Abstract
Fungi are found in a wide range of environments, and the ecological and host diversity of the fungus Nectria haematococca has been shown to be due in part to unique genes on different supernumerary chromosomes. These chromosomes have been called “conditionally dispensable” (CD) since they are not needed for axenic growth but are important for expanding the host range of individual isolates. From a biological perspective, the CD chromosomes can be compared to bacterial plasmids that carry unique genes that can define the habits of these microorganisms. The current study establishes that the N. haematococca PDA1-CD chromosome, which contains the genes for pea pathogenicity (PEP cluster) on pea roots, also carries a gene(s) for the utilization of homoserine, a compound found in large amounts in pea root exudates. Competition studies demonstrate that an isolate that lacks the PEP cluster but carries a portion of the CD chromosome which includes the homoserine utilization (HUT) gene(s) is more competitive in the pea rhizosphere than an isolate without the CD chromosome.
Plants release as much as 40% of their photosynthates into the soil through their roots (52). The excreted photosynthetic products have various functions, such as detoxifying soil toxins, sequestering nutrients, lubricating the roots for growth through soil, signaling between adjacent plants and between plants and beneficial microbes, and supporting the growth of different types of microbes in the rhizosphere (16). The phenomenon whereby microbial populations are increased in soil that is under the direct influence of root exudates is known as the “rhizosphere effect” (4). Most researchers agree that there is an enrichment of a specific spectrum of microorganisms in the rhizosphere and that this population is less diverse than that in the bulk soil, which is not under the direct influence of root exudates (4, 16). Less accepted are how often a specific microbial genotype is favored in the rhizosphere by a specific plant species or genotype and whether there are particular traits of plants and microbes that control this specificity.
Over the past ∼20 years, experiments combining the genetic dissection of rhizosphere colonization with other novel approaches have identified several bacterial traits for rhizosphere competence and colonization (23, 28, 34). For example, studies on the symbiotic bacteria that fix nitrogen in legume roots have demonstrated that the NOD genes, which induce nodulation of plant roots in response to specific plant signals, are found on the symbiosis (Sym) plasmid (25). Sym plasmids carry not only the genes for nodulating specific hosts (36) but also the genes for catabolizing unique compounds present in the root exudates of the host plants (1, 10). The correlation between nodulation specificity and the ability to catabolize specific host root exudates was originally suggested by VanEgeraat (45), who discovered that the pea-nodulating bacterium Rhizobium leguminosarum could catabolize homoserine (HS), a compound found in high concentrations in pea root exudates. It was shown later that the Sym plasmid of R. leguminosarum carries the gene for HS utilization as well as the NOD genes (6).
Although the diversity of fungi in the rhizosphere is well recognized, in contrast to the case for bacteria the genetic determinants that enable fungi to inhabit the rhizosphere of host plants remain, for the most part, unknown. Fungal interactions with plant roots are of major ecological and economic importance for the development of mycorrhizal symbiosis and the control of soilborne pathogens. In the current study, the understanding of the bacterial genes involved in symbiosis and in the colonization of the host rhizosphere (23, 28, 34) was used as a model to identify some of the genes that may play parallel roles in the root-pathogenic fungus Nectria haematococca (anamorph, Fusarium solani).
The fungus N. haematococca is found as a soil saprobe, a commensal organism in the rhizosphere, and a pathogen of many different plant species (26, 46). The genetic and habitat diversity of N. haematococca is due in part to the presence of supernumerary chromosomes (49). These “extra” chromosomes are called “conditionally dispensable” (CD) chromosomes because while they are not required for axenic growth, they may allow isolates to have an expanded host range (3). There are several different CD chromosomes, one of which, the PDA1-CD chromosome, carries a cluster of genes for pea pathogenicity (PEP cluster) (13). N. haematococca isolates with the PDA1-CD chromosome are highly virulent on pea plants (3, 27, 49). The PDA1 gene, from which the PDA1-CD chromosome takes its name, is a member of the PEP cluster and codes for a cytochrome P450 enzyme that detoxifies the pea phytoalexin (defense molecule) pisatin (24). The PDA1 gene is used routinely as a marker for the presence of this CD chromosome (49).
In this study, we show that isolates of N. haematococca that are pathogenic on pea plants can grow on HS as a sole carbon and nitrogen source but that isolates from other hosts and sources usually cannot. Furthermore, we show that the gene(s) for HS utilization (HUT) is on the PDA1-CD chromosome. We also report the development of a real-time PCR technique that overcomes one of the major hurdles in studying fungal rhizosphere competence and colonization, i.e., finding an accurate means to quantify fungal biomass (cell number) in the rhizosphere (32). Using this technique, we demonstrate that the portion of the PDA1-CD chromosome that contains the HUT gene(s) provides N. haematococca isolates with a competitive advantage in the pea rhizosphere.
MATERIALS AND METHODS
Fungal strains.
Isolates of N. haematococca were obtained from the culture collection of Hans VanEtten (Tables 1, 2, and 3). Stock cultures of N. haematococca were maintained as slant cultures on V-8 agar medium (M-29 medium) (39). Cultures were grown in the dark at 27°C.
TABLE 1.
Growth on HS of N. haematococca isolates that are pathogenic on pea plantsa
| Isolate | Dry wt (mg)c | Radial growth (mm)d | Geographical origin |
|---|---|---|---|
| T1 | 6.5 | 12/20 | New York |
| T2b | 6.5 | 24/24 | New York |
| T8b | 5.7 | 12/15 | New York |
| T9b | 6.8 | 18/18 | New York |
| T10 | 5.7 | 18/20 | New York |
| T17 | 5.2 | 15/20 | New York |
| T23b | 7 | ND | New York |
| T30b | 4.7 | 12/20 | Washington |
| T63 | 5.6 | 20/25 | New York |
| T70b | 6.9 | 20/20 | Michigan |
| T468b | 7.3 | 17/14 | England |
| T547b | 6.9 | 18/18 | New Zealand |
| T406 | 5.6 | 14/15 | Taiwan |
| T558e | 4.6 | 22/21 | Japan |
| T559e | 4 | 20/25 | Japan |
| T560e | 4.8 | 20/25 | Japan |
| T561e | 3.6 | 26/25 | Japan |
Isolates which are known to have a PDA1-CD chromosome, based on the presence of an ∼1.6-Mb chromosome containing the PDA1 gene, as determined by PFGE and/or Southern analysis (8, 27, 41).
After growth in 2 ml of 25-mg/ml l-HS for 14 days. Isolates grown on 2 ml of 25-mg/ml l-glutamic acid had dry weights ranging from 4.3 to 4.8 mg.
Diameter of the mycelial colony after growth for 4 days on semisolid medium containing 25 mg/ml of l-HS (first number) or 10 mg/ml of glucose (second number). ND, not determined.
Unpublished data.
TABLE 2.
