. fastidiosa isolates.
We used the only locus fixed in our study (lacF; see Results) to develop a host-specific scheme for differential PCR detection of citrus and coffee X. fastidiosa isolates. Our multiplex approach used three primers, a reverse primer shared by citrus and coffee isolates (5′ CCTCGGGTCATCACATAAGGC 3′; 10 μM, based on reference 44) and forward citrus (5′ GCGCTGATTTGGCGTTACT 3′; 10 μM)- and coffee (5′ GTATGCTCTGGGCAATCCG 3′; 10 μM)-specific primers. We noted that the reverse primer had one substitution compared to the genome sequence of 9a5C (a fully sequenced citrus isolate), but amplification results were not affected. The amplicon size obtained for CVC X. fastidiosa samples was 353 bp, whereas for CLS samples it was 597 bp. The PCR steps used were 94°C for 1 min, 66°C for 45 s, and 72°C for 1 min for 30 cycles.
Nucleotide sequence accession numbers.
Sequences obtained were deposited in the GenBank database under accession numbers EU496570 to EU496799.
RESULTS
Microsatellite data analyses.
We tested if X. fastidiosa isolates collected from symptomatic citrus and coffee plants in four Brazilian states were genetically grouped based on host plant or geographical location. AMOVA showed no statistical difference among groups of X. fastidiosa populations (Table 2), failing to support the hypothesis that either host plant or geography was associated with the genetic structure of the isolates. Grouping populations by host plant resulted in a lower (but nonsignificant) P value than grouping them by the region where the isolates were collected. Most of the variation observed occurred among individuals within populations (Table 2), indicating that the loci used may evolve quickly. Differences among populations within groups and isolates within populations were statistically significant, suggesting a genetic structure in the data. Populations within groups showed statistical differences when grouped by state, indicating that coffee and citrus X. fastidiosa isolates from the same region were genetically different (Table 2). FST pairwise comparisons showed an interesting trend in which coffee populations were different from citrus populations and vice versa (Table 3). The low number of samples (n = 2 isolates) in the Minas Gerais citrus population may have affected its pairwise comparisons. Because populations were not clearly grouped by host plant or geographic location (AMOVA), we used BAPS to infer the best structure for the citrus and coffee X. fastidiosa populations. BAPS partitioned the populations into three clusters with a higher probability (0.833) than that for two clusters (0.165). The three clusters were (i) citrus isolates from Sao Paulo and Minas Gerais; (ii) citrus isolates from Bahia; and (iii) coffee isolates from Sao Paulo, Distrito Federal, and Minas Gerais. Therefore, although AMOVA of microsatellite data did not support the grouping of X. fastidiosa isolates by host plant (P = 0.10) or region (P = 0.34), different tests performed suggest some level of genetic clustering for coffee and citrus X. fastidiosa populations. It must be emphasized that the small numbers of loci and samples used per population probably limited our conclusions with this analytical method.
TABLE 2.
AMOVA for microsatellite (SSR) and MLST data on citrus and coffee X. fastidiosa isolates grouped by host plant and geographical location (state)
| Groupinga | Method | df | Sum of squares | Variance component | % of variation | Fixation index (P) |
|---|---|---|---|---|---|---|
| Host plant | ||||||
| Among groups | SSR | 1 | 7.61 | 0.18 | 8.68 | FCT = 0.086 (0.103) |
| MLST | 1 | 254.76 | 10.90 | 67.89 | FCT = 0.679 (0.101) | |
| Among populations | SSR | 4 | 10.66 | 0.15 | 7.16 | FSC = 0.078 (0.018) |
| Within groups | MLST | 4 | 26.35 | 0.29 | 1.87 | FSC = 0.058 (0.281) |
| Among individuals | SSR | 40 | 71.55 | 1.78 | 84.16 | FST = 0.158 (<0.001) |
| Within populations | MLST | 40 | 194.35 | 4.85 | 30.24 | FST = 0.697 (<0.001) |
| Region (state) | ||||||
| Among groups | SSR | 3 | 11.48 | 0.06 | 3.25 | FCT = 0.032 (0.340) |
| MLST | 3 | 126.36 | −3.17 | −27.10 | FCT = −0.271 (0.610) | |
| Among populations | SSR | 2 | 6.78 | 0.22 | 10.71 | FSC = 0.110 (<0.001) |
| Within groups | MLST | 2 | 154.73 | 10.05 | 85.68 | FSC = 0.674 (<0.001) |
| Among individuals | SSR | 40 | 71.55 | 1.78 | 86.05 | FST = 0.139 (<0.001) |
| Within populations | MLST | 40 | 194.35 | 4.85 | 41.42 | FST = 0.585 (<0.001) |
Populations comprised isolates collected from citrus or coffee plants from different states.
TABLE 3.