Growth on HS of N. haematococca field isolates that are not pathogenic on pea plantsa
| Isolate | Dry wt (mg)b | Radial growth (mm)c | Geographical origin | Source |
|---|---|---|---|---|
| T34d | 0.5 | 0/15 | Australia | Cooling tower |
| T77d | 0.1 | 0/23 | Pennsylvania | Alfalfa |
| T78d | 0.4 | 0/20 | Pennsylvania | Alfalfa |
| T95d | 0.5 | 0/25 | South Carolina | Tulip tree |
| T110d | 0.4 | ND | Mississippi | Cottonwood |
| T213d | 0 | ND | Kentucky | Cottonwood |
| T215d | 0.4 | 0/13 | North Dakota | Potato |
| T217d | 0.5 | 0/25 | Pennsylvania | Carnation |
| T219d | 0 | 0/15 | New York | Soil |
| T272d | 5.1 | 10/20 | Utah | Unknown |
| T273d | 4.7 | 16/20 | Utah | Unknown |
| T288d | 1.3 | 0/25 | Pennsylvania | Soil |
| T300d | 0.7 | 0/20 | Pennsylvania | Red clover |
| T314d | 0.8 | 0/22 | Pennsylvania | Shrimp |
| T347d | 0.1 | 0/15 | New York | Alfalfa |
| T351d | 0.6 | 0/16 | New York | Alfalfa |
| T386d | 5.2 | ND | Spain | Chickpea |
| T562 | 0.6 | 0/20 | Japan | Alfalfa |
| T474d | 0.4 | 0/23 | England | Pea soil |
After growth in 2 ml of 25-mg/ml l-HS for 14 days. Isolates grown in 2 ml of 25-mg/ml l-glutamic acid had values ranging from 4.3 to 4.8 mg.
Diameter of the mycelial colony after growth for 4 days on semisolid medium containing 25 mg/ml of l-HS (first number) or 10 mg/ml of glucose (second number). ND = not determined.
TABLE 3.
Growth on HS of selected isolates of N. haematococca that do or do not contain the PDA1-CD chromosome or its marker gene, PDA1
| Isolate(s) | Description | Growth on HSh |
|---|---|---|
| 77-1-3, 77-5-5, 77-13-1, 94-6-1, 44-100,a 44-1, 44-22, 44-36, 44-46, 44-64, 44-75 | Meiotic progeny without the PDA1 geneg | − |
| 77-1-3, 77-5-7, 77-13-4,b 77-13-7,b 44-16, 44-20, 44-37, 94-1-6 | Meiotic progeny with the PDA1 geneg | + |
| B-13, B-32, B-33, B-34, B-35, B-36, B-37 | hph-tagged CD chromosome loss induced by benomyl treatment of Tr18.5 and isolates selected by loss of hygromycin resistancec | − |
| HT1, HT3, HT4, HT5, HT6, HT7, HT8, HT9, HT10 | CD chromosome loss induced by benomyl treatment of 77-13-4 and isolates selected by loss of HS utilization | − |
| Tr115.1,d Tr135.8,d Tr23.1d | CD chromosome lost during transformation of 77-13-4e | − |
| Tr78.2d | Portion of the CD chromosome was retained after transformation of 77-13-4e | + |
| N-15 | 100-kb truncation of the CD chromosome in 77-13-7f | + |
| Tr18.5d | PDA1 gene disruptant of 77-13-7e | + |
| Tr86.1d | PDA1 gene disruptant of 77-13-4e | + |
Lack of PDA1-CD chromosome demonstrated (8).
Presence of PDA1-CD chromosome demonstrated (51).
Isolates shown to lack a PDA1-CD chromosome (50).
Absence of PDA1-CD chromosome demonstrated (51).
Isolates from reference 51.
Isolate from reference 18.
Isolates from reference 17.
Growth was determined as described in the legend to Fig. 1. +, obvious growth after 4 days; −, no growth after 4 days on semisolid medium containing 25 mg/ml of l-HS.
Media.
Measurements of the growth of N. haematococca on different carbon and/or nitrogen sources were carried out on solidified Ustilago minimal medium (M-100 medium) (39). M-100 medium, which contains 10 mg/ml glucose as the carbon source and 3 mg/ml NH3(NO3)2 as the nitrogen source, was modified by replacing glucose and/or NH3(NO3)2 with the following chemicals: HS (Sigma-Aldrich, St. Louis, MO) (HS medium), trigonelline (Sigma-Aldrich, St. Louis, MO) (TGR medium), γ-glutamyl-d-alanine (a gift from Andrew Mort, Department of Biochemistry, Molecular and Cellular Biology, Oklahoma State University) (GAA medium), and glutamic acid (Sigma-Aldrich, St. Louis, MO). In some experiments, dl-HS was used instead of l-HS because dl-HS was more readily available. None of the isolates could grow on d-HS.
Spore production and collection.
N. haematococca spores were produced in petri dishes containing solidified V-8 agar (39). Cultures were incubated at 24°C ± 1°C under lighted conditions to encourage conidiation. After 1 to 2 weeks, the spores were harvested, rinsed with sterile water, suspended in water, and counted. For rhizosphere competence assays, the numbers of viable spores in the inocula were confirmed by dilution plating the spore suspensions on potato dextrose agar (Difco Laboratories, Detroit, MI) and counting fungal colonies.
Growth assays.
To measure growth as the change in culture turbidity, 100 μl of liquid medium containing the substrate being tested was placed into each well of a 96-well microtiter plate, and each well was inoculated with ∼500 spores. Plates were incubated in a moist chamber at 27°C. Turbidity was measured with a spectrophotometer (OpsysMR; ThermoLabsystem) as the optical densities at 540 nm and 620 nm at 12- to 24-h intervals for 5 days. To assay growth by measuring the change in dry weight, test tubes containing 2 ml of liquid medium were inoculated with 5 × 103 to 10 × 103 conidia and incubated at room temperature on a gyrorotatory shaker at 100 rpm for 13 days. Mycelium was collected by filtration, dried, and weighed. To assay radial growth, semisolid media containing Gelrite Gellan gum (3.5 mg/ml; Sigma-Aldrich, St. Louis, MO) as the solidifying agent and either 0.01 mg/ml glucose or 25 mg/ml l-HS were inoculated with 25 μl of a spore suspension containing 1,000 to 5,000 conidia. Colony diameters were measured after 4 days.
Benomyl treatment to induce loss of the PDA1-CD chromosome.
Fifty milliliters of M-100 medium containing 37 μg of benomyl/ml was inoculated with ∼3 × 104 spores of isolate 77-13-4 and incubated at room temperature with shaking at 190 rpm. After 8 days, the culture was filtered through two layers of cotton filter paper, and fresh liquid M-100 and benomyl were added to the filtrate to obtain a final volume of 75 ml with 37 μg of benomyl/ml. After another 6 days, the culture was filtered and the filtrate was centrifuged to collect the spores. The spores were washed three times with sterile water and used to inoculate 50 ml of liquid dl-HS medium (50 mg dl-HS/ml). The cultures were incubated overnight at room temperature with shaking at 50 rpm. The culture was then filtered through one layer of cotton filter paper to remove germinated spores, and the filtrate was centrifuged to collect the ungerminated spores. The spores were suspended in water and counted, and 150 spores were spread onto M-100 agar containing 0.8 mg/ml Triton X-100 (Fisher Scientific, Pittsburgh, PA). The plates (100 × 15 mm) were overlaid immediately with nylon membranes (curtain fabric; 3 fibers/mm) as described previously (50). After 24 h, the nylon membranes were transferred to HS agar (50 mg dl-HS/ml) containing 0.4 mg/ml Triton X-100. Spores were collected from those colonies on the M-100 agar that did (HUT+) and did not (HUT−) grow on HS agar. After single-spore isolation, the HUT phenotype of a single spore culture from each of the colonies was retested on HS agar.
Southern hybridization.
For Southern hybridization, 1 μg of genomic DNA was digested with XhoI according to standard protocols. The digested DNA was size fractionated by electrophoresis on a 0.7% agarose gel, followed by transfer to a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech Ltd.). The membranes were prehybridized for 4 h at 42°C in prehybridization solution (50% formamide, 5× Denhardt's solution, 1 M NaCl, 50 mM PIPES, 0.5% Sarkosyl, 500 μg/ml salmon sperm DNA, 25 μg/ml tRNA, and 10 mM EDTA) and then hybridized overnight with one of the probes at 42°C.
To obtain DNA templates for the synthesis of radioactively labeled probes, three cosmids (3B04, 6H10, and 8A10) from a PDA1-CD chromosome-specific library were digested with either EcoRI or EcoRI and EcoRV. The restriction fragments were resolved by electrophoresis in 1% low-melting-point agarose, all at 24 V/cm overnight. The portions of the gel containing a 1.7-kb EcoRI fragment from cosmid 3B04, a 1.5-kb EcoRI fragment from cosmid 6H10, and a 1.5-kb EcoRI/EcoRV fragment from cosmid 8A10 were excised, and the DNAs were radiolabeled using a Rad-Prime kit (Gibco-BRL) according to the manufacturer's instructions.
Following hybridization, membranes were washed twice in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) at room temperature for 30 min, twice in 2× SSPE containing 0.1% sodium dodecyl sulfate at 65°C for 30 min, and twice in 0.2× SSPE containing 0.1% sodium dodecyl sulfate at 65°C for 30 min.
PFGE analyses of chromosomal DNA.
The preparation of protoplasts for karyotypic chromosomal analysis was performed as described previously by Taga et al. (40), with the exception that 1.2 M MgSO4 was used as the osmotic medium instead of 1.2 M NaCl. Chromosomes were resolved in agarose gels (0.008 g/ml pulsed-field certified agarose; Bio-Rad Laboratories, Inc., Hercules, CA) prepared in running buffer (0.5× Tris-borate-EDTA) by pulsed-field gel electrophoresis (PFGE).
Fluorescence in situ hybridization.
Fixed specimens of mitotic nuclei and chromosomes for cytological observation and hybridizations were prepared as previously described (40, 43). The probe, genomic DNA from isolate 77-13-4, was labeled with biotin-14-dATP by nick translation using the BioNick labeling system (Invitrogen Corp., Carlsbad, CA). Observations were made with an Olympus BX60 epifluorescence microscope (Olympus America Inc., Center Valley, PA) equipped with an Olympus U-MWU2 excitation cube (Olympus America Inc.) for DAPI (4′,6′-diamidino-2-phenylindole), an Olympus U-MNB excitation cube (Olympus America Inc.) for Alexa Fluor 488, and a triple-band-pass filter (Chroma Technology Corp., Rockingham, VT) for DAPI and Alexa Fluor 488. Photographs were taken with an Optronics DEI-750D charge-coupled device camera (Optronics, Goleta, CA).
Rhizosphere competition assays.
A replacement series technique (5) was used to measure rhizosphere competition as a function of the relative abundances of the two competing N. haematococca isolates. Pea seeds were surface sterilized in 70% ethanol for 5 min and in a 2.5% sodium hypochlorite solution for 10 min and then rinsed thoroughly with sterile distilled water. Surface-sterilized seeds were soaked in sterile distilled water overnight at room temperature to allow the seeds to imbibe and then were planted (one seed per box) in Magenta GA-7 boxes (Magenta Corp., Chicago, IL) containing 100 g of sterile potting mixture (4 parts vermiculite to 1 part quartz sand wetted with 1 liter of sterile distilled water per 10 liter of mixture). After germination, plants were grown under a 12-hour-12-hour light-dark regimen at 24°C ± 1°C. After a 2-week growth period, the potting mixture was inoculated with spores of two isolates (Tr78.2 and either HT1 or HT5) prepared at a constant density of 105 N. haematococca spores per gram of potting mixture but at various ratios of the two isolates (0:100, 25:75, 50:50, 75:25, and 100:0). The plants were grown for another 2 or 3 weeks under the same conditions. During the growth period, the plants were watered with Hoagland's solution every 4 to 6 days. At the end of the 2- or 3-week period, the rhizospheres and roots were harvested by cutting the stem immediately above the uppermost roots and gently shaking off the loose potting mixture. The harvested roots and adhering potting mixture were lyophilized and ground to a fine powder in liquid nitrogen by use of a mortar and pestle. DNA for real-time PCR was extracted from 1 gram of ground rhizosphere material by use of an Ultraclean Soil DNA kit (Mo Bio Laboratories, Inc., Solana Beach, CA). Six replicates were performed for each ratio of N. haematococca isolates.
deWit replacement curves.
Competition between HUT+ and HUT− isolates is shown diagrammatically by plotting the results on a deWit replacement curve, on which dashed lines represent the growth of the isolates without competition and solid lines represent the actual growth under competition. If there is no competition between the two isolates, i.e., each can colonize the rhizosphere equally, then the ratios of the two isolates recovered from the rhizospheres of plants inoculated with HUT+/HUT− mixtures should be the same as the inoculation ratios.
Real-time quantitative reverse transcription-PCR.
The number of cells in each sample was determined from the target gene copy number, which was quantified by comparing the cycle threshold (CT) value of the samples to the CT value of the respective standard curve. Standard curves were constructed with serial dilutions of the PCR products obtained from the genomic DNAs of N. haematococca isolate 77-13-4 and the pea plant (cv. Little Marvel), using the same primer pairs as those used for real-time quantitative PCR with rhizosphere samples. The sequence for the N. haematococca actin gene was obtained from Liu et al. (22) and used for real-time PCR to determine the total number of fungal cells of isolates Tr78.2 and HT1 in the rhizosphere samples. A portion of the N. haematococca PDA1 sequence was used for real-time PCR to detect the number of fungal cells of Tr78.2. Tr78.2 lacks a wild-type copy of PDA1 but contains the hygromycin resistance gene (hph) flanked by 692 bp 5′ and 888 bp 3′ of the PDA1 gene (51). The PDA1 sequence from the 888-bp 3′-flanking region was used for real-time PCR. Real-time quantitative PCR using TaqMan technology was performed on an Abbott Prism system (Abbott Park, IL) according to the manufacturer's protocol. Sequences of the primers (Invitrogen Corporation, Carlsbad, CA) and TaqMan fluorescent probes (Applied Biosystems, Foster City, CA) used in the quantitative real-time PCR study were as follows: PDA1 forward primer, 5′-GATGAGCAGACTGAGGTTGGT3′; PDA1 reverse primer, 5′-CTGTGATGCCAAGGTCACTTA-3′; and PDA1 probe, 6-carboxyfluorescein (FAM)-AAGCGATCTTTGGCAACGATGCAAG-6-carboxytetramethylrhodamine (TAMRA); actin gene forward primer, 5′-ATCCACGTCACCACCTTCAA-3′; actin gene reverse primer, 5′-GTGCCAGAGTTAGAAATGATC-3′; and actin gene probe, FAM-ACATCGACATCACACTTCATGATGGAG-TAMRA.