FST pairwise differentiation among Xylella fastidiosa populations by use of microsatellite and MLST dataa
| Host and state (no. of isolates) | Result for indicated host and state
|
||||
|---|---|---|---|---|---|
| Coffee
|
Citrus
|
||||
| SP | MG | DF | SP | MG | |
| Coffee | |||||
| SP (8) | |||||
| MG (6) | −0.049/−0.158 | ||||
| DF (6) | 0.089/0.222 | 0.060/0.262 | |||
| Citrus | |||||
| SP (20) | 0.105*/0.633* | 0.111*/0.641* | 0.148*/0.809* | ||
| MG (2) | 0.023/0.573* | 0.044/0.555 | 0.221/1.000* | −0.060/0.999 | |
| BA (4) | 0.304*/0.653* | 0.282*/0.655* | 0.436*/1.000* | 0.205*/0.563 | 0.279/0.999 |
The numerator and the denominator represent results based on microsatellite and MLST data, respectively (*, P < 0.05). See Table 1 for state abbreviations.
Clustering analyses.
For the microsatellite data, we also used a clustering approach to infer the structure of X. fastidiosa isolates analyzed. This approach allowed for statistical methods for determining which isolates would be grouped together. These methods assume linkage equilibrium, which was violated (P < 0.001) when the whole dataset was used as well as when citrus and coffee isolates were tested independently; however, the standardized index of association (IAS) was <0.15 for those data (0 indicates linkage equilibrium, and 1 indicates clonality). Thus, although assumptions were violated, the association index observed was small. STRUCTURE reported the highest probability when the number of clusters (k) was three, which was confirmed when the ΔK value was determined to be larger with three populations (data not shown). STRUCTURAMA also estimated that clustering the isolates into three groups resulted in a higher posterior probability. The assignments made by STRUCTURAMA and STRUCTURE are compared to the STs obtained with MLST (Table 1).
MLST data analyses.
Sequencing of five loci identified seven X. fastidiosa STs in Brazil, two from citrus plants and five from coffee plants (Table 4). We found no citrus STs in coffee plants or vice versa (Table 1). A dominant ST was identified in citrus plants (ST1), accounting for 85% of the samples collected throughout Brazil. The citrus isolate that has been sequenced (50), 9a5c, belongs to the less common ST ST2. Similarly, ST4 was the most common ST obtained from symptomatic coffee plants, accounting for 65% of isolates collected. Citrus isolates had a lower index of association (IAS = 0.251) than coffee isolates (IAS = 0.578), and both were determined to be in linkage disequilibrium, as they were by the microsatellite data (as tested with LIAN). The allelic profile suggests evidence of recombination between citrus and coffee isolates, e.g., petC in ST2 and most coffee STs and rfbD for ST1 and ST3 (Table 4). We detected no positive selection for the loci sequenced (Table 5). AMOVA results for MLST data, which grouped isolates into populations, like microsatellite data, and treated each sequence as a locus, showed results similar to those for the microsatellite analysis (Table 2). Grouping of populations by host plant showed that 68% of the data variation occurred among groups (citrus versus coffee) and 30% among individuals within populations. Interestingly, only 2% of the variation in that case was accounted for by differences among populations (different locations, i.e., states) within groups (host plants), suggesting a lack of geographic structure for these pathogens in Brazil and a separation based on host plant. Similarly, FST pairwise comparisons also indicated that X. fastidiosa citrus populations are genetically different from those from coffee plants (Table 3). Although the MLST results identified STs specific to citrus and coffee plants without any overlap, the AMOVA did not separate X. fastidiosa isolates solely on the basis of host plant. However, grouping populations by host plant (P = 0.10) better explained the data than grouping them by geographic region (state) (P = 0.61). Nevertheless, comparisons among populations identified differences between X. fastidiosa isolates causing disease in coffee and citrus plants.
TABLE 4.
ST assignment of coffee and citrus Xylella fastidiosa isolates based on MLST allelic profile
| Host and MLST | Result for:
|
No. of isolates | ||||
|---|---|---|---|---|---|---|
| lacF | petC | rfbD | cysG | leuA | ||
| Citrus | ||||||
| 1 | 1 | 1 | 1 | 1 | 1 | 22 |
| 2 | 1 | 2 | 2 | 1 | 1 | 4 |
| Coffee | ||||||
| 3 | 2 | 2 | 1 | 2 | 1 | 2 |
| 4 | 2 | 2 | 3 | 2 | 1 | 13 |
| 5 | 2 | 2 | 4 | 2 | 1 | 1 |
| 6 | 2 | 3 | 4 | 3 | 2 | 1 |
| 7 | 2 | 3 | 4 | 3 | 3 | 3 |
TABLE 5.
Summary of MLST data of coffee and citrus Xylella fastidiosa isolates
| Locus | Length (bp) | No. of alleles | No. of polymorphic sites | dN/dSa |
|---|---|---|---|---|
| lacF | 564 | 2 | 11 | 0.0695 |
| petC | 534 | 3 | 11 | 0.4048 |
| rfbD | 437 | 4 | 17 | 0.0625 |
| cysG | 1,227 | 3 | 10 | 0.0889 |
| leuA | 1,260 | 3 | 13 | 0.0589 |
dN/dS represents the ratio of nonsynonymous to synonymous substitutions, which are indicative of positive selective pressure on loci (none was observed here).
Phylogenetic analyses.
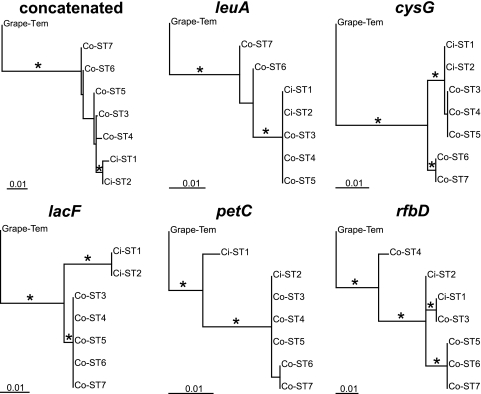
We conducted phylogenetic analyses for all loci and concatenated sequences separately to determine congruency among trees (Fig. 1). Concatenated sequences grouped STs into a citrus group and a coffee group. A third group (coffee ST6 and ST7) had 97% bootstrap support with the maximum parsimony method but only 73% bootstrap support with the maximum likelihood method. The inferences with individual loci, however, produced a series of incongruent trees with different relationships among STs, suggesting the presence of recombination between citrus and coffee X. fastidiosa isolates. The Shimodaira-Hasegawa test, which in essence compares the topologies of trees, confirmed a general lack of congruency among all single-locus trees and among those in relation to the concatenated tree (Table 6). These results indicate that the use of phylogenetic trees to reconstruct the evolutionary history of X. fastidiosa in Brazil is problematic. The use of single (or few) loci for specific detection of X. fastidiosa strains may have serious limitations, given the results that we obtained, at least for CVC and CLS. lacF (ABC transporter sugar permease) was the only locus examined that was fixed in coffee and citrus isolates. The lack of support for locus congruence is indicative of recombination among the STs. In addition, we used tests available in RDP3.14 (Recombination Detection Program) (28) to identify recombination events in the concatenated dataset of the coffee and citrus STs (defaults were used). The tests identified zero to three recombinant regions in the dataset, depending on the method used (data not shown).
FIG. 1.
Concatenated and single-locus phylogenetic maximum likelihood trees of citrus (Ci) and coffee (Co) Xylella fastidiosa STs identified in our survey, with a grape isolate (Temecula) as the outgroup. Asterisks represent branches supported by >80% bootstrap support with maximum parsimony and maximum likelihood tree searching methods. Note the lack of congruency among trees for the citrus and coffee STs.
TABLE 6.
Shimodaira-Hasegawa test for congruency among tree topologies for the five loci used in this study and their concatenated sequencea
| Locus | Result for:
|
|||||
|---|---|---|---|---|---|---|
| Concatenation | petC | cysG | lacF | leuA | rfbD | |
| Concatenation | 0.001* | 0.444 | 0.621 | 1.000 | 0.021* | |
| petC | 0.002* | 0.047* | 0.001* | 0.656 | 0.001* | |
| cysG | 0.114 | 0.003* | 0.001* | 0.851 | 0.001* | |
| lacF | 0.001* | 0.003* | 0.001* | 0.007* | 0.001* | |
| leuA | 0.007* | 0.001* | 0.209 | 0.001* | 0.001* | |
| rfbD | 0.001* | 0.004* | 0.001* | 0.001* | 0.003* | |
P values (*, P < 0.05) represent differences in likelihood score between the maximum likelihood topology of each locus (column) and that of the same locus constrained by the maximum likelihood topology obtained for other loci (row).
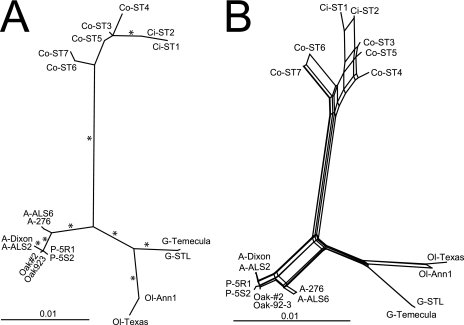
We used different approaches to determine the genetic relationships among the concatenated sequences of South and North American X. fastidiosa isolates. Different reconstruction methods suggest that diversity in Brazil, as determined by branch length in trees, is larger for coffee isolates than for isolates from any other X. fastidiosa-susceptible crop plant previously studied. However, a more inclusive analysis is warranted to determine if that is in fact true. It is interesting to note that citrus X. fastidiosa was represented by a small clade in the reconstructions, similarly to STs from other host plants but not coffee plants. The maximum likelihood reconstruction using concatenated sequences divided Brazilian X. fastidiosa isolates into citrus and coffee clades. However, inferences on the evolutionary history of these taxa are not possible due to the presence of recombination among them. Thus, concatenation of few locus sequences (Fig. 2A) may have applied importance (i.e., clustering of genetically and biologically similar sequences) but limited value for evolutionary inference. The presence of a network among Brazilian STs (Fig. 2B) is further indication of recombination among those taxa. It also suggests that recombination may occur more often among those taxa than among others in North America. Interestingly, the neighbor-net tree also shows evidence of recombination among South and North American taxa and a lack of monophyly between these geographic regions (Fig. 2B). Although ClonalFrame generated relationships similar to those for the other methods (with runs that converged for all parameters tested), it broke down the host plant subdivisions in the X. fastidiosa subsp. multiplex clade (almond, peach, and oak; data not shown). Those host plant relationships (peach and oak) within X. fastidiosa subsp. multiplex also had no branch support in other analyses (Fig. 2A).