The rubisco activase (RCA) gene from the pea was used to measure the number of pea cells in the sample and to normalize the amount and quality of the genomic DNA. RCA mRNA sequences were obtained from the GenBank sequence database for 12 plant species and aligned using the Clustal function of MacVector software. The region with the highest identity across species was used to design the forward (5′-CATTATGATGAGTGCTGGAGA-3′) and reverse (5′-TCCATACGACCATCACGGAT-3′) PCR primers. These primers were used to amplify the corresponding, ∼350-bp region in the pea RCA gene, using genomic DNA of the pea as the template. The pea RCA DNA sequence was used similarly to design the PCR primers and the TaqMan probe. The following were used for RCA: forward primer, 5′-CCTCTTCATCAACGATCTCGAT-3′; reverse primer, 5′-GGTTGTCAGCAATGTTCATGAG-3′; and probe, tetrachloro-6-carboxyfluorescein (TET)-CACCGTCAACAACCAGATGGTGA-ATG-TAMRA.
Each TaqMan probe was designed to anneal to a specific sequence between the forward and reverse primers of its target gene and to have at least a 5°C higher melting temperature than that of the PCR primers. Individual probes contained a reporter fluorochrome (FAM for PDA1 and the actin gene and TET for RCA) at the 5′ end and a quencher fluorochrome (TAMRA) at the 3′ end. PCR analyses were performed in duplicate, with duplicate reactions on each run. Each reaction mix had a total volume of 25 μl containing 12.5 μl of qPCR Mastermix Plus (contains reaction buffer deoxynucleoside triphosphates [including dUTP], Hot Goldstar DNA polymerase, 5 mM MgCl2, uracil-N-glycosylase, stabilizers, and a passive reference; Invitrogen Corp., Carlsbad, CA), 300 nM forward primer, 300 nM reverse primer, 200 nM TaqMan probe, 2 μl rhizosphere DNA template, and 9 μl of sterile distilled water. The PCR parameters were as follows: an initial uracil-N-glycosylase step at 50°C for 2 min, followed by an initial denaturation and Hot Goldstar DNA polymerase activation step at 95°C for 10 min and 40 cycles at 95°C for 15 s and 60°C for 30 s. Real-time PCRs for PDA1, the actin gene, and pea RCA were performed at least three times.
RESULTS
Growth of N. haematococca on compounds found in pea root exudates.
Previous studies have shown that all isolates of N. haematococca, independent of their pathogenicity and CD chromosome content, can grow on pea root exudates (11). In the present study, chemicals which are found in pea root exudates (19), i.e., HS, GAA, and TGR, were tested for the ability to support the growth of N. haematococca. Growth on HS and TGR was measured as an increase in biomass in liquid media and as an increase in colony diameter on semisolid media. Due to the small amounts of compound available, growth on GAA was measured only by the increase in turbidity.
All four isolates (T77, T219, T347, and 77-13-4) used in an initial screen were able to use GAA as a sole C and N source. However, only 77-13-4, the sole isolate of the four that is a pathogen on pea plants and carries the PDA1-CD chromosome (51), could grow on HS medium. TGR was inhibitory to growth when it was added to M-100 medium at concentrations above 0.05%, and no isolate could use this compound as a sole C and/or N source (data not shown).
Relationship between an isolate's pathogenicity and its ability to use HS.
To test further whether the pathogenicity of a field isolate is correlated with its ability to use HS as a sole C and N source, 36 field isolates from different hosts and geographic locations were examined. The 17 field isolates in Table 1 are pathogenic on pea plants, and some are known to contain a PDA1-CD chromosome based on Southern hybridization and PFGE analyses (27, 41). All of these isolates grew on HS, regardless of their geographic origin (Table 1). None of the 19 isolates in Table 2 are pathogenic on pea plants, and although a few of them, based on PFGE and Southern hybridization analyses, contain an ∼1.6-Mb chromosome, they do not contain a PDA1 gene (27). The isolates in Table 2 were obtained from a variety of habitats other than pea plants, and only three (T272, T273, and T386) were able to use HS as the sole C and N source (Table 2). Isolates T272 and T273 were obtained from soils with unknown plant associations. Isolate T386 came from a chickpea plant. All of the isolates in Tables 1 and 2 could grow on glutamic acid as a sole C and N source, whereas pea-pathogenic isolates had an additional metabolic capability, i.e., the ability to use the amino acid HS as a sole C and N source.
Location of the HS utilization (HUT) gene(s) on the PDA1-CD chromosome.
Previous studies have shown that the CD chromosomes of N. haematococca can be lost during sexual crosses, during transformation, and following treatment with benomyl (49, 50, 51). To determine if HUT is on the PDA1-CD chromosome, a series of related isolates, which differed with respect to the presence of the PDA1-CD chromosome or its marker gene, PDA1, were examined for HS utilization. A total of 18 progeny from three different crosses (crosses 44, 77, and 94) (17) in which PDA1 segregated were assayed for the HUT phenotype. The 8 progeny that inherited PDA1 grew on HS, whereas the 11 progeny that did not inherit PDA1 did not grow on HS (Table 3; Fig. 1). These results further support the hypothesis that the PDA1 gene and the gene(s) conferring the ability to grow on HS are located in the same linkage group, i.e., the PDA1-CD chromosome.
FIG. 1.
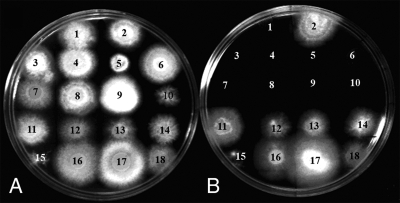
Growth of N. haematococca isolates on glucose (A) and HS (B) media. Isolates lacking the PDA1-CD chromosome or its marker gene, PDA1, included the following: 1, isolate Tr23.1; 2, isolate Tr78.2; 3, isolate Tr115.1; 4, isolate Tr135.8; 5, isolate B-13; 6, isolate 44-100; 7, isolate 44-75; 8, isolate 44-22; 9, isolate 44-1; and 10, isolate 94-6-1. Isolates with the PDA1-CD chromosome or its marker gene, PDA1, included the following: 11, isolate Tr86.1; 12, isolate 77-2-3; 13, isolate 77-13-4; 14, isolate 77-13-7; 15, isolate N15; 16, isolate 44-16; 17, isolate 44-37; and 18, isolate 94-1-6. Plates were photographed 4 days after inoculation.
This linkage was also tested with isolates that had lost the PDA1-CD chromosome during experimental manipulations. It had been observed that the PDA1-CD chromosome of N. haematococca isolate 77-13-4 was occasionally lost during transformation (51). Three of the four transformants of 77-13-4 (Tr115.1, Tr135.8, and Tr23.1) that had lost the PDA1-CD chromosome (51) could not grow on HS (Table 3; Fig. 1). Kistler et al. (18) also generated a transformant of 77-13-7, transformant N15, which lacked the PEP cluster due to an ∼100-kb truncation in the PDA1-CD chromosome. However, transformant N15 can grow on HS, indicating that the HUT gene(s) is located on the remaining portion of the PDA1-CD chromosome and is not part of the previously characterized PEP cluster.
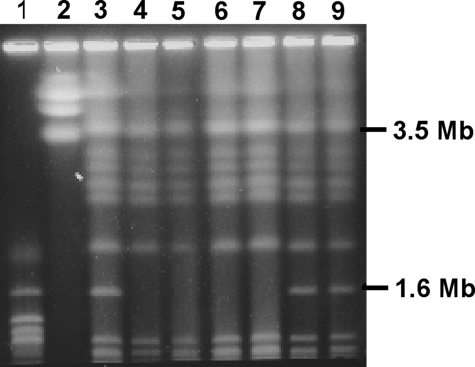
Previous studies evaluating the role of PDA1 in pea pathogenicity used site-directed mutagenesis to disrupt the PDA1 gene in isolate 77-13-7 (51). The resulting PDA-negative (PDA−) transformants had the PDA1-CD chromosome tagged with a hygromycin resistance gene (51) and were HUT+ (Table 3). One of these transformants (Tr18.5) was treated with benomyl, which can cause aneuploidy during vegetative growth in fungi, and the loss of the CD chromosome (50) was detected by a loss of hygromycin resistance. All seven hygromycin-sensitive isolates identified in those experiments (B-13, B-32, B-33, B-34, B-35, B-36, and B-37) lack the PDA1-CD chromosome (50) and cannot grow on HS (Table 3; Fig. 1). A similar benomyl treatment was performed on 77-13-4 in the current study, but in this case isolates were selected for a loss of the ability to grow on HS. Nine HUT− isolates were obtained (Table 3), and the chromosomes of four (HT1, HT3, HT4, and HT5) were resolved by PFGE; all had lost the PDA1-CD chromosome (Fig. 2). As controls, two isolates (HT11 and HT12) that retained the ability to grow on HS after the benomyl treatment were also analyzed and shown to have the PDA1-CD chromosome (Fig. 2).
FIG. 2.
PFGE karyotypes of isolate 77-13-4 and of mutants of 77-13-4 that lost the ability to grow on HS after treatment with benomyl. Lane 1, chromosomes of Saccharomyces cerevisiae; lane 2, chromosomes of Saccharomyces pombe; lane 3, chromosomes of 77-13-4; lanes 4 to 7, chromosomes of isolates that were HUT− (lane 4, HT1; lane 5, HT3; lane 6, HT4; and lane 7, HT5); lanes 8 and 9, chromosomes of isolates that were HUT+ after benomyl treatment (lane 8, HT11; and lane 9, HT12).
Isolate Tr78.2 contains the portion of the PDA1-CD chromosome that confers HUT.
The only result that appears to conflict with the hypothesis that the HUT gene(s) is on the PDA1-CD chromosome is that transformant Tr78.2, which was shown by Southern analysis to lack a wild-type PDA1 gene and by PFGE analysis to lack the PDA1-CD chromosome (51), grew on HS (Table 3; Fig. 1). However, it is possible that the translocation of a portion of the PDA1-CD chromosome occurred during transformation and was missed in the previous study, since it is difficult to resolve chromosomes larger than ∼4 Mb by PFGE (49).
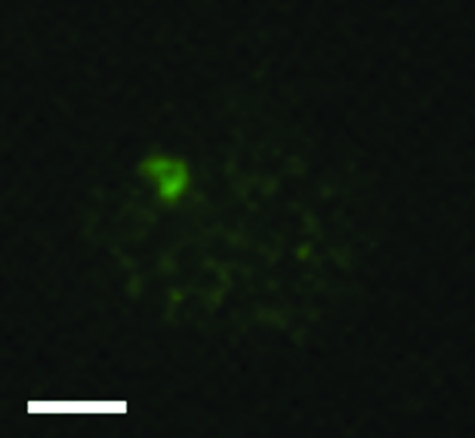
To determine whether a portion of the PDA1-CD chromosome might have been retained in isolate Tr78.2, interphase nuclei from Tr78.2 were examined using fluorescence in situ hybridization (Fig. 3). Nuclei from Tr78.2 were hybridized first with genomic DNA from isolate HT1 and subsequently with biotin-labeled DNA from isolate 77-13-4. This type of experiment has previously been shown to detect CD chromosome-specific DNA in isolates that differ only in the presence of a CD chromosome (40). A strongly hybridizing signal was detected (Fig. 3), which is consistent with Tr78.2 carrying the portion of the PDA1-CD chromosome that contains the HUT gene(s).
FIG. 3.
Visualization of interphase chromosomes of isolate Tr78.2. Interphase nuclei of Tr78.2 were hybridized with genomic DNA from isolate HT1, followed by hybridization with biotin-labeled DNA from isolate 77-13-4. The biotin-labeled DNA was detected with goat anti-biotin antibody, followed by staining with Alexa Fluor 488-conjugated rabbit anti-goat antibody. Scale bar = 2 μm.
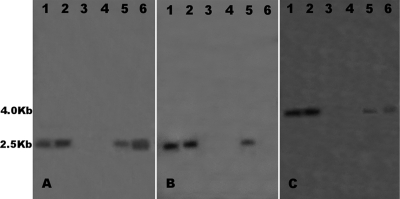
To verify that Tr78.2 contained a portion of the PDA1-CD chromosome, Southern blots of Tr78.2 and other isolates, with and without the PDA1-CD chromosome, were probed with DNAs from cosmid clones from the PDA1-CD chromosome. A previously constructed partial physical map of the PDA1-CD chromosome (35) allowed the selection of cosmid clones spanning ∼800 kb of the PDA1-CD chromosome. DNA fragments from cosmid clones (3B04 and 8A10) that contain DNA from the ends of this 800-kb region and from a cosmid (6H10) that contains DNA from the interior of the 800-kb region were hybridized to genomic DNAs of Tr78.2, 77-13-4 (the source of Tr78.2), and 77-13-7, another isolate from cross 77 that has the PDA1-CD chromosome. These fragments hybridized to Tr78.2, 77-13-4, and 77-13-7 but did not hybridize to HT1 and HT5 (Fig. 4), a result consistent with the presence of a portion of the PDA1-CD chromosome in Tr78.2.
FIG. 4.
Southern hybridization analysis of DNAs from isolates 77-13-4, 77-13-7, HT1, HT5, and Tr78.2 (lanes 1 to 5, respectively) for the presence of an ∼800-kb portion of the PDA1-CD chromosome. Fragments used as probes were a 1.7-kb EcoRI fragment from cosmid 3B04, which contains DNA from one end of the 800-kb region (A); a 1.5-kb EcoRI fragment from cosmid 6H10, which contains DNA from an internal portion of the 800-kb region (B); and a 1.5-kb EcoRI/EcoRV fragment from cosmid 8A10, which contains DNA from the opposite end of the 800-kb region (C).
Analysis of the rhizosphere competitive ability of isolate Tr78.2.