FIG. 2.
Concatenated locus phylogeny of Xylella fastidiosa STs from coffee (Co) and citrus (Ci) plants in Brazil and two representative STs of each North American genetic cluster previously identified (47). Plant hosts of origin: A, almond; Oak, oak; P, peach; Ol, oleander; G, grape. (A) Maximum likelihood tree. The maximum parsimony search yielded similar topology. Bootstrap support (>90%) is indicated by an asterisk where supported by both the maximum likelihood and the maximum parsimony methods. (B) Neighbor-net reconstruction of relationships among X. fastidiosa STs.
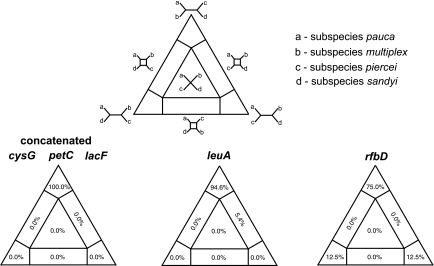
In addition, we looked at the possibility of recombination among the four proposed X. fastidiosa subspecies by using TREE-PUZZLE. We assigned the isolates used for Fig. 2 to their respective X. fastidiosa subspecies (following references 45 and 47). No conflicting phylogenetic signal was observed for the concatenated sequence or for that of locus petC, cysG, or lacF. However, that was not the case for leuA or rfbD. These results indicate that recombination in X. fastidiosa may be occurring not only within subspecies, such as X. fastidiosa subsp. pauca (citrus and coffee isolates), but also among subspecies. Recombination among isolates used for this analysis was also detected using PHItest (6). Intriguingly, we noted that the rfbD allele for coffee ST4 can be divided into two fragments, with part of the sequence having overall homology to other alleles in X. fastidiosa subsp. pauca and one fragment at the end of the sequence having strong homology to subspecies in North America. Recombination between an X. fastidiosa subsp. piercei isolate (Temecula) and ST4 was detected with the software package RDP3.14 when the concatenated sequences of the X. fastidiosa subsp. piercei isolate were added to an alignment of the citrus and coffee STs analyzed in this study (data not shown).
Host colonization assays.
We conducted two experiments to determine if citrus and coffee X. fastidiosa isolates collected from symptomatic plants were capable of colonizing both host plants under greenhouse conditions. In the first experiment, we looked at long-term colonization by sampling inoculated plants 11 months after inoculation (Table 7). We found no host plant cross-infection for any of the STs tested, suggesting that the citrus and coffee isolates tested are biologically distinct. Furthermore, CVC symptoms were observed only in plants inoculated with the citrus STs; typical CLS symptoms were not observed within the evaluation period of this experiment. In the second experiment, we determined patterns of colonization 4 months after inoculation (Table 8). We found no coffee isolates colonizing citrus plants, but a few citrus isolates colonized and moved within coffee plants. Taken together, the results for these tests suggest that coffee X. fastidiosa does not colonize or sustain long-term infections in citrus plants. In addition, the data suggest that citrus isolates can colonize coffee plants under greenhouse conditions but that eventually those infections will probably die off (Tables 7 and 8). More-detailed studies with additional sampling dates should be conducted to test the latter hypothesis.
Multiplex PCR detection of coffee and citrus X. fastidiosa isolates.
The presence of recombination among citrus and coffee STs is problematic for phylogenetic inference and development of reliable markers for detection of isolates causing disease in either host plant. The multiplex PCR approach that we developed correctly separated all 46 isolates used in this study on the basis of the host plant that they were collected from (data not shown). Although these results validate this fast and easy typing scheme, we tested only the isolates used in this study, for which we knew that lacF would differentiate the CVC and CLS strains. Thus, we caution that this approach may occasionally generate incorrect strain assignment if large numbers of samples are tested, as there is probably much more X. fastidiosa diversity in Brazil that has not been appropriately analyzed. In addition, our data show the presence of recombination among citrus- and coffee-colonizing X. fastidiosa isolates, which may affect the usefulness of this diagnostic tool.
DISCUSSION
We used microsatellites and a five-locus MLST scheme to study the genetic structure of X. fastidiosa isolates causing disease in citrus and coffee plants in Brazil and conducted biological tests to determine colonization patterns of isolates in both plants. Diversity estimates for microsatellite data (citrus H = 0.613 ± 0.117; coffee H = 0.632 ± 0.117) were higher than those observed using MLST (citrus H = 0.108 ± 0.066; coffee H = 0.311 ± 0.086). These are not unexpected findings but show the usefulness of microsatellites for population level studies of X. fastidiosa, while MLST may be more appropriate for strain/subspecies studies. Although citrus and coffee isolates tended to cluster separately in our study, evidence for recombination among those clusters indicates that further work is necessary to define the evolutionary relationships between X. fastidiosa isolates causing disease in these hosts. That research may also identify consistent differences among those isolates that are associated with pathogen-host specificity, an important question not addressed so far for X. fastidiosa. Nevertheless, independently of the method used, citrus and coffee X. fastidiosa isolates were usually grouped in different genetic clusters and were biologically distinct in cross-inoculation experiments.
Although both microsatellite and MLST provided similar general results, MLST was more informative for our purposes. In this study, STRUCTURAMA analysis of the microsatellite data better matched the MLST results than did STRUCTURE analysis. An increased number of loci for the microsatellite study would probably enhance the resolution power of the clustering algorithms used here (as already suggested [32]); large numbers of microsatellites have been previously shown to successfully group X. fastidiosa isolates (27). Nevertheless, microsatellite data assigned most isolates to groups based on host plant of origin, with recombinant citrus ST2 occasionally grouping with coffee STs. Despite the fact that the overall assignment of isolates suggested that CVC and CLS are caused by different strains of X. fastidiosa, AMOVA did not support that interpretation (P = 0.1). The presence of widespread recombination in the dataset and the small number of loci tested may have been important factors affecting this analysis. Similar factors may underlie contradictory results previously published on the genetic relationships among CVC and CLS X. fastidiosa isolates (29, 30, 42). The lack of geographical structure in the samples is not surprising. Coffee and citrus nurseries in Sao Paulo state ship plant material to growers throughout the country and are located in the region where both diseases were first diagnosed. Strict regulations have been recently implemented to control the production and shipment of healthy citrus nursery trees in Sao Paulo and other Brazilian states (J. R. S. Lopes, personal communication).
Our finding that the sequenced isolate 9a5c (ST2) is not a common ST and is recombinant with coffee X. fastidiosa provides an explanation for why some citrus isolates were occasionally not clustered together within the major CVC genetic group (e.g., reference 29); we would have concluded similarly if the MLST component of this work had not been performed. This observation is of relevance as the genome sequence of 9a5c is the basis for methods of detection of the causal agent of CVC, despite the fact that it is not the most common ST causing the disease. Because the CVC strain is on the U.S. government list of selected foreign threatening agents (5), adequate genetic data on this strain of X. fastidiosa is of importance. Similarly, better biological characterization of CLS isolates is necessary, as STs identified here may be biologically distinct and explain differences in CLS symptoms observed in Brazil (J. R. S. Lopes, personal observations). In addition to causing disease in citrus and coffee plants in Brazil, both strains have been reported to cause disease in grape plants (25).
The biological host range of X. fastidiosa strains is generally poorly understood. This bacterium shows a broad host range when nonsymptomatic systemic hosts are included in studies (40), as disease is often not the outcome of X. fastidiosa-plant encounters. It was observed here and in a previous study (37) that X. fastidiosa CLS STs do not colonize citrus plants, whereas citrus-X. fastidiosa STs can multiply in coffee plants when higher inoculum concentrations are used, but the infection likely dies off in the latter host (26, 37). Our results match those of Prado et al. (37), indicating that citrus and coffee X. fastidiosa isolates are biologically distinct. Isolates used in that study were typed here and belong to ST1 and ST4, the most common citrus and coffee X. fastidiosa STs detected by us. However, our results differ from those of Li et al. (26), who used one isolate each from coffee and citrus plants to infect coffee plants and observed CLS with both isolates. Possible reasons for such discrepancy have been previously discussed (37). We did not observe typical CLS symptoms, such as those described previously (26), in our biological tests. It is possible that symptoms of CLS appear only years after infection in the laboratory or field and also that our greenhouse conditions were not appropriate for the presence of CLS symptoms on plants. We observed disease symptoms in citrus but not coffee plants.
Scally et al. (44) were the first to show the presence of homologous recombination in X. fastidiosa, although they primarily analyzed North American isolates of this bacterium. We were able to obtain data supporting the presence of recombination events among coffee and citrus X. fastidiosa isolates. The network-like representation of the relationships among coffee and citrus STs (Fig. 2B) compared to what was found for North American isolates is also indicative of higher recombination rates in South America. That may be explained ecologically by the fact that citrus and coffee plants are grown sympatrically in Brazil and share leafhopper vectors of X. fastidiosa (43), increasing the number of opportunities for recombination events to occur. The diversity of species in the vector taxon (Cicadellinae) is much higher in South America (43). Thus, X. fastidiosa isolates infecting crop plants in North America usually have more-restricted geographical, vector, and host plant ranges, potentially limiting recombination. Although high recombination rates may maintain the integrity of genetic clusters, reducing divergence (17), X. fastidiosa with ecological opportunities for recombination (sympatric host plants and shared vector species) may speciate in the presence of a strong selective force. We propose that the major force maintaining the integrity of these clusters of sympatric, recombinogenic X. fastidiosa in Brazil is their respective host plants, with selection for pathogenicity (i.e., multiplication and movement within hosts in this case, which are both required for pathogenicity and vector transmission) driving speciation. It will be interesting to compare genome sequences of CVC- and CLS-causing X. fastidiosa isolates, as those may provide insights into what drives host-pathogen specificity.