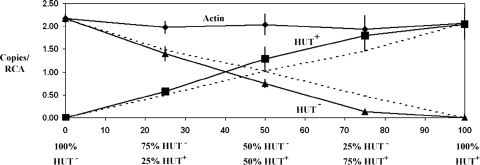
To determine if the portion of the PDA1-CD chromosome containing the HUT gene(s) confers a competitive advantage in the pea rhizosphere, we used a replacement series approach (5). Pea plants were grown for 2 weeks and then inoculated with different ratios of Tr78.2 and HT1 and of Tr78.2 and HT5. Two (for HT1) or three (for HT5) weeks after inoculation, the rhizosphere (roots with adhering potting mix) was removed and the DNA extracted. Real-time PCR analyses of the single-copy actin gene of N. haematococca and of a region of the PDA1 gene from the vector used to produce Tr78.2 were used to determine the total number of fungal cells and the relative number of Tr78.2 cells, respectively. Real-time PCR analyses of the pea RCA gene were used to measure the number of plant cells and to normalize the data. When the inoculum was 100% of either Tr78.2 (HUT+) or HT1 (HUT−), the same quantity of fungal cells (2.06 ± 0.32 versus 2.17 ± 0.09 actin genes/RCA gene) was present in the rhizospheres of pea plants, indicating that the pea rhizosphere had the same carrying capacity for both isolates (Fig. 5). Therefore, when there is no competition between the two isolates, both isolates would be expected to colonize the rhizosphere equally and to be present at the same relative ratios as those present in the initial inoculum. However, the real-time PCR results showed that 2 weeks after the pea rhizosphere was inoculated with 25:75, 50:50, and 75:25 ratios of Tr78.2 to HT1, the HUT+ isolate Tr78.2 made up 31%, 60%, and 90% of the total fungal biomass, while the HUT− isolate HT1 made up only 69%, 40%, and 10% of the fungal biomass (Fig. 5). Repeat experiments with isolates HT1 and HT5 gave similar results, with isolate Tr78.2 accounting for up to 87% of the fungal biomass 2 weeks (for HT1) or 3 weeks (for HT5) after inoculation with a 50:50 mixture of both isolates (data not shown). The inoculated roots did not show any disease symptoms, and fungal DNA was not detected inside the roots, which demonstrates, as expected, that the fungi were not growing inside the roots but in the rhizosphere.
FIG. 5.
Relative amounts of isolate Tr78.2 (HUT+) and isolate HT1 (HUT−) recovered from the rhizospheres of pea plants 2 weeks after inoculation of 2-week-old seedlings with different ratios of these isolates. Two-week-old pea seedlings were inoculated with spore suspensions containing mixtures (100:0, 75:25, 50:50, 25:75, and 0:100) of Tr78.2 and HT1. A rhizosphere sample was obtained from each treatment (six samples per treatment) 2 weeks after inoculation, and the DNA was extracted. The amounts of the pea RCA gene, the N. haematococca actin gene, and the 888-bp portion of the PDA1 gene in each sample were determined by real-time PCR. Broken lines show the expected relative ratios of both isolates if they are not competing. Solid lines show the experimental values for the HUT+ and HUT− isolates. The means are significantly different from expected results for the 75:25, 50:50, and 25:75 treatments (Newman-Keuls multiple comparison test; P < 0.05).
DISCUSSION
A priori, it seems likely that a selective ability to utilize a nutrient in the rhizosphere would benefit a rhizosphere-inhabiting organism. This study shows that the ability of N. haematococca to utilize the pea root exudate HS is correlated with its ability to be a pea root pathogen and that the gene(s) for HS utilization (HUT) is located on the same PDA1-CD chromosome as the genes for pea pathogenicity (PEP cluster). Furthermore, the portion of the chromosome that contains the HUT gene(s) endows an isolate with an increased competitive advantage in the pea rhizosphere. In support of the possibility that the competitive advantage might be due to the HUT gene(s), several research groups have engineered plants to produce novel nutrients in their root exudates and have shown that bacteria transformed with the ability to utilize these nutrients have a competitive advantage in the rhizosphere (29, 38). Though not yet tested, one might logically predict that it would also be advantageous for a root pathogen to be more competitive in the rhizosphere prior to its entry into the roots of its host.
The identification on a CD chromosome of another trait which enhances the ability of N. haematococca to expand its habitat further supports the notion that fungal CD chromosomes are analogous to host-specifying plasmids in plant-associated bacteria. For example, the different allelic variants of the Ti plasmids in Agrobacterium tumefaciens and the sym plasmids in Rhizobium spp. determine the host specificity for each bacterial strain (2). Furthermore, the symbiotic and nonsymbiotic plasmids of bacteria isolated from the rhizosphere of their host plants often contain genes for the utilization of host-specific root exudates (6, 30, 34). For some bacteria, these utilization genes have been shown to confer an increased competitive ability in the rhizosphere of their respective host plants (15, 34). For example, the sym plasmid of the Sinorhizobium meliloti bacterium, which nodulates alfalfa roots, carries genes for the catabolism of the alfalfa seed exudate stachydrine (10, 34). These genes for stachydrine catabolism also increase the competitive ability of S. meliloti in the rhizosphere of alfalfa (34). In addition, stachydrine not only supports the growth of S. meliloti but also induces the expression of its NOD genes, which are carried on the sym plasmid (33). A parallel situation may exist between the fungus N. haematococca and its host, the pea. HS, a root exudate of the pea, supports the growth of pea-pathogenic isolates of this fungus. Others (53) have shown that HS induces the expression of a pectin-degrading gene, pelD, which is a pathogenicity gene in this fungus.
The transfer of plasmids between bacteria has long been known to change the properties of the recipient bacterium and is another form of the horizontal transfer of large “genomic islands” of DNA which serves as a major force in the evolution of bacteria, allowing them to inhabit new environments (21). The clustering of genes for the colonization of certain habitats may facilitate their transfer and improve their retention via positive selection in those environments (12, 21). The current work is among the first to provide an example of a specific habitat, the rhizosphere of pea plants, for which the clustering of genes on a CD chromosome might be beneficial not only for host specificity but also for inhabiting an environment conducive to horizontal gene transfer.
Many lines of evidence have implied that all or part of the CD chromosomes might have originated through horizontal gene transfer. First, the PEP genes on the PDA1-CD chromosome have a different codon usage and GC content from those of genes on the other chromosomes (13, 22). Second, a supernumerary chromosome in another plant-pathogenic fungus, Colletotrichum gloeosporioides, can be transferred laterally (14). Third, CD chromosomes are present in some, but not all, isolates of N. haematococca, and this DNA is not found in other portions of the genome (3). Finally, the PEP genes have also been shown to have a discontinuous phylogenetic distribution (42), another feature of horizontally transferred DNA. Since it has been demonstrated repeatedly that the rhizosphere is conducive to horizontal gene transfer between bacteria (7), we hypothesize that the PDA1-CD chromosome could have been obtained through horizontal gene transfer in the rhizosphere and maintained by environmental selection acting on clustered host-specifying genes.