Reconstructing the evolutionary history and defining species of recombinogenic bacteria is a challenging endeavor (16, 34). In the case of X. fastidiosa, the use of nonrepresentative datasets and the reliance on individual markers or markers with inadequate resolution have limited our understanding of the biology and evolution of this pathogen. We used data from Schuenzel et al. (47) on North American X. fastidiosa to determine the relationship of those STs with those from Brazil. X. fastidiosa isolates from North and South America formed distinct clades, without intermediate taxa. Four main clades were identified in our study: (i) X. fastidiosa subsp. multiplex, (ii) X. fastidiosa subsp. piercei, (iii) X. fastidiosa subsp. sandyi, and (iv) X. fastidiosa subsp. pauca. CLS isolates were not included in the studies that described these subspecies, but we assume that those isolates would belong to X. fastidiosa subsp. pauca. From our results with coffee STs, it seems clear that increasing the number of X. fastidiosa isolates tested using an MLST scheme will improve our understanding of the evolutionary relationships among strains of this pathogen. In addition, it would permit researchers to determine the degrees of clonality and recombination among these taxa, which would be useful for estimating the robustness of pathogen-specific detection markers. The study of isolates from Central America (32) and Taiwan (24) with an MLST approach would provide important information on the evolutionary history of X. fastidiosa. A recent genetic study of isolates from Costa Rica showed that they cluster with North instead of South American X. fastidiosa, although specific relationships were poorly resolved with the methods used (32). It was intriguing to identify a probable recombination event between a South American isolate from a coffee plant and North American isolates (one grape isolate was tested, but others have similar sequences for that locus as well). A previous report indicated that an isolate of X. fastidiosa from plum plants in Brazil had more genome similarity, as determined by microarray hybridization studies, to strains in North America than to those in Brazil (33). Those authors suggested that the introduction of contaminated plant material may explain that observation. It will be interesting to genetically analyze plum leaf scald-causing X. fastidiosa from Brazil and compare those data with data for samples from other host plants. More research is needed to address this question, but our results support the lack of monophyly between North and South American X. fastidiosa isolates.
The combination of microsatellite- and MLST-based approaches in the present study increased our understanding of the genetic relationships among X. fastidiosa isolates collected from symptomatic citrus and coffee plants in Brazil. Although we argue that further characterization of this system is necessary, our results allow us to conclude that CVC and CLS are caused by genetically distinct groups of X. fastidiosa that are frequently recombining. In addition, results from this and a previous study (37) indicate that CVC isolates may multiply but do not sustain infection in coffee plants, while CLS isolates do not colonize citrus plants. However, because recombination is an integral component of this pathosystem and our sample was relatively small, future studies should increase the number of isolates tested genetically and biologically to confirm or reject some of the hypotheses we have proposed. Furthermore, recombination may be widespread in X. fastidiosa and is potentially very important to its evolution, adaptation to new host plants, and speciation. In addition, horizontal gene transfer of pathogenicity factors may drive the emergence of new X. fastidiosa diseases (13).
FIG. 3.
Maximum likelihood mapping analysis with concatenated sequence and single-locus data by grouping of X. fastidiosa by subspecies: X. fastidiosa subsp. pauca (citrus and coffee), X. fastidiosa subsp. piercei (grape), X. fastidiosa subsp. sandyi (oleander), and X. fastidiosa subsp. multiplex (oak, peach, and almond). Values in the different regions of the equilateral triangle indicate support for possible tree topologies. Strong support for specific topologies is indicated by high values at single vertices of the triangle.
Acknowledgments
We acknowledge the researchers cited in Table 1 for providing some of the isolates used for this study. We also thank Mate Lopes, Cristiane Cava, and Érica Olandini for technical assistance with the bioassays and S. Purcell, L. Nunney, J. Lozier, and two anonymous reviewers for helpful discussions and comments on the manuscript.
This work was partly funded by the UC-AES and Fundecitrus. The last author acknowledges CAPES/Brazil for a postdoctoral scholarship that made possible the collaboration that resulted in this publication.
Footnotes
Published ahead of print on 18 April 2008.
REFERENCES
- 1.Almeida, R. P. P., M. J. Blua, J. R. S. Lopes, and A. H. Purcell. 2005. Vector transmission of Xylella fastidiosa: applying fundamental knowledge to generate disease management strategies. Ann. Entomol. Soc. Am. 98:775-786. [Google Scholar]
- 2.Almeida, R. P. P., E. F. Pereira, A. H. Purcell, and J. R. S. Lopes. 2001. Multiplication and movement of a citrus strain of Xylella fastidiosa within sweet orange. Plant Dis. 85:382-386. [DOI] [PubMed] [Google Scholar]
- 3.Almeida, R. P. P., and A. H. Purcell. 2003. Biological traits of Xylella fastidiosa strains from grapes and almonds. Appl. Environ. Microbiol. 69:7447-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, P., A. Cunningham, N. Patel, F. Morales, P. Epstein, and P. Daszak. 2004. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19:535-544. [DOI] [PubMed] [Google Scholar]
- 5.Anonymous. 2002. Agricultural bioterrorism protection act of 2002; possession, use and transfer of biological agents and toxins. Fed. Regist. 67:76908-76937. [Google Scholar]
- 6.Bruen, T., H. Philippe, and D. Bryant. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, C. J., M. Garnier, L. Zreik, V. Rossetti, and J. M. Bove. 1993. Culture and serological detection of the xylem-limited bacterium causing citrus variegated chlorosis and its identification as a strain of Xylella fastidiosa. Curr. Microbiol. 27:137-142. [DOI] [PubMed] [Google Scholar]
- 8.Coletta-Filho, H. D., M. A. Takita, A. A. Souza, C. I. Aguilar-Vildoso, and M. A. Machado. 2001. Differentiation of strains of Xylella fastidiosa by a variable number of tandem repeat analysis. Appl. Environ. Microbiol. 67:4091-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corander, J., P. Waldmann, and M. J. Sillanpää. 2003. Bayesian analysis of genetic differentiation between populations. Genetics 163:367-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didelot, X., and D. Falush. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evanno, G., S. Regnaut, and J. Goudet. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14:2611-2620. [DOI] [PubMed] [Google Scholar]
- 12.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 13.Friesen, T. L., E. H. Stukenbrock, Z. Liu, S. Meinhardt, H. Ling, J. D. Faris, J. B. Rasmussen, P. S. Solomon, B. A. McDonald, and R. P. Oliver. 2006. Emergence of a new disease as a result of interspecific virulence locus transfer. Nat. Genet. 38:953-956. [DOI] [PubMed] [Google Scholar]
- 14.Gelman, A., and D. B. Rubin. 1992. Inference from iterative simulation using multiple sequences. Stat. Sci. 7:457-511. [Google Scholar]
- 15.Hall, T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 16.Hanage, W., C. Fraser, and B. Spratt. 2005. Fuzzy species among recombinogenic bacteria. BMC Biol. 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanage, W., B. Spratt, K. Turner, and C. Fraser. 2006. Modeling bacterial speciation. Philos. Trans. R. Soc. Lond. B 361:2039-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartung, J. S., J. Beretta, R. H. Brlansky, J. Spisso, and R. F. Lee. 1994. Citrus variegated chlorosis bacterium: axenic culture, pathogenicity, and serological relationships with other strains of Xylella fastidiosa. Phytopathology 84:591-597. [Google Scholar]
- 19.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 20.Hill, B. L., and A. H. Purcell. 1995. Multiplication and movement of Xylella fastidiosa within grapevine and four other plants. Phytopathology 85:1368-1372. [Google Scholar]
- 21.Hopkins, D. L., and A. H. Purcell. 2002. Xylella fastidiosa: cause of Pierce's disease of grapevine and other emergent diseases. Plant Dis. 86:1056-1066. [DOI] [PubMed] [Google Scholar]
- 22.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 23.Jolley, K., E. Feil, M. Chan, and M. Maiden. 2001. Sequence type analysis and recombinational tests. Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 24.Leu, L. S., and C. C. Su. 1993. Isolation, cultivation, and pathogenicity of Xylella fastidiosa, the causal bacterium of pear leaf scorch disease in Taiwan. Plant Dis. 77:642-646. [Google Scholar]
- 25.Li, W. B., C. H. Zhou, W. D. Pria, D. C. Teixeira, V. S. Miranda, E. O. Pereira, A. J. Ayres, C. X. He, P. I. Costa, and J. S. Hartung. 2002. Citrus and coffee strains of Xylella fastidiosa induce Pierce's disease in grapevine. Plant Dis. 86:1206-1210. [DOI] [PubMed] [Google Scholar]
- 26.Li, W. B., W. D. Pria, D. C. Teixeira, V. S. Miranda, A. J. Ayres, C. F. Franco, M. G. Costa, C. S. He, P. I. Costa, and J. S. Hartung. 2001. Coffee leaf scorch caused by a strain of Xylella fastidiosa from citrus. Plant Dis. 85:501-505. [DOI] [PubMed] [Google Scholar]
- 27.Lin, H., E. L. Civerolo, R. Hu, S. Barros, M. Francis, and A. W. Walker. 2005. Multilocus simple sequence repeat markers for differentiating strains and evaluating genetic diversity of Xylella fastidiosa. Appl. Environ. Microbiol. 71:4888-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 29.Mehta, A., R. Leite, and Y. Rosato. 2001. Assessment of the genetic diversity of Xylella fastidiosa isolated from citrus in Brazil by PCR-RFLP of the 16S rDNA and 16S-23S intergenic spacer and rep-PCR fingerprinting. Antonie van Leeuwenhoek 79:53-59. [DOI] [PubMed] [Google Scholar]