The ability to use HS in fungi other than pea pathogens is rare; however, some fungi which are pathogenic on other legumes (e.g., F. solani f. sp. phaseoli) could also grow on this amino acid (35). HS is also present in the root exudates of chickpea plants (20), and it is apparently present in the vegetative tissue of the jack bean (37). Therefore, the HUT+ phenotype might be beneficial for pathogenicity on other plants.
It will be interesting to examine whether the initial site of infection by pea-pathogenic fungi on pea roots occurs at a site of HS release, the lateral roots in the pea plant (44). Since in pea plants HS has also been shown to accumulate in large amounts in the vegetative tissue because it is used for the storage and transport of carbon and nitrogen (31), it is also possible that HS utilization might be important in nutrient acquisition even after the fungus has infected the plant. Testing the effect of the HUT gene(s) on pathogenicity or rhizosphere competency will be possible once the genes for HS utilization are characterized, a feat apparently not yet accomplished for any microorganisms.
Acknowledgments
We thank N. P. Huong Tan for generating the benomyl-treated isolates; Catherine Wasmann for scientific, technical, and editorial advice; Esteban Temporini for scientific and technical advice; Andrew Mort for providing γ-glutamyl-d-alanine; and Elizabeth Pierson for advice on the deWit replacement series.
This research was supported in part by grant 9900717 from the Soils and Soil Biology Program, USDA/NRA.
Footnotes
Published ahead of print on 11 April 2008.
REFERENCES
- 1.Boivin, C., L. R. Barran, C. A. Malpica, and C. Rosenberg. 1991. Genetic analysis of a region of the Rhizobium meliloti pSym plasmid specifying catabolism of trigonelline, a secondary metabolite present in legumes. J. Bacteriol. 173:2809-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brencic, A., and S. C. Winans. 2005. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 69:155-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Covert, S. F. 1998. Supernumerary chromosomes in filamentous fungi. Curr. Genet. 33:311-319. [DOI] [PubMed] [Google Scholar]
- 4.Curl, E. A., and B. Truelove. 1985. The rhizosphere. Springer-Verlag, Berlin, Germany.
- 5.DeWit, C. T. 1960. On competition. Versl. Landbouwkd. Onderz. 66:1-82. [Google Scholar]
- 6.Economou, A., F. K. L. Hawkins, and A. W. B. Johnston. 1988. pRL1JI specifies the catabolism of l-homoserine and contains a gene, rhi, whose transcription is reduced in the presence of nod gene inducer molecules, p. 462-482. In H. Bothe, F. J. de Bruijn, and W. E. Newton (ed.), Nitrogen fixation: hundred years after. Gustav Fischer, Stuttgart, Germany.
- 7.Espinosa-Urgel, M. 2004. Plant-associated Pseudomonas populations: molecular biology, DNA dynamics, and gene transfer. Plasmid 52:139-150. [DOI] [PubMed] [Google Scholar]
- 8.Funnell, D. L., and H. D. VanEtten. 2002. Pisatin demethylase genes are on dispensable chromosomes while genes for pathogenicity on carrot and ripe tomato are on other chromosomes in Nectria haematococca. Mol. Plant-Microbe Interact. 15:840-846. [DOI] [PubMed] [Google Scholar]
- 9.Funnell, D. L., P. S. Matthews, and H. D. VanEtten. 2002. Identification of new pisatin demethylase genes (PDA5 and PDA7) in Nectria haematococca and non-Mendelian segregation of pisatin demethylating ability and virulence on pea due to loss of chromosomal elements. Fungal Genet. Biol. 37:121-133. [DOI] [PubMed] [Google Scholar]
- 10.Goldmann, A., L. Lecoeur, B. Message, M. Delarue, E. Schoonejans, and D. Tepfer. 1994. Symbiotic plasmid genes essential to the catabolism of proline betaine, or stachydrine, are also required for efficient nodulation by Rhizobium meliloti. FEMS Microbiol. Lett. 115:305-311. [Google Scholar]
- 11.Gunawardena, U., M. Rodriguez, D. Straney, J. T. Romeo, H. D. VanEtten, and M. C. Hawes. 2005. Tissue-specific localization of pea root infection by Nectria haematococca. Mechanisms and consequences. Plant Phys. 137:1363-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity: a Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han, Y., X. Liu, U. L. Benny, H. C. Kistler, and H. D. VanEtten. 2001. Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria haematococca. Plant J. 25:305-314. [DOI] [PubMed] [Google Scholar]
- 14.He, C., A. G. Rusu, A. M. Poplawski, J. A. G. Irwin, and J. M. Manners. 1998. Transfer of a supernumerary chromosome between vegetatively incompatible biotypes of the fungus Colletotrichum gloeosporioides. Genetics 150:1459-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hynes, M. F., and M. O'Connell. 1990. Host plant effect on competition among strains of Rhizobium leguminosarum. Can. J. Microbiol. 36:864-869. [Google Scholar]
- 16.Kent, A., and E. W. Triplett. 2002. Microbial communities and their interaction in soil and rhizosphere ecosystems. Annu. Rev. Microbiol. 56:211-236. [DOI] [PubMed] [Google Scholar]
- 17.Kistler, H. C., and H. D. VanEtten. 1984. Three non-allelic genes for pisatin demethylation in the fungus Nectria haematococca. J. Gen. Microbiol. 130:2595-2603. [Google Scholar]
- 18.Kistler, H. C., L. W. Meinhard, and U. Benny. 1996. Mutants of Nectria haematococca created by a site-directed chromosome breakage are greatly reduced in virulence toward pea. Mol. Plant-Microbe Interact. 9:804-809. [Google Scholar]
- 19.Kuo, Y. H., F. Lambein, F. Igekami, and R. Van Parijis. 1982. Isoxazolin-5-ones and amino acids in root exudates of pea and sweet pea seedlings. Plant Physiol. 70:1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kush, K., and K. R. Dadarwall. 1980. Root exudates as pre-invasive factors in the nodulation of chickpea varieties. Soil Biochem. 13:51-55. [Google Scholar]
- 21.Lawrence, J. G. 2005. Common themes in the genome strategies of pathogens. Curr. Opin. Gene Dev. 15:584-588. [DOI] [PubMed] [Google Scholar]
- 22.Liu, X. G., M. Inlow, and H. D. VanEtten. 2003. Expression profiles of pea pathogenicity (PEP) genes in vivo and in vitro, characterization of the flanking regions of the PEP cluster and evidence that the PEP cluster region resulted from horizontal gene transfer in the fungal pathogen Nectria haematococca. Curr. Genet. 44:95-103. [DOI] [PubMed] [Google Scholar]
- 23.Lugtenberg, B. J. J., L. Dekkers, and G. V. Bloemberg. 2001. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 39:461-490. [DOI] [PubMed] [Google Scholar]
- 24.Maloney, A. P., and H. D. VanEtten. 1994. A gene from the fungal plant pathogen Nectria haematococca that encodes the phytoalexin-detoxifying enzyme pisatin demethylase defines a new cytochrome P450 family. Mol. Gen. Genet. 243:506-514. [DOI] [PubMed] [Google Scholar]
- 25.Martínez-Romero, E., and J. Caballero-Mellado. 1996. Rhizobium phylogenies and bacterial genetic diversity. Crit. Rev. Plant Sci. 15:113-140. [Google Scholar]
- 26.Matuo, T., and W. C. Snyder. 1973. Use of morphology and mating populations in identification of formae speciales in Fusarium solani. Phytopathology 63:562-565. [Google Scholar]
- 27.Miao, V. P. W., D. E. Matthews, and H. D. VanEtten. 1991. Identification and chromosomal locations of a family of cytochrome P-450 genes for pisatin detoxification in the fungus Nectria haematococca. Mol. Gen. Genet. 226:214-223. [DOI] [PubMed] [Google Scholar]
- 28.Morris, C. E., and J. M. Monier. 2003. The ecological significance of biofilm formation by plant-associated bacteria. Annu. Rev. Phytopathol. 41:429-453. [DOI] [PubMed] [Google Scholar]
- 29.Oger, P. M., H. Mansouri, X. Nesme, and Y. Desauux. 2004. Engineering root exudation of Lotus toward the production of two novel carbon compounds leads to the selection of distinct microbial populations in the rhizosphere. Microb. Ecol. 47:96-103. [DOI] [PubMed] [Google Scholar]
- 30.Oresnik, I. J., L. Faas, S. O'Brian, and M. F. Hynes. 1998. Plasmid-encoded catabolic genes in Rhizobium leguminosarum bv. trifolii: evidence for a plant-inducible rhamnose locus involved in competition for nodulation. Mol. Plant-Microbe Interact. 11:1175-1185. [Google Scholar]
- 31.Pate, J. S. 1980. Transport and partitioning of nitrogenous solutes. Annu. Rev. Plant Physiol. 31:313-340. [Google Scholar]
- 32.Paulitz, T. C. 2000. Population dynamics of biocontrol agents and pathogens in soils and rhizospheres. Eur. J. Plant Pathol. 106:401-413. [Google Scholar]
- 33.Phillips, D. A., M. C. Joseph, and C. A. Maxwell. 1992. Trigonelline and stachydrine released from alfalfa seeds activated NodD2 protein in Rhizobium meliloti. Plant Physiol. 99:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips, D. A., W. R. Streit, H. Volpin, J. D. Palumbo, C. M. Joseph, E. S. Sande, F. J. deBruijn, and C. I. Kado. 1996. Plant regulation of bacterial root colonization, p. 481-486. In G. Stacey, B. Mullin, and P. M. Gresshoff (ed.), Biology of plant-microbe interactions. International Society of Molecular Plant-Microbe Interactions, St. Paul, MN.
- 35.Rodriguez-Carres, M. 2006. Habitat-defining genes and synteny of conditionally dispensable (CD) chromosomes in the fungus Nectria haematococca. Ph.D. dissertation. The University of Arizona, Tucson.
- 36.Rosenberg, C., P. Boistard, J. Dénarié, and F. Casse-Delbart. 1981. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol. Gen. Genet. 184:326-333. [DOI] [PubMed] [Google Scholar]
- 37.Rosenthal, G. A. L. 1982. l-Canavanine metabolism in jack bean, Canavalia ensiformis (L) DC (Leguminosae). Plant Physiol. 69:1066-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savka, M. A., and S. K. Farrand. 1997. Modification of rhizobacterial populations by engineering bacterium utilization of a novel plant-produced resource. Nat. Biotechnol. 15:363-368. [DOI] [PubMed] [Google Scholar]
- 39.Stephens, R. B. 1974. Mycology guidebook. University of Washington Press, Seattle.
- 40.Taga, M., M. Murata, and H. D. VanEtten. 1999. Visualization of a conditionally dispensable chromosome in the filamentous ascomycete Nectria haematococca by fluorescence in situ hybridization. Fungal Genet. Biol. 26:169-177. [DOI] [PubMed] [Google Scholar]
- 41.Temporini, E. D., and H. D. VanEtten. 2002. Distribution of the pea pathogenicity (PEP) genes in the fungus Nectria haematococca mating population VI. Curr. Genet. 41:107-114. [DOI] [PubMed] [Google Scholar]
- 42.Temporini, E. D., and H. D. VanEtten. 2004. An analysis of the phylogenetic distribution of the pea pathogenicity genes of Nectria haematococca MPVI supports the hypothesis of their origin by horizontal transfer and uncovers a potentially new pathogen of garden pea: Neocosmospora boniensis. Curr. Genet. 46:29-36. [DOI] [PubMed] [Google Scholar]
- 43.Tsuchiya, D., and M. Taga. 2001. Cytological karyotyping of three Cochliobolus spp. by the germ tube burst method. Phytopathology 91:354-360. [DOI] [PubMed] [Google Scholar]
- 44.Van Egeraat, A. W. S. M. 1975. Exudation of ninhydrin-positive compounds by pea-seedling roots: a study of the sites of exudation and of the composition of the exudate. Plant Soil 42:37-47. [Google Scholar]
- 45.Van Egeraat, A. W. S. M. 1975. The possible role of homoserine in the development of Rhizobium leguminosarum in the rhizospheres of pea seedlings. Plant Soil 42:381-386. [Google Scholar]
- 46.VanEtten, H. D. 1978. Identification of additional habitats of Nectria haematococca mating population VI. Phytopathology 68:1552-1556. [Google Scholar]
- 47.VanEtten, H. D., and P. S. Matthews. 1984. Naturally occurring variation in the inducibility of pisatin demethylating activity in Nectria haematococca mating population VI. Physiol. Plant Pathol. 25:149-160. [Google Scholar]
- 48.VanEtten, H. D., P. S. Matthews, K. J. Tegtmeier, M. F. Dietert, and J. I. Stein. 1980. The association of pisatin tolerance and demethylation with virulence on pea in Nectria haematococca. Physiol. Plant Pathol. 16:257-268. [Google Scholar]
- 49.VanEtten, H. D., D. Straney, S. Covert, and C. Kistler. 2001. The genetics of Nectria haematococca mating population VI with special emphasis on its conditionally dispensible (CD) chromosomes: a source of habitat specific genes, p. 97-112. In B. A. Summerell, J. F. Leslie, D. Backhouse, W. L. Bryden, and L. W. Burgess (ed.), Fusarium. APS Press, St. Paul, MN.
- 50.VanEtten, H. D., S. Jorgensen, J. Enkerli, and S. F. Covert. 1998. Inducing the loss of conditionally dispensable chromosomes in Nectria haematococca during vegetative growth. Curr. Genet. 33:299-303. [DOI] [PubMed] [Google Scholar]
- 51.Wasmann, C. C., and H. D. VanEtten. 1996. Transformation-mediated chromosome loss and disruption of a gene for pisatin demethylase decrease the virulence of Nectria haematococca. Mol. Plant-Microbe Interact. 9:793-803. [Google Scholar]
- 52.Whipps, J. M. 1990. Carbon economy, p. 59-97. In J. M. Lynch (ed.), The rhizosphere. Wiley Interscience, West Sussex, United Kingdom.
- 53.Yang, Z., L. M. Rogers, Y. Song, W. Guo, and P. E. Kolattukudy. 2005. Homoserine and asparagine are host signals that trigger in planta expression of a pathogenesis gene in Nectria haematococca. Proc. Natl. Acad. Sci. USA 102:4197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]