- 30.Mehta, A., and Y. Rosato. 2001. Phylogenetic relationships of Xylella fastidiosa strains from different hosts, based on 16S rDNA and 16S-23S intergenic spacer sequenceS. Int. J. Syst. Evol. Microbiol. 51:311-318. [DOI] [PubMed] [Google Scholar]
- 31.Minsavage, G. V., C. M. Thompson, D. L. Hopkins, R. M. V. B. C. Leite, and R. E. Stall. 1994. Development of a polymerase chain reaction protocol for detection of Xylella fastidiosa in plant tissue. Phytopathology 84:456-461. [Google Scholar]
- 32.Montero-Astua, M., J. Hartung, E. Aguilar, C. Chacon, W. Li, F. Albertazzi, and C. Rivera. 2007. Genetic diversity of Xylella fastidiosa strains from Costa Rica, Sao Paulo, Brazil, and United States. Phytopathology 97:1338-1347. [DOI] [PubMed] [Google Scholar]
- 33.Nunes, L. R., Y. B. Rosato, N. H. Muto, G. M. Yanai, V. S. Silva, D. B. Leite, E. R. Goncalves, A. A. Souza, H. D. Colleta-Filho, M. A. Machado, S. A. Lopes, and R. C. Oliveira. 2003. Microarray analyses of Xylella fastidiosa provide evidence of coordinated transcription control of laterally transferred elements. Genome Res. 13:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papke, R., O. Zhaxybayeva, E. Feil, K. Sommerfeld, D. Muise, and W. Doolittle. 2007. Searching for species in haloarchaea. Proc. Natl. Acad. Sci. USA 104:14092-14097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paradela Filho, O., M. Sugimori, I. Ribeiro, A. Garcia, M. Beretta, R. Harakava, M. Machado, F. Laranjeira, J. Rodrigues Neto, and L. Beriam. 1997. Constatacao de Xylella fastidiosa em cafeeiro no Brasil. Summa Phytopathol. 23:46-49. [Google Scholar]
- 36.Posada, D., and K. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 37.Prado, S., J. R. S. Lopes, C. Demetrio, A. Borgatto, and R. P. P. Almeida. 2008. Host colonization differences between citrus and coffee isolates of Xylella fastidiosa in reciprocal inoculation. Sci. Agricola 65:251-258. [Google Scholar]
- 38.Pritchard, J. K., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purcell, A. H. 1997. Xylella fastidiosa, a regional problem or global threat? J. Plant Pathol. 79:99-105. [Google Scholar]
- 40.Purcell, A. H., and S. R. Saunders. 1999. Fate of Pierce's disease strains of Xylella fastidiosa in common riparian plants in California. Plant Dis. 83:825-830. [DOI] [PubMed] [Google Scholar]
- 41.Purcell, A. H., S. R. Saunders, M. Hendson, M. E. Grebus, and M. J. Henry. 1999. Causal role of Xylella fastidiosa in oleander leaf scorch disease. Phytopathology 89:53-58. [DOI] [PubMed] [Google Scholar]
- 42.Qin, X., V. S. Miranda, M. A. Machado, E. G. M. Lemos, and J. S. Hartung. 2001. An evaluation of the genetic diversity of Xylella fastidiosa isolated from diseased citrus and coffee in Sao Paulo, Brazil. Phytopathology 91:599-605. [DOI] [PubMed] [Google Scholar]
- 43.Redak, R. A., A. H. Purcell, J. R. S. Lopes, M. J. Blua, R. F. Mizell, and P. C. Andersen. 2004. The biology of xylem fluid-feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology. Annu. Rev. Entomol. 49:243-270. [DOI] [PubMed] [Google Scholar]
- 44.Scally, M., E. L. Schuenzel, R. Stouthamer, and L. Nunney. 2005. Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Appl. Environ. Microbiol. 71:8491-8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaad, N. W., E. Postnikova, G. Lacy, M. Fatmi, and C. J. Chang. 2004. Xylella fastidiosa subspecies: X. fastidiosa subsp. piercei subsp. nov., X. fastidiosa subsp. multiplex subsp. nov., X. fastidiosa subsp. pauca subsp. nov. Syst. Appl. Microbiol. 27:290-300. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt, H. A. K., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 47.Schuenzel, E., M. Scally, R. Stouthamer, and L. Nunney. 2005. A multilocus phylogenetic study of clonal diversity and divergence in North American strains of the plant pathogen Xylella fastidiosa. Appl. Environ. Microbiol. 71:3832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 49.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Sluys, M. A., M. C. de Oliveira, C. B. Monteiro-Vitorello, C. Y. Miyaki, L. R. Furlan, L. E. A. Camargo, A. C. R. da Silva, D. H. Moon, M. A. Takita, E. G. M. Lemos, M. A. Machado, M. I. T. Ferro, F. R. da Silva, M. H. S. Goldman, G. H. Goldman, M. V. F. Lemos, H. El-Dorry, S. M. Tsai, H. Carrer, D. M. Carraro, R. C. de Oliveira, L. R. Nunes, W. J. Siqueira, L. L. Coutinho, E. T. Kimura, E. S. Ferro, R. Harakava, E. E. Kuramae, C. L. Marino, E. Giglioti, I. L. Abreu, L. M. C. Alves, A. M. do Amaral, G. S. Baia, S. R. Blanco, M. S. Brito, F. S. Cannavan, A. V. Celestino, A. F. da Cunha, R. C. Fenille, J. A. Ferro, E. F. Formighieri, L. T. Kishi, S. G. Leoni, A. R. Oliveira, V. E. Rosa, F. T. Sassaki, J. A. D. Sena, A. A. de Souza, D. Truffi, F. Tsukumo, G. M. Yanai, L. G. Zaros, E. L. Civerolo, A. J. G. Simpson, N. F. Almeida, J. C. Setubal, and J. P. Kitajima. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woolhouse, M., and S. Gowtage-Sequeira. 2005. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 11:1842-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]